Ustilago maydis Yeast Mutant Produces Cytosolic Melanin by Tyrosine-Tyrosinase Activity with Stain Biosorption Capability
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Maintenance
2.2. Mutant Construction
2.2.1. Plasmid Construction for H⁺-ATPase Mutation
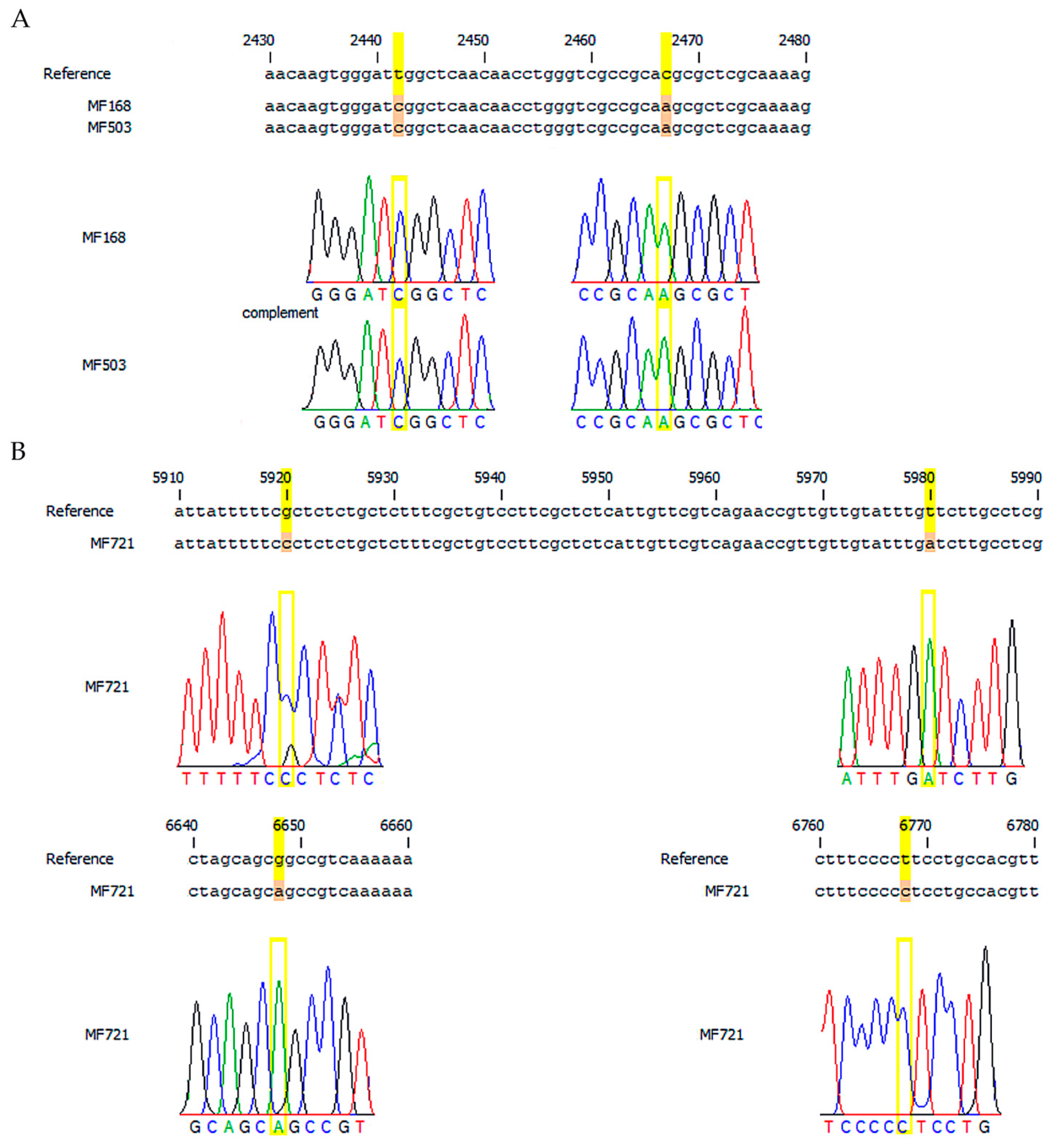
2.2.2. Sequencing
2.3. Macroscopic and Microscopic Morphology of the Mutants
2.3.1. Macroscopic and Microscopic Analysis
2.3.2. Morphology Analysis of Mutant Strain by Electron Microscopy
2.4. Pigment Extraction and Quantification
2.5. Identification of Melanin by Thin Layer Chromatography, UV, and IR Spectrophotometry
2.6. Assay of Physiological Response
Growth Assay
2.7. Assay of Tyrosinase Activity
2.7.1. Monophenolase and Diphenolase Activity
2.7.2. Effect of Ions on the Enzyme Activity
2.8. Biosorption of Methylene Blue with U. maydis Biomass
3. Results
3.1. Sequencing Analysis
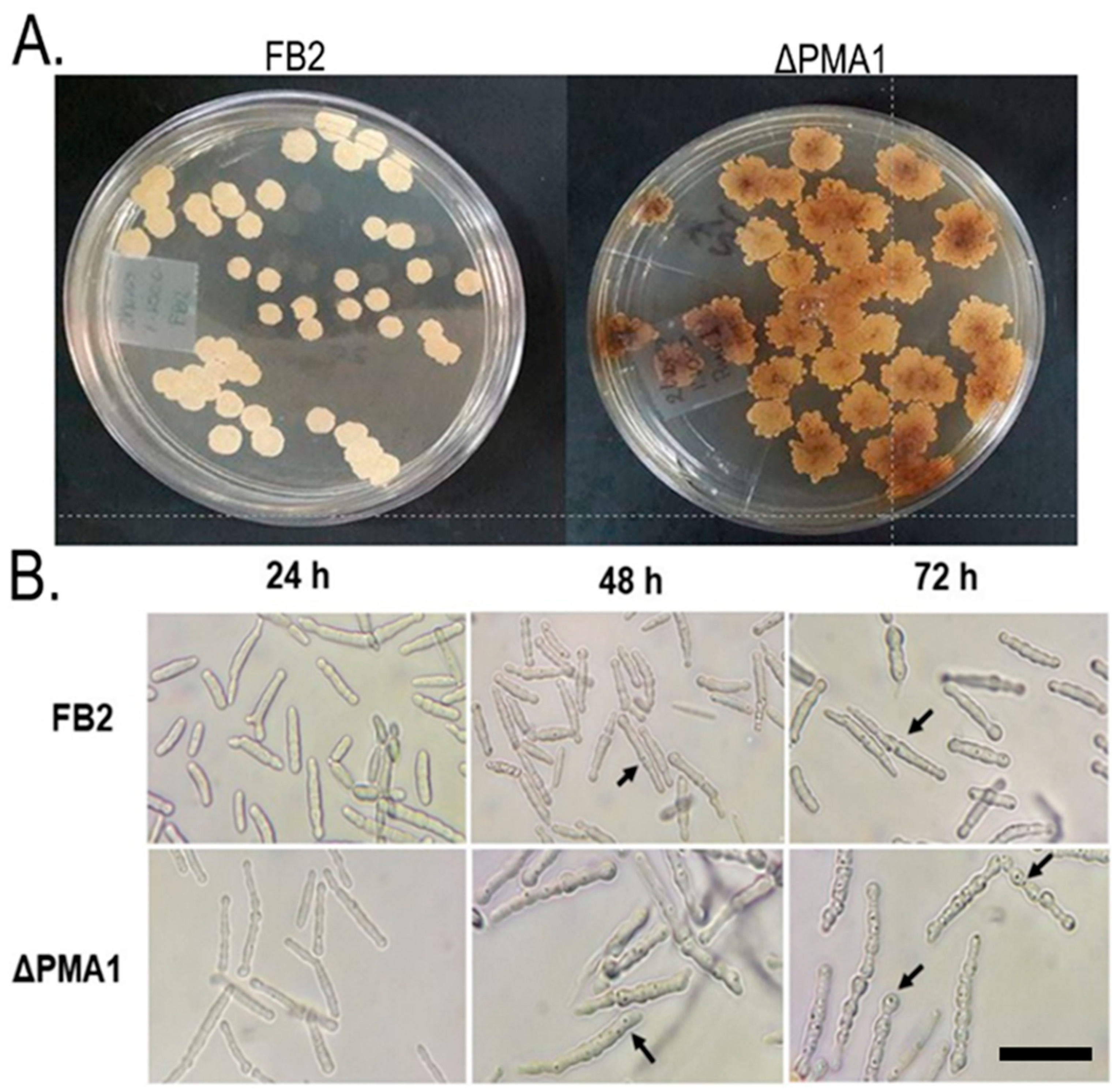
3.2. Morphological Characterization of ∆PMA1 Strain
3.2.1. Growth on Solid Media and Optical Microscopy
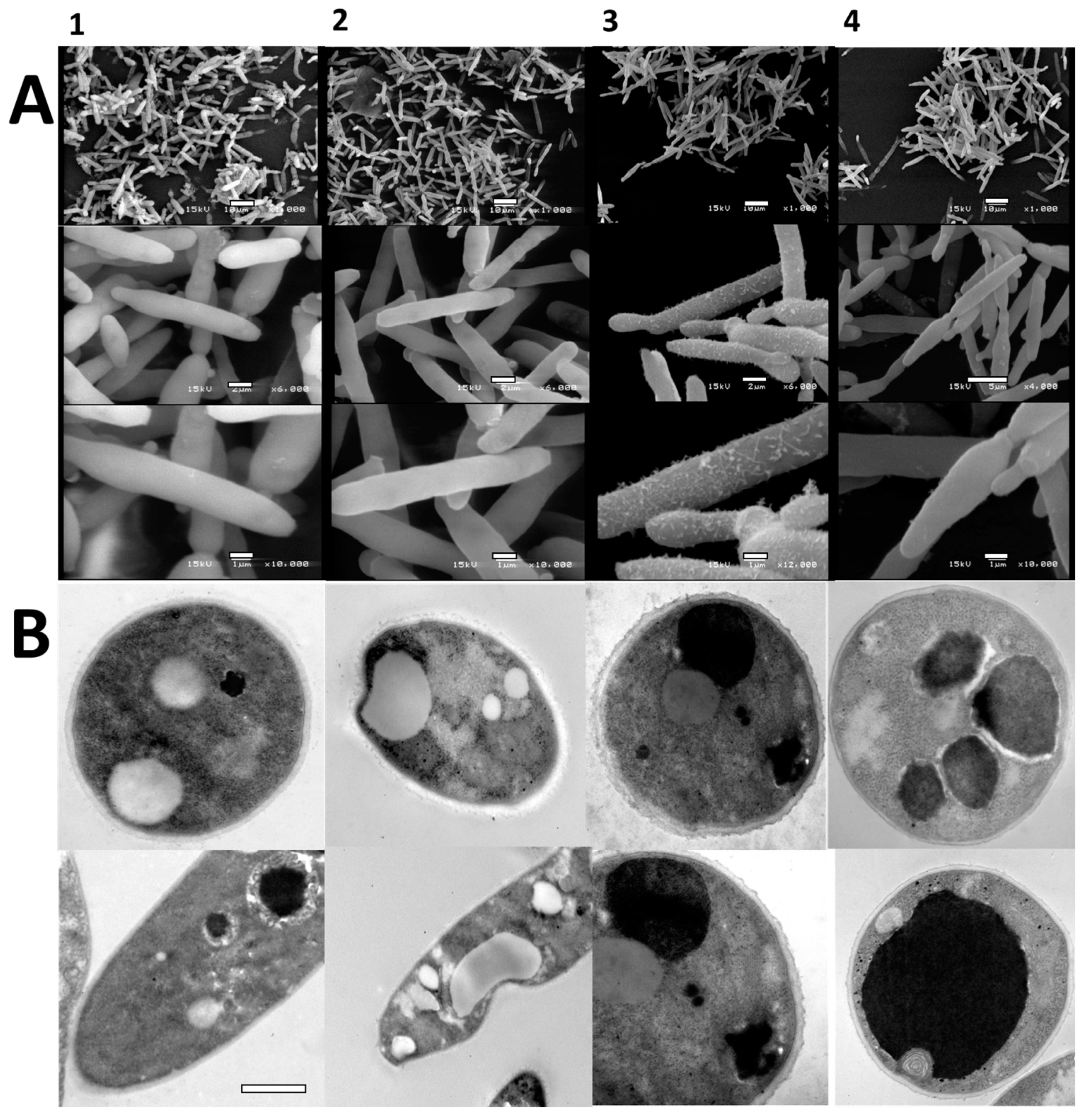
3.2.2. Scanning and Electron Microscopy
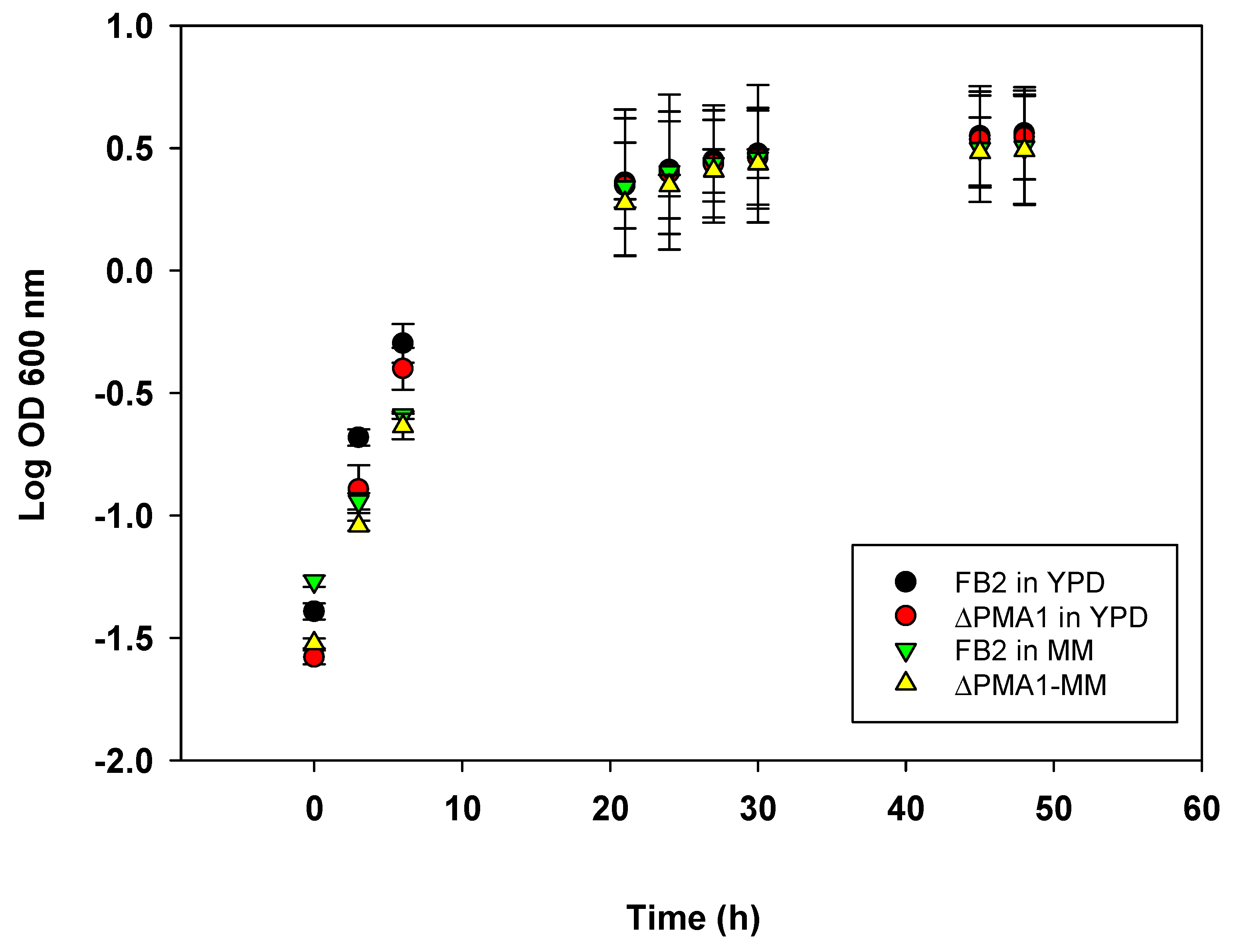
3.3. Growth Analysis
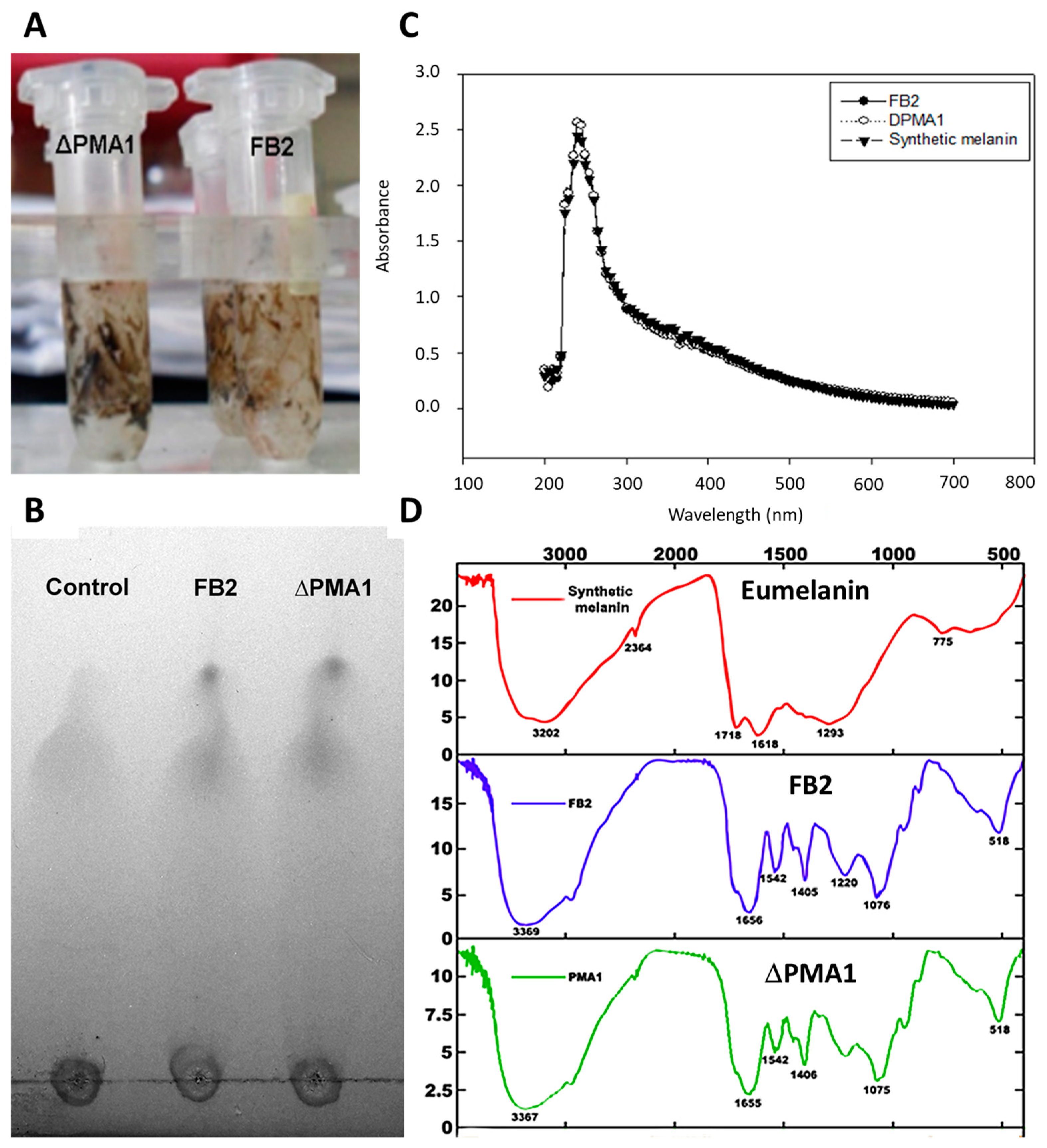
3.4. Isolation and Identification of the Intracellular Pigment
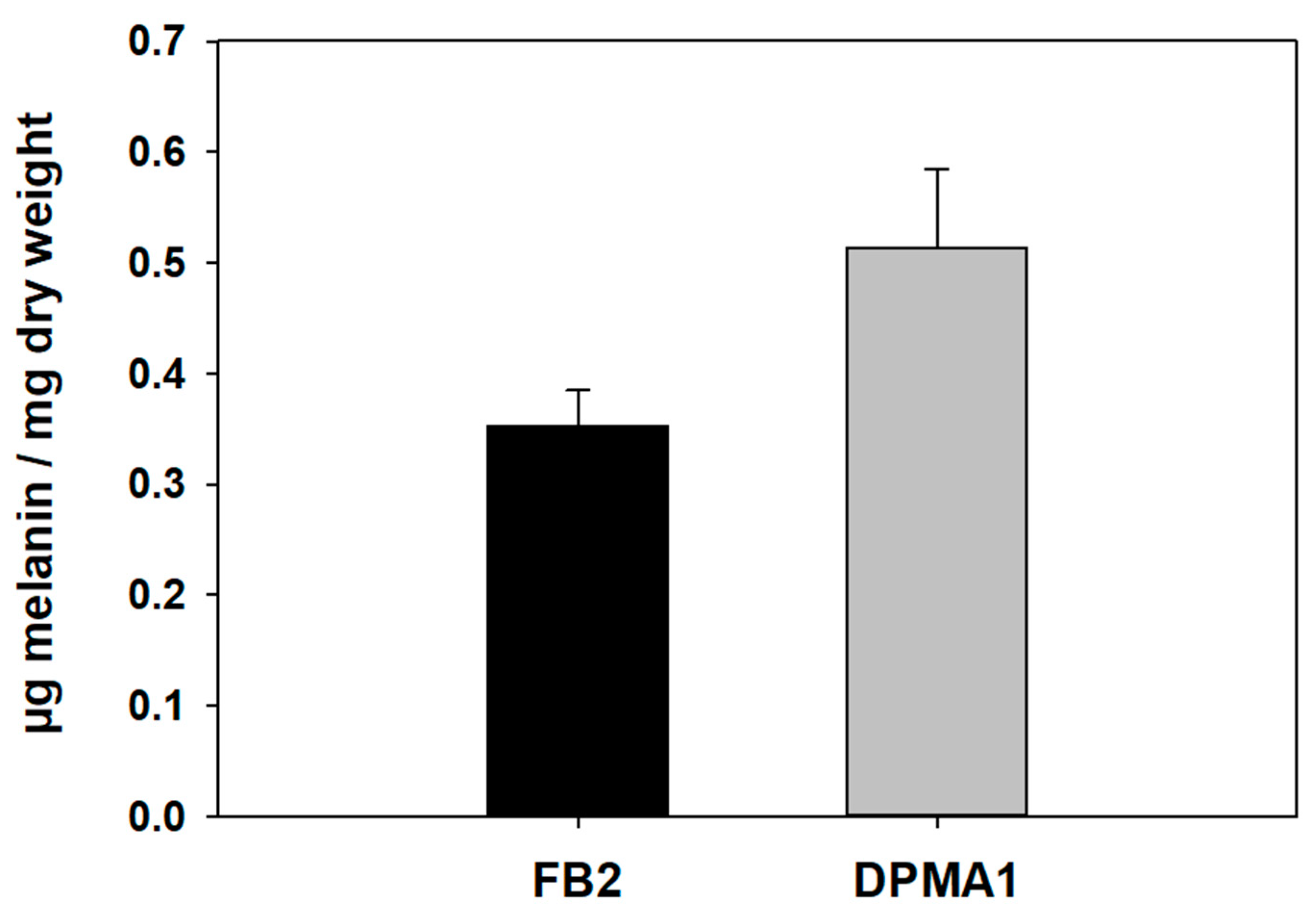
3.4.1. Content of Eumelanin in Wild-Type FB2 and ΔPMA1 Strains
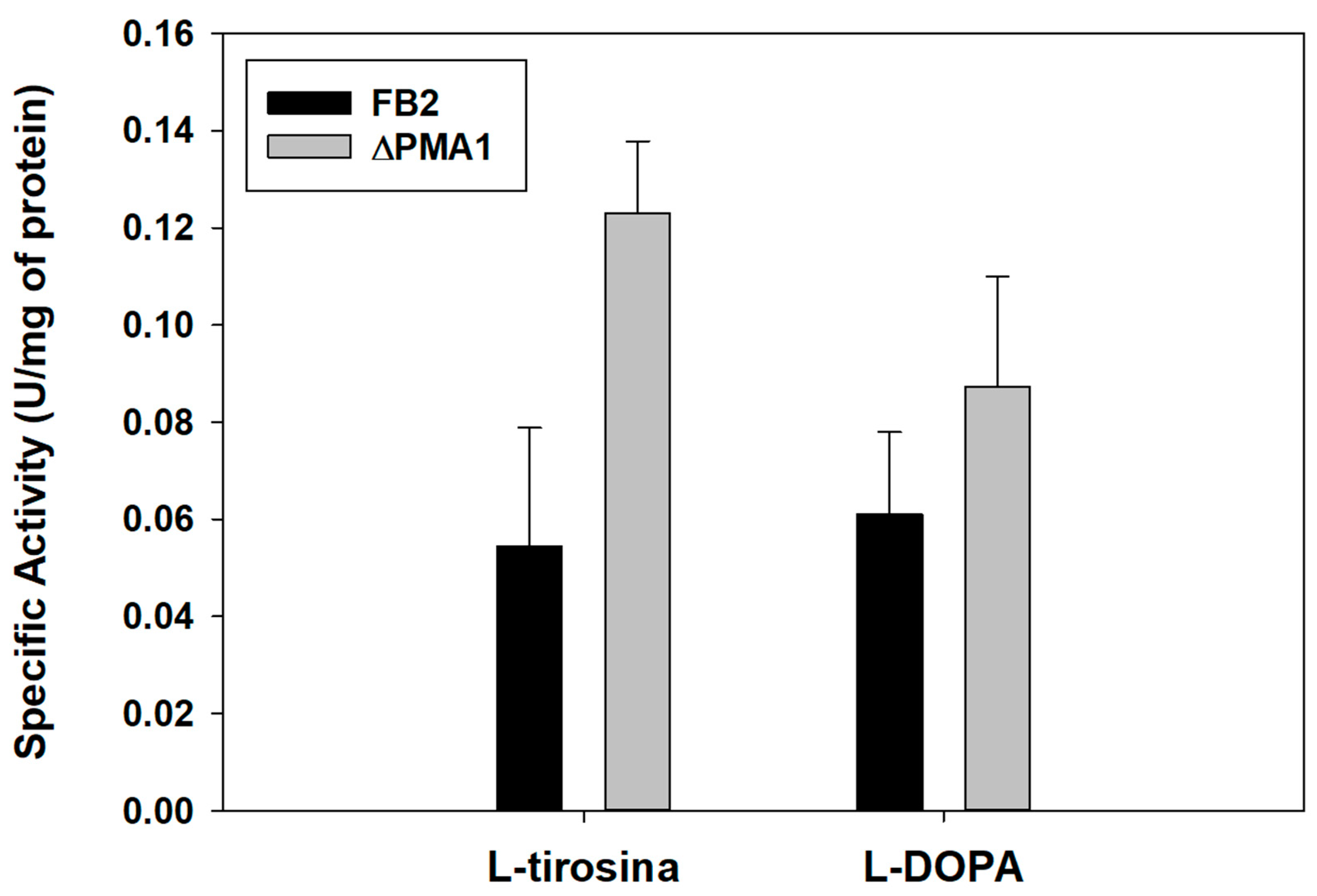
3.4.2. Assay of Tyrosinase Activity with L-Tyrosine and L-DOPA as Substrate
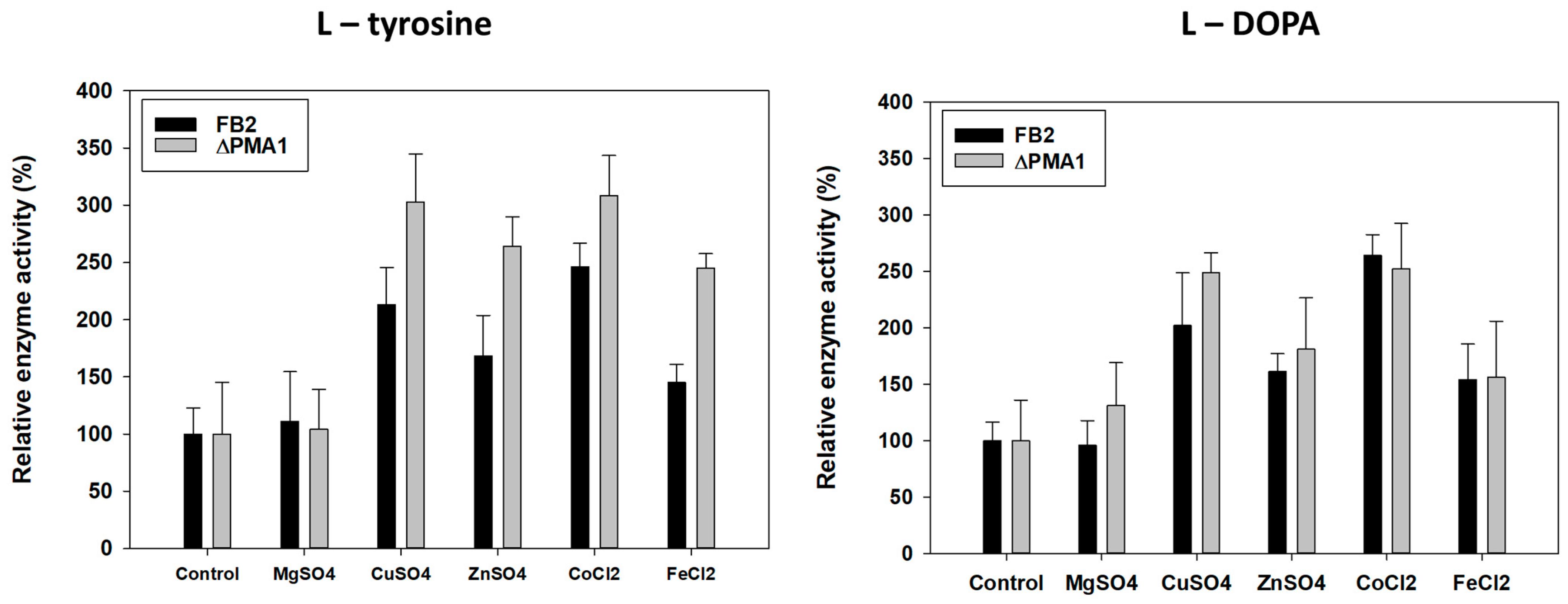
3.5. Tyrosinase Activity Assay with Metal Ions
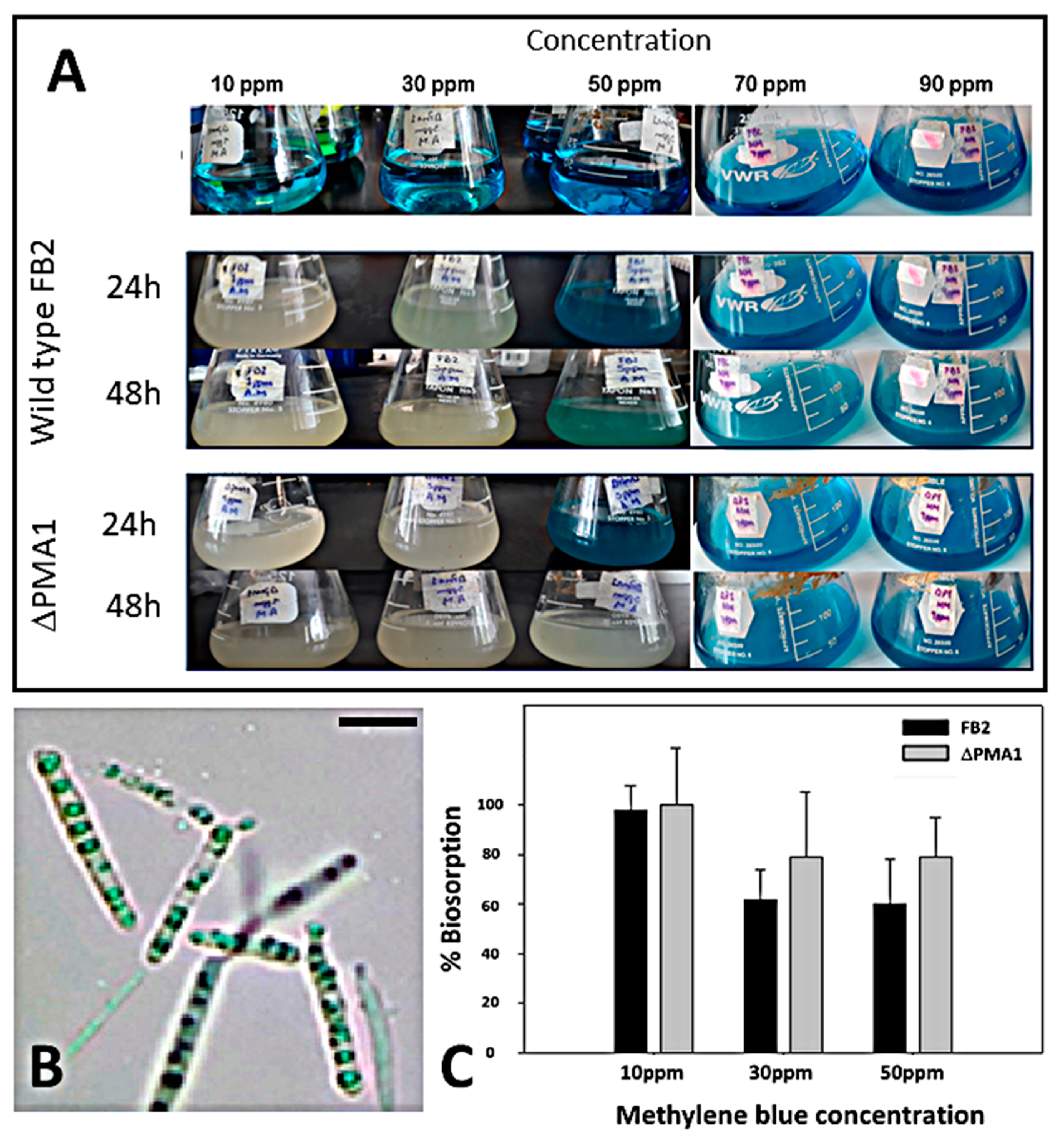
3.6. Removal of Methylene Blue from Water Solutions of FB2 and Mutant Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olicón-Hernández, D.R.; Araiza-Villanueva, M.G.; Pardo, J.P.; Aranda, E.; Guerra-Sánchez, G. New Insights of Ustilago maydis as Yeast Model for Genetic and Biotechnological Research: A Review. Curr. Microbiol. 2019, 76, 917–926. [Google Scholar] [CrossRef]
