The Investigation of the Effect of a-Tomatine as a Novel Matrix Metalloproteinase Inhibitor on the Bond Strength of Sound and Eroded Dentine through In Vitro and In Silico Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Size Calculation
2.2. Formation of Erosion on the Dentine Surface
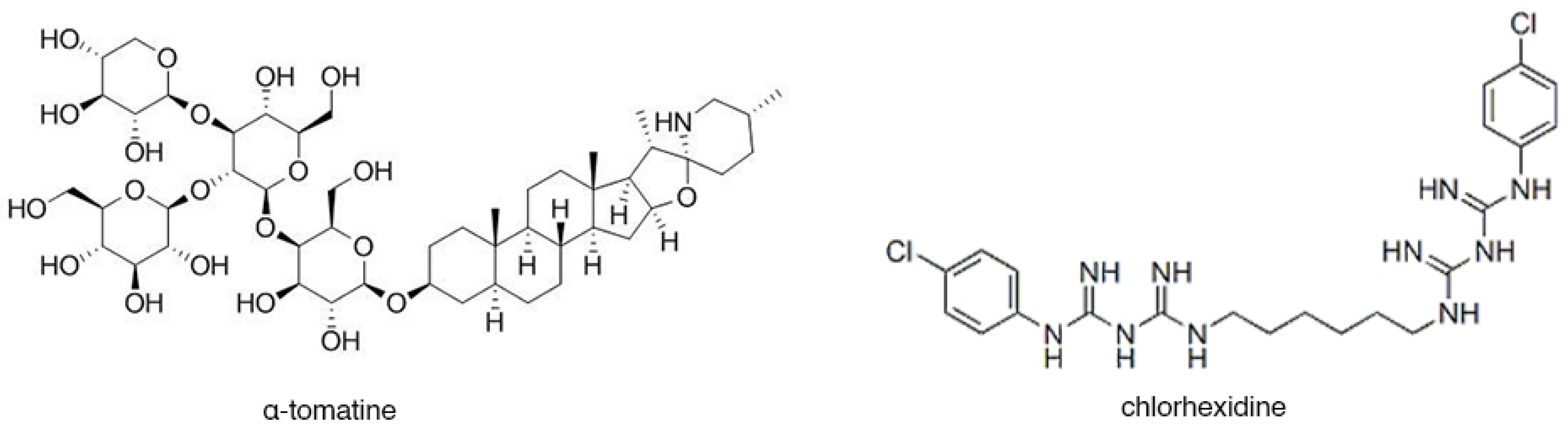
2.3. Preparation of a-tomatine
2.4. Restoration Stage and Bond Strength Measurement
2.5. Computational Method
2.5.1. Molecular Docking
Ligand Preparation
Protein Preparation
Ligand Docking
2.5.2. Molecular Dynamics Study
Molecular Dynamics Simulation System Setup
Molecular Dynamic Simulation Protocols
2.6. Statistical Analysis
3. Results
3.1. Microtensile Bond Strength
3.2. Molecular Docking
3.3. Molecular Dynamics
3.4. Root-Mean-Square Deviation (RMSD) Analysis
3.5. Root-Mean-Square Fluctuation (RMSF) Analysis
4. Discussion
4.1. Molecular Docking
4.2. Molecular Dynamics
4.3. Root-Mean-Square Deviation (RMSD) Analysis
4.4. Root-Mean-Square Fluctuation (RMSF) Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buzalaf, M.A.R.; Kato, M.T.; Hannas, A.R. The Role of Matrix Metalloproteinases in Dental Erosion. Adv. Dent. Res. 2012, 24, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Viana, Í.E.L.; Alania, Y.; Feitosa, S.; Borges, A.B.; Braga, R.R.; Scaramucci, T. Bioactive Materials Subjected to Erosion/Abrasion and Their Influence on Dental Tissues. Oper. Dent. 2020, 45, E114–E123. [Google Scholar] [CrossRef] [PubMed]
- Tezvergil-Mutluay, A.; Pashley, D.; Mutluay, M.M. Long-Term Durability of Dental Adhesives. Curr. Oral. Health Rep. 2015, 2, 174–181. [Google Scholar] [CrossRef]
- Carrilho, M.R.O.; Carvalho, R.M.; de Goes, M.F.; di Hipólito, V.; Geraldeli, S.; Tay, F.R.; Pashley, D.H.; Tjäderhane, L. Chlorhexidine Preserves Dentin Bond In Vitro. J. Dent. Res. 2007, 86, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Breschi, L.; Martin, P.; Mazzoni, A.; Nato, F.; Carrilho, M.; Tjäderhane, L.; Visintini, E.; Cadenaro, M.; Tay, F.R.; Dorigo, E.D.S. Use of a Specific MMP-Inhibitor (Galardin) for Preservation of Hybrid Layer. Dent. Mater. 2010, 26, 571–578. [Google Scholar] [CrossRef]
- Do Amaral, S.F.; Scaffa, P.M.C.; Rodrigues, R.D.S.; Nesadal, D.; Marques, M.M.; Nogueira, F.N.; Sobral, M.A.P. Dynamic Influence of PH on Metalloproteinase Activity in Human Coronal and Radicular Dentin. Caries Res. 2018, 52, 113–118. [Google Scholar] [CrossRef]
- Gendron, R.; Grenier, D.; Sorsa, T.; Mayrand, D. Inhibition of the Activities of Matrix Metalloproteinases 2, 8, and 9 by Chlorhexidine. Clin. Diagn. Lab. Immunol. 1999, 6, 437–439. [Google Scholar] [CrossRef]
- Siqueira, F.S.F.; Cardenas, A.M.; Ocampo, J.B.; Hass, V.; Bandeca, M.C.; Gomes, J.C.; Reis, A.; Loguercio, A.D. Bonding Performance of Universal Adhesives to Eroded Dentin. J. Adhes. Dent. 2018, 20, 121–132. [Google Scholar] [CrossRef]