- Liebal, U.W.; Ullmann, L.; Lieven, C.; Kohl, P.; Wibberg, D.; Zambanini, T.; Blank, L.M. Ustilago maydis Metabolic Characterization and Growth Quantification with a Genome-Scale Metabolic Model. J. Fungi 2022, 8, 524. [Google Scholar] [CrossRef]
- Horst, R.J.; Doehlemann, G.; Wahl, R.; Hofmann, J.; Schmiedl, A.; Kahmann, R.; Kämper, J.; Voll, L.M. A model of Ustilago maydis leaf tumor metabolism. Plant Signal Behav. 2010, 5, 1446–1449. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Paul, J.A.; Wallen, R.M.; Zhao, C.; Shi, T.; Perlin, M.H. Coordinate regulation of Ustilago maydis ammonium transporters and genes involved in mating and pathogenicity. Fungal Biol. 2018, 122, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Lanver, D.; Tollot, M.; Schweizer, G.; Lo Presti, L.; Reissmann, S.; Ma, L.-S.; Schuster, M.; Tanaka, S.; Liang, L.; Ludwig, N.; et al. Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 2017, 15, 409–421. [Google Scholar] [CrossRef]
- Yu, C.; Qi, J.; Han, H.; Wang, P.; Liu, C. Progress in pathogenesis research of Ustilago maydis, and the metabolites involved along with their biosynthesis. Mol. Plant Pathol. 2023, 24, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Heucken, N.; Tang, K.; Hüsemann, L.; Heßler, N.; Müntjes, K.; Feldbrügge, M.; Göhre, V.; Zurbriggen, M.D. Engineering and Implementation of Synthetic Molecular Tools in the Basidiomycete Fungus Ustilago maydis. J. Fungi 2023, 9, 480. [Google Scholar] [CrossRef]
- Djamei, A.; Depotter, J.; Saridis, G.; Prokchorchik, M.; Barghahn, S.; Silva, N.D.S.T.E.; Zuo, W.; Villamil, J.M.; Doehlemann, G. Modulation of Host Immunity and Development by Ustilago maydis. In Plant Relationships: Fungal-Plant Interactions; Scott, B., Mesarich, C., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 3–30. [Google Scholar] [CrossRef]
- Lin, J.-S.; Happel, P.; Kahmann, R. Nuclear status and leaf tumor formation in the Ustilago maydis–maize pathosystem. New Phytol. 2021, 231, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Li, Y.; Zhang, W.; Jia, Y.; Wang, Y.; Ma, Y.; Lv, X.; Xuan, Y.; Du, W. Early infection response of fungal biotroph Ustilago maydis in maize. Front. Plant Sci. 2022, 13, 970897. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.J.B.; Casadevall, A. Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; Saber, W.I.A. Natural Melanin: Current Trends, and Future Approaches, with Especial Reference to Microbial Source. Polymers 2022, 14, 1339. [Google Scholar] [CrossRef] [PubMed]
- Kyrmizi, I.; Ferreira, H.; Carvalho, A.; Figueroa, J.A.L.; Zarmpas, P.; Cunha, C.; Akoumianaki, T.; Stylianou, K.; Deepe, G.S.; Samonis, G.; et al. Calcium sequestration by fungal melanin inhibits calcium–calmodulin signalling to prevent LC3-associated phagocytosis. Nat. Microbiol. 2018, 3, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Meng, X.; Mo, C.; Wei, X.; Ma, A. Melanin of fungi: From classification to application. World J. Microbiol. Biotechnol. 2022, 38, 228. [Google Scholar] [CrossRef] [PubMed]
- Gow Neil, A.R.; Latge, J.-P.; Munro Carol, A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Rodríguez-Piña, A.L.; Juárez-Montiel, M.; Hernández-Sánchez, I.E.; Rodríguez-Hernández, A.A.; Bautista, E.; Becerra-Flora, A.; López-Villegas, E.O.; Jiménez-Bremont, J.F. The Ustilago maydis null mutant strains of the RNA-binding protein UmRrm75 accumulate hydrogen peroxide and melanin. Sci. Rep. 2019, 9, 10813. [Google Scholar] [CrossRef]
- Reyes-Fernández Esmeralda, Z.; Shi, Y.-M.; Grün, P.; Bode Helge, B.; Bölker, M. An Unconventional Melanin Biosynthesis Pathway in Ustilago maydis. Appl. Environ. Microb. 2021, 87, e01510–e01520. [Google Scholar] [CrossRef]
- Fei, W.; Liu, Y. Biotrophic Fungal Pathogens: A Critical Overview. Appl. Biochem. Biotech. 2023, 195, 1–16. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Jayawardena, R.S.; Chen, Y.-J.; Konta, S.; Tibpromma, S.; Abeywickrama, P.D.; Gomdola, D.; Balasuriya, A.; Xu, J.; Lumyong, S.; et al. Diversity and Function of Appressoria. Pathogens 2021, 10, 746. [Google Scholar] [CrossRef]
- Rassabina, A.; Khabibrakhmanova, V.; Babaev, V.; Daminova, A.; Minibayeva, F. Melanins from the Lichens Lobaria pulmonaria and Lobaria retigera as Eco-Friendly Adsorbents of Synthetic Dyes. Int. J. Mol. Sci. 2022, 23, 15605. [Google Scholar] [CrossRef]