- Yelken, B.Ö.; Balcı, T.; Süslüer, S.Y.; Kayabaşı, Ç.; Avcı, Ç.B.; Kırmızıbayrak, P.B.; Gündüz, C. The Effect of Tomatine on Metastasis Related Matrix Metalloproteinase (MMP) Activities in Breast Cancer Cell Model. Gene 2017, 627, 408–411. [Google Scholar] [CrossRef]
- Shi, M.-D.; Shih, Y.-W.; Lee, Y.-S.; Cheng, Y.-F.; Tsai, L.-Y. Suppression of 12-O-Tetradecanoylphorbol-13-Acetate-Induced MCF-7 Breast Adenocarcinoma Cells Invasion/Migration by α-Tomatine through Activating PKCα/ERK/NF-ΚB-Dependent MMP-2/MMP-9 Expressions. Cell Biochem. Biophys. 2013, 66, 161–174. [Google Scholar] [CrossRef]
- Maravic, T.; Breschi, L.; Paganelli, F.; Bonetti, G.A.; Martina, S.; Di Giorgio, G.; Bossù, M.; Polimeni, A.; Checchi, V.; Generali, L. Endogenous Enzymatic Activity of Primary and Permanent Dentine. Materials 2021, 14, 4043. [Google Scholar] [CrossRef]
- Ahmet, B.S.O.; Seseogullari-Dirihan, R.; Tezvergil-Mutluay, A. Activation of Matrix-Bound Endogenous Proteases by Self-Etch Adhesives. Dent. Mater. J. 2020, 39, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Lim-Wilby, M. Molecular Docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for Molecular Docking: A Review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; de Magalhães, C.S.; Dardenne, L.E. Receptor-Ligand Molecular Docking. Biophys. Rev. 2014, 6, 75–87. [Google Scholar] [CrossRef]
- Bakir Boga, O.; Bugra Ortaakarsu, A.; Kadırustaoglu, B.; Basaran Kurbanoglu, E. Phytochemical Profiling, in Vitro Biological Activities and in Silico Molecular Docking Studies of the Crude Extract of Crambe Orientalis, an Endemic Plant in Turkey. Chem. Biodivers. 2023, 20, e202201142. [Google Scholar] [CrossRef] [PubMed]
- Tunc, T.; Ortaakarsu, A.B.; Hatipoglu, S.M.; Kazancı, U.; Karabocek, S.; Karabocek, N.; Dege, N.; Karacan, N. New Schiff Bases with a 2,6-Bis(2-Aminophenylthio)Pyridine Moiety Acting as Glutathione Reductase Activator and Inhibitors: Synthesis and Molecular Docking Studies. J. Mol. Struct. 2022, 1254, 132299. [Google Scholar] [CrossRef]
- Shoichet, B.K.; McGovern, S.L.; Wei, B.; Irwin, J.J. Lead Discovery Using Molecular Docking. Curr. Opin. Chem. Biol. 2002, 6, 439–446. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Carvalho, C.; Fernandes, F.P.; da Freitas, V.P.; França, F.M.G.; Basting, R.T.; Turssi, C.P.; do Amaral, F.L.B. Effect of Green Tea Extract on Bonding Durability of an Etch-and-Rinse Adhesive System to Caries-Affected Dentin. J. Appl. Oral. Sci. 2016, 24, 211–217. [Google Scholar] [CrossRef]
- Zimmerli, B.; De Munck, J.; Lussi, A.; Lambrechts, P.; Van Meerbeek, B. Long-Term Bonding to Eroded Dentin Requires Superficial Bur Preparation. Clin. Oral. Investig. 2012, 16, 1451–1461. [Google Scholar] [CrossRef]
- Lee, S.T.; Wong, P.F.; Cheah, S.C.; Mustafa, M.R. Alpha-Tomatine Induces Apoptosis and Inhibits Nuclear Factor-Kappa B Activation on Human Prostatic Adenocarcinoma PC-3 Cells. PLoS ONE 2011, 6, e18915. [Google Scholar] [CrossRef]
- Schrödinger Release 2023-2; Maestro, Schrödinger, LLC: New York, NY, USA, 2023.