- Almeida-Paes, R.; Figueiredo-Carvalho, M.H.G.; da Silva, L.B.R.; Gerfen, G.; de S Araújo, G.R.; Frases, S.; Zancopé-Oliveira, R.M.; Nosanchuk, J.D. Candida glabrata produces a melanin-like pigment that protects against stress conditions encountered during parasitism. Future Microbiol. 2021, 16, 509–520. [Google Scholar] [CrossRef] [PubMed]
- García-Carnero, L.C.; Clavijo-Giraldo, D.M.; Gómez-Gaviria, M.; Lozoya-Pérez, N.E.; Tamez-Castrellón, A.K.; López-Ramírez, L.A.; Mora-Montes, H.M. Early Virulence Predictors during the Candida Species–Galleria mellonella Interaction. J. Fungi 2020, 6, 152. [Google Scholar] [CrossRef]
- Yuan, Z.; Druzhinina, I.S.; Gibbons, J.G.; Zhong, Z.; Van de Peer, Y.; Rodriguez, R.J.; Liu, Z.; Wang, X.; Wei, H.; Wu, Q.; et al. Divergence of a genomic island leads to the evolution of melanization in a halophyte root fungus. ISME J. 2021, 15, 3468–3479. [Google Scholar] [CrossRef]
- Chrissian, C.; Lin, C.P.-C.; Camacho, E.; Casadevall, A.; Neiman, A.M.; Stark, R.E. Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae. J. Fungi 2020, 6, 329. [Google Scholar] [CrossRef]
- Szilágyi, M.; Anton, F.; Pócsi, I.; Emri, T. Autolytic enzymes are responsible for increased melanization of carbon stressed Aspergillus nidulans cultures. J. Basic. Microb. 2018, 58, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.; Vij, R.; Chrissian, C.; Prados-Rosales, R.; Gil, D.; O’Meally, R.N.; Cordero, R.J.B.; Cole, R.N.; McCaffery, J.M.; Stark, R.E.; et al. The structural unit of melanin in the cell wall of the fungal pathogen Cryptococcus neoformans. J. Biol. Chem. 2019, 294, 10471–10489. [Google Scholar] [CrossRef]
- Smith, D.F.Q.; Casadevall, A. The Role of Melanin in Fungal Pathogenesis for Animal Hosts. In Fungal Physiology and Immunopathogenesis; Rodrigues, M.L., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–30. [Google Scholar] [CrossRef]
- Azumi, J.; Takeda, T.; Shimada, Y.; Aso, H.; Nakamura, T. The Organogermanium Compound THGP Suppresses Melanin Synthesis via Complex Formation with L-DOPA on Mushroom Tyrosinase and in B16 4A5 Melanoma Cells. Int. J. Mol. Sci. 2019, 20, 4785. [Google Scholar] [CrossRef]
- Pacelli, C.; Cassaro, A.; Maturilli, A.; Timperio, A.M.; Gevi, F.; Cavalazzi, B.; Stefan, M.; Ghica, D.; Onofri, S. Multidisciplinary characterization of melanin pigments from the black fungus Cryomyces antarcticus. Appl. Microbiol. Biot. 2020, 104, 6385–6395. [Google Scholar] [CrossRef]
- Vázquez-Carrada, M.; Feldbrügge, M.; Olicón-Hernández, D.R.; Guerra-Sánchez, G.; Pardo, J.P. Functional Analysis of the Plasma Membrane H+-ATPases of Ustilago maydis. J. Fungi 2022, 8, 550. [Google Scholar] [CrossRef]
- Terfrüchte, M.; Joehnk, B.; Fajardo-Somera, R.; Braus, G.H.; Riquelme, M.; Schipper, K.; Feldbrügge, M. Establishing a versatile Golden Gate cloning system for genetic engineering in fungi. Fungal Genet. Biol. 2014, 62, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Olicón-Hernández, D.R.; Hernández-Lauzardo, A.N.; Pardo, J.P.; Peña, A.; Velázquez-del Valle, M.G.; Guerra-Sánchez, G. Influence of chitosan and its derivatives on cell development and physiology of Ustilago maydis. Int. J. Biol. Macromol. 2015, 79, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Harki, E.; Talou, T.; Dargent, R. Purification, characterisation and analysis of melanin extracted from Tuber. melanosporum Vitt. Food Chem. 1997, 58, 69–73. [Google Scholar] [CrossRef]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein Measurement With The Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Santiago, M.; Strobel, S. Chapter Twenty-Four—Thin Layer Chromatography. In Methods in Enzymology; Lorsch, J., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 533, pp. 303–324. [Google Scholar]
- Goldfeder, M.; Kanteev, M.; Adir, N.; Fishman, A. Influencing the monophenolase/diphenolase activity ratio in tyrosinase. BBA—Proteins Proteom. 2013, 1834, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Liang, Q.; Li, Y.; Li, X.; Liu, X.; Zhang, D.; Li, J. Effects of metal ions on activity and structure of phenoloxidase in Penaeus vannamei. Int. J. Biol. Macromol. 2021, 174, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Carreira, A.; Ferreira, L.M.; Loureiro, V. Production of brown tyrosine pigments by the yeast Yarrowia lipolytica. J. Appl. Microbiol. 2001, 90, 372–379. [Google Scholar] [CrossRef]