- Schrödinger Schrödinger Release 2023-2. In Maestro-Desmond Interoperability Tools, Desmond Molecular Dynamics System; Maestro, Schrödinger, LLC: New York, NY, USA, 2023.
- Baliga, S.; Muglikar, S.; Kale, R. Salivary PH: A Diagnostic Biomarker. J. Indian. Soc. Periodontol. 2013, 17, 461–465. [Google Scholar] [CrossRef]
- Schrödinger LigPrep. Schrödinger Release 2023-2; Schrödinger, LLC: New York, NY, USA, 2023. [Google Scholar]
- Schrödinger Epik. Schrödinger Release 2023-2; Schrödinger, LLC: New York, NY, USA, 2023. [Google Scholar]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the Comprehensive, Rapid, and Accurate Prediction of the Favorable Tautomeric States of Drug-like Molecules in Aqueous Solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.L.C. Schrödinger, L.L.C. Schrödinger Release 2023-2: Protein Preparation Wizard. In Epic Impact Prime Schrödinger; Schrödinger, LLC: New York, NY, USA, 2023. [Google Scholar]
- Feng, Y.; Likos, J.J.; Zhu, L.; Woodward, H.; Munie, G.; Mcdonald, J.J.; Stevens, A.M.; Howard, C.P.; De Crescenzo, G.A.; Welsch, D.; et al. Solution Structure and Backbone Dynamics of the Catalytic Domain of Matrix Metalloproteinase-2 Complexed with a Hydroxamic Acid Inhibitor. Biochim. Biophys. Acta BBA Proteins Proteom. 2002, 1598, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Campestre, C.; Agamennone, M.; Tortorella, P.; Preziuso, S.; Biasone, A.; Gavuzzo, E.; Pochetti, G.; Mazza, F.; Hiller, O.; Tschesche, H.; et al. N-Hydroxyurea as Zinc Binding Group in Matrix Metalloproteinase Inhibition: Mode of Binding in a Complex with MMP-8. Bioorg. Med. Chem. Lett. 2006, 16, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Tochowicz, A.; Maskos, K.; Huber, R.; Oltenfreiter, R.; Dive, V.; Yiotakis, A.; Zanda, M.; Bode, W.; Goettig, P. Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-Chain to Selectivity. J. Mol. Biol. 2007, 371, 989–1006. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; SØndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical p K a Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Users Manual. Schrödinger Release 2023-2: Glide, Schrödinger, LLC: New York, NY, 2023. In Schrödinger Release 2023-2: LigPrep; Schrödinger, LLC: New York, NY, USA, 2023.
- Gopal, S.M.; Kuhn, A.B.; Schäfer, L.V. Systematic Evaluation of Bundled SPC Water for Biomolecular Simulations. Phys. Chem. Chem. Phys. 2015, 17, 8393–8406. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Desmond Molecular Dynamics System, D.E. Shaw Research, New York, NY, 2023; Maestro-Desmond Interoperability Tools, Schrödinger: New York, NY, USA, 2023.