- Singh, S.; Nimse, S.B.; Mathew, D.E.; Dhimmar, A.; Sahastrabudhe, H.; Gajjar, A.; Ghadge, V.A.; Kumar, P.; Shinde, P.B. Microbial melanin: Recent advances in biosynthesis, extraction, characterization, and applications. Biotechnol. Adv. 2021, 53, 107773. [Google Scholar] [CrossRef] [PubMed]
- Walker Claire, A.; Gómez Beatriz, L.; Mora-Montes Héctor, M.; Mackenzie Kevin, S.; Munro Carol, A.; Brown Alistair, J.P.; Gow Neil, A.R.; Kibbler Christopher, C.; Odds Frank, C. Melanin Externalization in Candida albicans Depends on Cell Wall Chitin Structures. Eukaryotic Cell 2010, 9, 1329–1342. [Google Scholar] [CrossRef]
- Zhan, F.; He, Y.; Zu, Y.; Li, T.; Zhao, Z. Characterization of melanin isolated from a dark septate endophyte (DSE), Exophiala pisciphila. World J. Microbiol. Biotechnol. 2011, 27, 2483–2489. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Nosanchuk, J.D.; Webber, J.B.W.; Emerson, R.J.; Camesano, T.A.; Casadevall, A. Microstructure of Cell Wall-Associated Melanin in the Human Pathogenic Fungus Cryptococcus neoformans. Biochemistry 2005, 44, 3683–3693. [Google Scholar] [CrossRef] [PubMed]
- Schlesser, A.; Ulaszewski, S.; Ghislain, M.; Goffeau, A. A second transport ATPase gene in Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 19480–19487. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.R.; Sá-Correia, I. Transcription patterns of PMA1 and PMA2 genes and activity of plasma membrane H+-ATPase in Saccharomyces cerevisiae during diauxic growth and stationary phase. Yeast 2003, 20, 207–219. [Google Scholar] [CrossRef]
- Robles-Martínez, L.; Pardo, J.P.; Miranda, M.; Mendez, T.L.; Matus-Ortega, M.G.; Mendoza-Hernández, G.; Guerra-Sánchez, G. The basidiomycete Ustilago maydis has two plasma membrane H+-ATPases related to fungi and plants. J. Bioenerg. Biomembr. 2013, 45, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, K.; Streibel, M.; Jahn, B.; Haase, G.; Brakhage, A.A. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 2003, 38, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. [Google Scholar] [CrossRef]