- Evans, D.J.; Holian, B.L. The Nose-Hoover Thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of Molecular Docking Programs for Virtual Screening against Dihydropteroate Synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.B.; Bonini, G.; Lenzi, T.L.; Imparato, J.C.P.; Raggio, D.P. Bonding Stability of Adhesive Systems to Eroded Dentin. Braz. Oral. Res. 2015, 29, S1806-83242015000100290. [Google Scholar] [CrossRef] [PubMed]
- Forgerini, T.V.; Ribeiro, J.F.; Rocha, R.O.; Soares, F.Z.M.; Lenzi, T.L. Role of Etching Mode on Bonding Longevity of a Universal Adhesive to Eroded Dentin. J. Adhes. Dent. 2017, 19, 69–75. [Google Scholar] [CrossRef]
- Shellis, R.P.; Ganss, C.; Ren, Y.; Zero, D.T.; Lussi, A. Methodology and Models in Erosion Research: Discussion and Conclusions. Caries Res. 2011, 45, 69–77. [Google Scholar] [CrossRef]
- Zero, D.T. Etiology of Dental Erosion-Extrinsic Factors. Eur. J. Oral. Sci. 1996, 104, 162–177. [Google Scholar] [CrossRef]
- Murrell, S.; Marshall, T.A.; Moynihan, P.J.; Qian, F.; Wefel, J.S. Comparison of in Vitro Erosion Potentials between Beverages Available in the United Kingdom and the United States. J. Dent. 2010, 38, 284–289. [Google Scholar] [CrossRef]
- Hannig, C.; Hamkens, A.; Becker, K.; Attin, R.; Attin, T. Erosive Effects of Different Acids on Bovine Enamel: Release of Calcium and Phosphate in Vitro. Arch. Oral. Biol. 2005, 50, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Lussi, A. Erosive Tooth Wear—A Multifactorial Condition of Growing Concern and Increasing Knowledge. Dental Erosion: From Diagnosis to Therapy. Monogr. Oral. Sci. 2006, 20, 1–8. [Google Scholar]
- Young, A.; Tenuta, L.M.A. Initial Erosion Models. Caries Res. 2011, 45, 33–42. [Google Scholar] [CrossRef] [PubMed]
- de Siqueira, F.S.F.; Hilgemberg, B.; Araujo, L.C.R.; Hass, V.; Bandeca, M.C.; Reis, A.; Gomes, J.C.; Cardenas, A.F.M.; Loguercio, A.D. Effect of Phosphoric Acid Containing MMP-Inactivator on the Properties of Resin Bonding to Eroded Dentin. J. Adhes. Dent. 2019, 21, 149–158. [Google Scholar] [CrossRef]
- de Siqueira, F.S.F.; Hilgemberg, B.; Araujo, L.C.R.; Hass, V.; Bandeca, M.C.; Gomes, J.C.; Reis, A.; Loguercio, A.D.; Cardenas, A.F.M. Improving Bonding to Eroded Dentin by Using Collagen Cross-Linking Agents: 2 Years of Water Storage. Clin. Oral. Investig. 2020, 24, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Machado, C.M.; Zamuner, A.C.; Modena, K.C.D.S.; Ishikiriama, S.K.; Wang, L. How Erosive Drinks and Enzyme Inhibitors Impact Bond Strength to Dentin. Braz. Oral. Res. 2015, 29, S1806-83242015000100300. [Google Scholar] [CrossRef]
- Francisconi-Dos-Rios, L.F.; Casas-Apayco, L.C.; Calabria, M.P.; Francisconi, P.A.; Borges, A.F.; Wang, L. Role of Chlorhexidine in Bond Strength to Artificially Eroded Dentin over Time. J. Adhes. Dent. 2015, 17, 133–139. [Google Scholar]
- Francisconi-Dos-Rios, L.F.; Calabria, M.P.; Casas-Apayco, L.C.; Honório, H.M.; De Oliveira Carrilho, M.R.; Pereira, J.C.; Wang, L. Chlorhexidine Does Not Improve but Preserves Bond Strength to Eroded Dentin. Am. J. Dent. 2015, 28, 28–32. [Google Scholar]
- Deari, S.; Wegehaupt, F.J.; Tauböck, T.T.; Attin, T. Influence of Different Pretreatments on the Microtensile Bond Strength to Eroded Dentin. J. Adhes. Dent. 2017, 19, 147–155. [Google Scholar]
- Krithi, B.; Vidhya, S.; Mahalaxmi, S. Microshear Bond Strength of Composite Resin to Demineralized Dentin after Remineralization with Sodium Fluoride, CPP-ACP and NovaMin Containing Dentifrices. J. Oral. Biol. Craniofac. Res. 2020, 10, 122–127. [Google Scholar] [CrossRef]
- Loguercio, A.D.; Stanislawczuk, R.; Polli, L.G.; Costa, J.A.; Michel, M.D.; Reis, A. Influence of Chlorhexidine Digluconate Concentration and Application Time on Resin-Dentin Bond Strength Durability. Eur. J. Oral. Sci. 2009, 117, 587–596. [Google Scholar] [CrossRef]
- Ozan, G.; Sar Sancakli, H.; Yucel, T. Effect of Black Tea and Matrix Metalloproteinase Inhibitors on Eroded Dentin in Situ. Microsc. Res. Technol. 2020, 83, 834–842. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, J.; Chen, L.; Li, D.; Tan, Y. The Incorporation of Chlorhexidine in a Two-Step Self-Etching Adhesive Preserves Dentin Bond in Vitro. J. Dent. 2009, 37, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Ozsoy, A.; Erdemir, U.; Yucel, T.; Yildiz, E. Effects of Cavity Disinfectants on Bond Strength of an Etch-and-Rinse Adhesive to Water- or Ethanol-Saturated Sound and Caries-Affected Dentin. J. Adhes. Sci. Technol. 2015, 29, 2551–2564. [Google Scholar] [CrossRef]
- de Menezes, L.R.; da Silva, E.O.; Maurat da Rocha, L.V.; Ferreira Barbosa, I.; Rodrigues Tavares, M. The Use of Clays for Chlorhexidine Controlled Release as a New Perspective for Longer Durability of Dentin Adhesion. J. Mater. Sci. Mater. Med. 2019, 30, 132. [Google Scholar] [CrossRef] [PubMed]
- Hamdan-Nassar, T.; Bellot-Arcís, C.; Paredes-Gallardo, V.; García-Sanz, V.; Pascual-Moscardó, A.; Almerich-Silla, J.M.; Montiel-Company, J.M. Effect of 2% Chlorhexidine Following Acid Etching on Microtensile Bond Strength of Resin Restorations: A Meta-Analysis. Medicine 2019, 55, 769. [Google Scholar] [CrossRef]
- de Castro, F.L.A.; de Andrade, M.F.; Duarte Júnior, S.L.L.; Vaz, L.G.; Ahid, F.J.M. Effect of 2% Chlorhexidine on Microtensile Bond Strength of Composite to Dentin. J. Adhes. Dent. 2003, 5, 129–138. [Google Scholar]
- Dalkilic, E.E.; Arisu, H.D.; Kivanc, B.H.; Uctasli, M.B.; Omurlu, H. Effect of Different Disinfectant Methods on the Initial Microtensile Bond Strength of a Self-Etch Adhesive to Dentin. Lasers Med. Sci. 2012, 27, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, T.L.; Soares, F.Z.M.S.; de Rocha, R.O. Degradation of Resin-Dentin Bonds of Etch-and-Rinse Adhesive System to Primary and Permanent Teeth. Braz. Oral. Res. 2012, 26, 511–515. [Google Scholar] [CrossRef]
- Soares, C.J.; Pereira, C.A.; Pereira, J.C.; Santana, F.R.; Do Prado, C.J. Effect of Chlorhexidine Application on Microtensile Bond Strength to Dentin. Oper. Dent. 2008, 33, 183–188. [Google Scholar] [CrossRef]
- Sodhi, R.N.S.; Grad, H.A.; Smith, D.C. Examination by x-Ray Photoelectron Spectroscopy of the Adsorption of Chlorhexidine on Hydroxyapatite. J. Dent. Res. 1992, 71, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Perdigao, J.; Swift, E.J.; Denehy, G.E.; Wefel, J.S.; Donly, K.J. In Vitro Bond Strengths and SEM Evaluation of Dentin Bonding Systems to Different Dentin Substrates. J. Dent. Res. 1994, 73, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.N.; Hobson, G.E. The Constituents of Tomato Fruit—The Influence of Environment, Nutrition, and Genotype. C. R. C. Crit. Rev. Food Sci. Nutr. 1981, 15, 205–280. [Google Scholar] [CrossRef] [PubMed]