- Williamson Peter, R.; Wakamatsu, K.; Ito, S. Melanin Biosynthesis in Cryptococcus neoformans. J. Bacteriol. 1998, 180, 1570–1572. [Google Scholar] [CrossRef]
- Toledo, A.V.; Franco, M.E.E.; Yanil Lopez, S.M.; Troncozo, M.I.; Saparrat, M.C.N.; Balatti, P.A. Melanins in fungi: Types, localization and putative biological roles. Physiol. Mol. Plant Pathol. 2017, 99, 2–6. [Google Scholar] [CrossRef]
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure–function correlations in tyrosinases. Protein Sci. 2015, 24, 1360–1369. [Google Scholar] [CrossRef]
- Fenoll, L.G.; Peñalver, M.J.; Rodríguez-López, J.N.; Varón, R.; García-Cánovas, F.; Tudela, J. Tyrosinase kinetics: Discrimination between two models to explain the oxidation mechanism of monophenol and diphenol substrates. Int. J. Biochem. Cell Biol. 2004, 36, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Misuraca, G.; D’Ischia, M.; Prota, G. Effect of metal ions on the kinetics of tyrosine oxidation catalysed by tyrosinase. Biochem. J. 1985, 228, 647–651. [Google Scholar] [CrossRef]
- Sargın, İ.; Arslan, G.; Kaya, M. Microfungal spores (Ustilago maydis and U. digitariae) immobilised chitosan microcapsules for heavy metal removal. Carbohyd Polym. 2016, 138, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Mattoon, E.R.; Cordero, R.J.B.; Casadevall, A. Fungal Melanins and Applications in Healthcare, Bioremediation and Industry. J. Fungi 2021, 7, 488. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sequence (5′-3′) | Use |
|---|---|---|
| ΔPma1 | ||
| UF FP | GGTCTCGCCTGCAATATTCCCATTTCCATTTCCATTC | Flank construction |
| UF RP | GGTCTCCAGGCCGATGAAAGAAAAAAGACTACCGAG | Flank construction |
| DF FP | GGTCTCCGGCCATCCTCATTCTCGACGAAGTGATTC | Flank construction |
| DF RP | GGTCTCGCTGCAATATTCGACCTCTGAATCGCTAGC | Flank construction |
| P1 | CGGTGTTGCCATGAACACCGATGGCCAGTG | Diagnostic PCR |
| P2 | GAGGGCAACGGATTCGAGCTTCTTGGTCTT | Diagnostic PCR |
| ΔPma2 | ||
| UF FP | GGTCTCGCCTGCAATATTCAACCTCTAAGACTCGCTT | Flank construction |