- Shieh, J.-M.; Cheng, T.-H.; Shi, M.-D.; Wu, P.-F.; Chen, Y.; Ko, S.-C.; Shih, Y.-W. α-Tomatine Suppresses Invasion and Migration of Human Non-Small Cell Lung Cancer NCI-H460 Cells Through Inactivating FAK/PI3K/Akt Signaling Pathway and Reducing Binding Activity of NF-ΚB. Cell Biochem. Biophys. 2011, 60, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Huang, H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef]
- Lee, K.R.; Kozukue, N.; Han, J.S.; Park, J.H.; Chang, E.Y.; Baek, E.J.; Chang, J.S.; Friedman, M. Glycoalkaloids and Metabolites Inhibit the Growth of Human Colon (HT29) and Liver (HepG2) Cancer Cells. J. Agric. Food Chem. 2004, 52, 2832–2839. [Google Scholar] [CrossRef] [PubMed]
- Sucha, L.; Hroch, M.; Rezacova, M.; Rudolf, E.; Havelek, R.; Sispera, L.; Cmielova, J.; Kohlerova, R.; Bezrouk, A.; Tomsik, P. The Cytotoxic Effect of α-Tomatine in MCF-7 Human Adenocarcinoma Breast Cancer Cells Depends on Its Interaction with Cholesterol in Incubation Media and Does Not Involve Apoptosis Induction. Oncol. Rep. 2013, 30, 2593–2602. [Google Scholar] [CrossRef]
- Benson, C.S.; Babu, S.D.; Radhakrishna, S.; Selvamurugan, N.; Sankar, B.R. Expression of Matrix Metalloproteinases in Human Breast Cancer Tissues. Dis. Mrk. 2013, 34, 395–405. [Google Scholar] [CrossRef]
- Shih, Y.W.; Shieh, J.M.; Wu, P.F.; Lee, Y.C.; Chen, Y.Z.; Chiang, T.A. α-Tomatine Inactivates PI3K/Akt and ERK Signaling Pathways in Human Lung Adenocarcinoma A549 Cells: Effect on Metastasis. Food Chem. Toxicol. 2009, 47, 1985–1995. [Google Scholar] [CrossRef]
- Chao, M.W.; Chen, C.H.; Chang, Y.L.; Teng, C.M.; Pan, S.L. α-Tomatine-Mediated Anti-Cancer Activity In Vitro and In Vivo through Cell Cycle- and Caspase-Independent Pathways. PLoS ONE 2012, 7, e44093. [Google Scholar] [CrossRef]
- Thorne, H.V.; Clarke, G.F.; Skuce, R. The Inactivation of Herpes Simplex Virus by Some Solanaceae Glycoalkaloids. Antivir. Res. 1985, 5, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Tomato Glycoalkaloids: Role in the Plant and in the Diet. J. Agric. Food Chem. 2002, 50, 5751–5780. [Google Scholar] [CrossRef] [PubMed]
- Simons, V.; Morrissey, J.P.; Latijnhouwers, M.; Csukai, M.; Cleaver, A.; Yarrow, C.; Osbourn, A. Dual Effects of Plant Steroidal Alkaloids on Saccharomyces Cerevisiae. Antimicrob. Agents Chemother. 2006, 50, 2732–2740. [Google Scholar] [CrossRef]
- Collares, F.M.; Rodrigues, S.B.; Leitune, V.C.; Celeste, R.K.; Borba de Araújo, F.; Samuel, S.M.; Borba de Araujo, F.; Samuel, S.M. Chlorhexidine Application in Adhesive Procedures: A Meta-Regression Analysis. J. Adhes. Dent. 2013, 15, 11–18. [Google Scholar] [CrossRef]
- Zhao, B.; Zhou, B.; Bao, L.; Yang, Y.; Guo, K. Alpha-Tomatine Exhibits Anti-Inflammatory Activity in Lipopolysaccharide-Activated Macrophages. Inflammation 2015, 38, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Milner, S.E.; Brunton, N.P.; Jones, P.W.; O’Brien, N.M.; Collins, S.G.; Maguire, A.R. Bioactivities of Glycoalkaloids and Their Aglycones from Solanum Species. J. Agric. Food Chem. 2011, 59, 3454–3484. [Google Scholar] [CrossRef] [PubMed]
- Chowański, S.; Adamski, Z.; Marciniak, P.; Rosiński, G.; Büyükgüzel, E.; Büyükgüzel, K.; Falabella, P.; Scrano, L.; Ventrella, E.; Lelario, F.; et al. A Review of Bioinsecticidal Activity of Solanaceae Alkaloids. Toxins 2016, 8, 60. [Google Scholar] [CrossRef]
- Zimmerley, M.; McClure, R.A.; Choi, B.; Potma, E.O. Following Dimethyl Sulfoxide Skin Optical Clearing Dynamics with Quantitative Nonlinear Multimodal Microscopy. Appl. Opt. 2009, 48, D79–D87. [Google Scholar] [CrossRef]
- Tjäderhane, L.; Mehtälä, P.; Scaffa, P.; Vidal, C.; Pääkkönen, V.; Breschi, L.; Hebling, J.; Tay, F.R.; Nascimento, F.D.; Pashley, D.H.; et al. The Effect of Dimethyl Sulfoxide (DMSO) on Dentin Bonding and Nanoleakage of Etch-and-Rinse Adhesives. Dent. Mater. 2013, 29, 1055–1062. [Google Scholar] [CrossRef]
- Stape, T.H.S.; Tjäderhane, L.; Marques, M.R.; Aguiar, F.H.B.; Martins, L.R.M. Effect of Dimethyl Sulfoxide Wet-Bonding Technique on Hybrid Layer Quality and Dentin Bond Strength. Dent. Mater. 2015, 31, 676–683. [Google Scholar] [CrossRef]
- Marren, K. Dimethyl Sulfoxide: An Effective Penetration Enhancer for Topical Administration of NSAIDs. Phys. Sport. 2011, 39, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Stape, T.H.S.; Tjäderhane, L.; Tezvergil-Mutluay, A.; Yanikian, C.R.F.; Szesz, A.L.; Loguercio, A.D.; Martins, L.R.M. Dentin Bond Optimization Using the Dimethyl Sulfoxide-Wet Bonding Strategy: A 2-Year in Vitro Study. Dent. Mater. 2016, 32, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Tekbas Atay, M.; Seseogullari-Dirihan, R.; Mutluay, M.M.; Tezvergil-Mutluay, A. Long-Term Effect of Curcuminoid Treatment on Resin-to-Dentin Bond Strength. Eur. J. Oral. Sci. 2022, 130, e12837. [Google Scholar] [CrossRef] [PubMed]
- de Mello, R.M.M.; Alcântara, B.A.R.; França, F.M.G.; do Amaral, F.L.B.; Basting, R.T. Dimethyl Sulfoxide Dentin Pretreatments Do Not Improve Bonding of a Universal Adhesive in Etch-and-Rinse or Self-Etch Modes. J. Adhes. Dent. 2022, 24, 49–56. [Google Scholar] [CrossRef]










| Dentine Type | Different Amounts of a-Tomatine * | Bond Strength (24-h) | |
|---|---|---|---|
| 20 s | 30 s | ||
| Mean ± Sd | Mean ± Sd | ||
| Sound dentine (n = 1) | 0.75 µM | 14.07 ± 4.31 | 14.12 ± 6.01 |
| 1 µM | 25.43 ± 8.96 | 25.38 ± 8.48 | |
| 1.5 µM | 43.17 ± 7.40 | 42.55 ± 16.86 | |
| p | 0.001 ** | 0.001 ** | |
| Material | Composition | Manufacturer and Batch Numbers |
|---|---|---|
| Citric acid | Citric acid monohydrate C6H8O7H2O | Merck, KGaA, Darmstadt, Germany (5949-29-1) |
| Dimethyl sulfoxide extra pure | Dimethyl sulfoxide C2H6OS | Merck, Sigma Aldrich, Germany |
| Saliva | 0.002 g of ascorbic acid, 0.58 g of NaCl, 0.17 g of CaCl2, 0.16 g of NH4Cl, 1.27 g of KCl, 0.16 g of NaSCN, 0.33 g of KH2PO4, and 0.34 g of Na2HPO4 for 1 L | Produced in the lab as artificial |
| Etching dental gel | 37.5% phosphoric acid gel | Kerr, Gel Etchant, Orange, USA (5887888) |
| Cavity cleanser | 2% chlorhexidine digluconate | Bisco, Inc., Schaumburg, IL, USA (1900000744) |
| Optibond FL | Primer: HEMA, PAMM, GPDM, water, ethanol, photoinitiator Adhesive: TEGDMA, DMA, GPDM, HEMA, BIS-GMA, filler, photoinitiator | Kerr, Orange, CA, USA (26684) |