| UF RP | GGTCTCCAGGCCTCTGCCTCTTATCTTGCTCTCTTAG | Flank construction |
| DF FP | GGTCTCCGGCCGGGGAAACGTGGAGAAGGTCGCGAAA | Flank construction |
| DF RP | GGTCTCGCTGCAATATTACCACCCTGTGCCCTCTAG | Flank construction |
| P1 | ACGCTTGACAATCTCGTACTTGTGCTCGGGG | Diagnostic PCR |
| P2 | GAGGGCAACGGATTCGAGCTTCTTGGTCTT | Diagnostic PCR |
| Plasmid | ||
| pUma2342 | Transforming plasmid to delete Pma1 gen | |
| pUma2343 | Transforming plasmid to delete Pma2 gen | |
| p1507 | Hyg resistance cassette | |
| Strain | Culture Medium | Growth Rate (µ) | Generation Time (g) |
|---|---|---|---|
| FB2 | YPD | 0.297 ± 0.04 h−1 | 2.34 h |
| MM | 0.271 ± 0.08 h−1 | 2.55 h | |
| ∆PMA1 | YPD | 0.282 ± 0.09 h−1 | 2.45 h |
| MM | 0.247 ± 0.09 h−1 | 2.80 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-López, C.; Vázquez-Carrada, M.; Flores-Herrera, O.; Pardo, J.P.; Olicón-Hernández, D.R.; Guerra-Sánchez, G. Ustilago maydis Yeast Mutant Produces Cytosolic Melanin by Tyrosine-Tyrosinase Activity with Stain Biosorption Capability. Appl. Sci. 2023, 13, 11288. https://doi.org/10.3390/app132011288
Martínez-López C, Vázquez-Carrada M, Flores-Herrera O, Pardo JP, Olicón-Hernández DR, Guerra-Sánchez G. Ustilago maydis Yeast Mutant Produces Cytosolic Melanin by Tyrosine-Tyrosinase Activity with Stain Biosorption Capability. Applied Sciences. 2023; 13(20):11288. https://doi.org/10.3390/app132011288
Chicago/Turabian StyleMartínez-López, Cinthya, Melissa Vázquez-Carrada, Oscar Flores-Herrera, Juan Pablo Pardo, Dario Rafael Olicón-Hernández, and Guadalupe Guerra-Sánchez. 2023. "Ustilago maydis Yeast Mutant Produces Cytosolic Melanin by Tyrosine-Tyrosinase Activity with Stain Biosorption Capability" Applied Sciences 13, no. 20: 11288. https://doi.org/10.3390/app132011288
APA StyleMartínez-López, C., Vázquez-Carrada, M., Flores-Herrera, O., Pardo, J. P., Olicón-Hernández, D. R., & Guerra-Sánchez, G. (2023). Ustilago maydis Yeast Mutant Produces Cytosolic Melanin by Tyrosine-Tyrosinase Activity with Stain Biosorption Capability. Applied Sciences, 13(20), 11288. https://doi.org/10.3390/app132011288