| FiltekTM Z250 | Organic matrix: BIS-GMA, UDMA, BIS-EMA Inorganic matrix: zirconia/silica as a filler, the loading of the inorganic filler (without treatment with silane) was 60% by volume with a particle size in the range of 0.01–3.5 μm | 3M ESPE, St Paul, MN, USA (6020A2) |
| Tomatine | a-tomatine (powdered) | PhyProof, PhytoLab GmbH Dutendorfer, Germany (89905) |
| Aging Time | Source | F | p |
|---|---|---|---|
| 24 h | MMP inhibitor | 10.220 | 0.049 * |
| Type of dentin | 536.727 | 0.002 ** | |
| MMP inhibitor * type of dentin | 3.607 | 0.037 * | |
| 6 months | MMP inhibitor | 131.422 | 0.008 ** |
| Type of dentin | 4236.352 | 0.000 ** | |
| MMP inhibitor * type of dentin | 0.295 | 0.746 |
| Dentin Surface | MMP Inhibitor Agent | 24 h | 6 Months | |||
|---|---|---|---|---|---|---|
| Min–Max (Median) | Mean ± SD | Min–Max (Median) | Mean ± Sd | p | ||
| Sound dentin | 1 Control | 28.8–53.6 (35.8) | 36.30 ± 5.07 a | 18.2–36.2 (28.2) | 27.94 ± 3.93 b | a > b ** |
| 2 Chlorhexidine | 22.3–44.6 (32.9) | 33.01 ± 5.02 a | 22.2–44.1 (30.4) | 31.43 ± 4.46 a | >0.05 | |
| 3 a-Tomatine | 23.6–55.2 (38.1) | 39.03 ± 6.97 a | 18.7–49.2 (33.3) | 34.00 ± 6.24 b | a > b ** | |
| p | 0.001 ** | 0.001 ** | ||||
| Post hoc test | 1 > 2 ** 3 > 1,2 ** | 1 < 2,3 ** 2 < 3 * | ||||
| Eroded dentin | 1 Control | 10.4–26.2 (16.3) | 16.65 ± 3.89 a | 3.4–18.2 (9.8) | 9.89 ± 3.64 b | a > b ** |
| 2 Chlorhexidine | 6.6–27.1 (14.6) | 15.27 ± 4.71 a | 4.2–22.1 (11.9) | 13.13 ± 3.92 b | a > b ** | |
| 3 a-Tomatine | 7.5–32.2 (17.8) | 18.50 ± 5.19 a | 5.8–24.5 (15.6) | 15.01 ± 3.84 b | a > b ** | |
| p | 0.001 ** | 0.001 ** | ||||
| Post hoc test | 3 > 1 * 3 > 2 ** | 1 < 2 * 1 < 3 ** 2 < 3 * | ||||
| Sound | Eroded | |||
|---|---|---|---|---|
| 24 h | 6 Months | 24 h | 6 Months | |
| a-Tomatine | 65/3/2 | 69/1/0 | 67/2/1 | 67/3/0 |
| Chlorhexidine | 64/5/1 | 67/2/1 | 64/4/2 | 69/1/0 |
| Control | 63/5/2 | 64/3/3 | 62/5/3 | 67/2/1 |
| MMP-2 | MMP-8 | MMP-9 | |
|---|---|---|---|
| a-Tomatine | −9.358 | −9.663 | −7.996 |
| Chlorhexidine | −7.673 | −7.132 | −5.102 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ucuncu, M.K.; Ortaakarsu, A.B.; Batu, S.; Yildiz, E. The Investigation of the Effect of a-Tomatine as a Novel Matrix Metalloproteinase Inhibitor on the Bond Strength of Sound and Eroded Dentine through In Vitro and In Silico Methods. Appl. Sci. 2023, 13, 10322. https://doi.org/10.3390/app131810322
Ucuncu MK, Ortaakarsu AB, Batu S, Yildiz E. The Investigation of the Effect of a-Tomatine as a Novel Matrix Metalloproteinase Inhibitor on the Bond Strength of Sound and Eroded Dentine through In Vitro and In Silico Methods. Applied Sciences. 2023; 13(18):10322. https://doi.org/10.3390/app131810322
Chicago/Turabian StyleUcuncu, Musa Kazim, Ahmet Bugra Ortaakarsu, Sule Batu, and Esra Yildiz. 2023. "The Investigation of the Effect of a-Tomatine as a Novel Matrix Metalloproteinase Inhibitor on the Bond Strength of Sound and Eroded Dentine through In Vitro and In Silico Methods" Applied Sciences 13, no. 18: 10322. https://doi.org/10.3390/app131810322
APA StyleUcuncu, M. K., Ortaakarsu, A. B., Batu, S., & Yildiz, E. (2023). The Investigation of the Effect of a-Tomatine as a Novel Matrix Metalloproteinase Inhibitor on the Bond Strength of Sound and Eroded Dentine through In Vitro and In Silico Methods. Applied Sciences, 13(18), 10322. https://doi.org/10.3390/app131810322






