Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI
Abstract
:1. Introduction
2. Materials and Methods
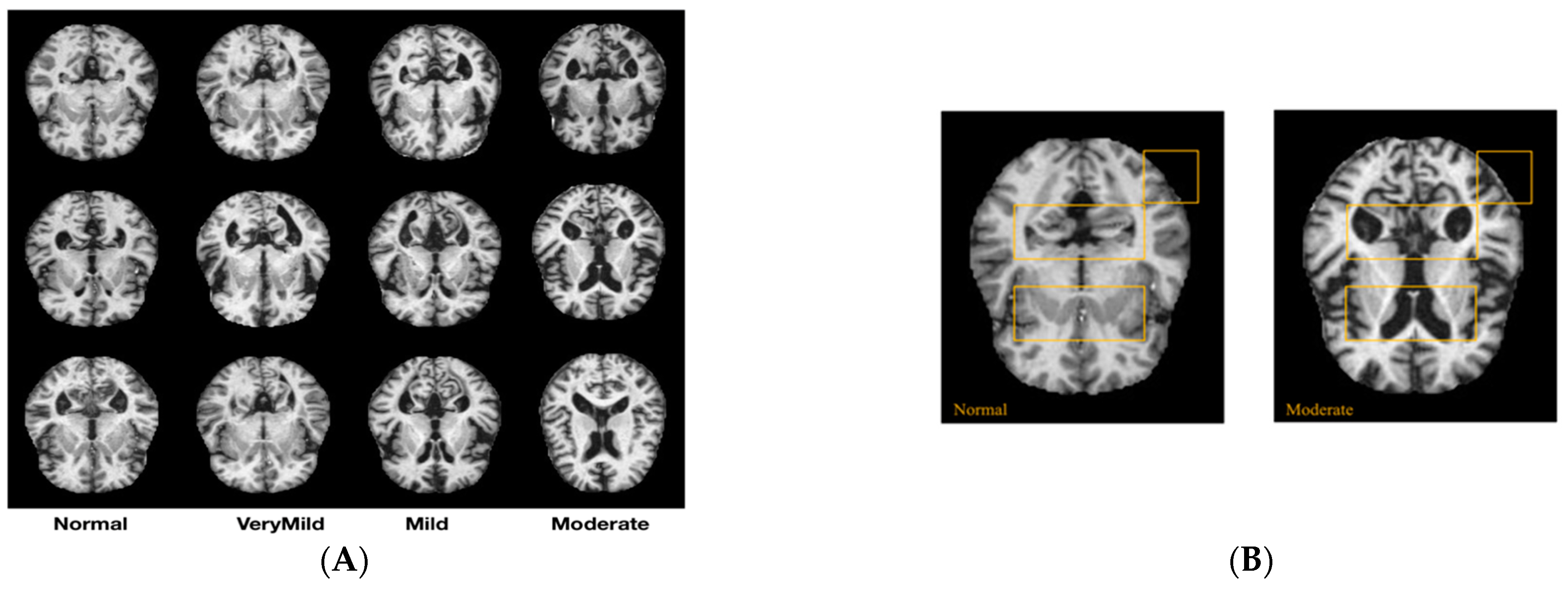
2.1. Dataset Description
2.2. Proposed Method for AD Diagnosis
- For a consistent starting point, identical weights were assigned to each model during initialization.
- During training, the designated hyper-parameters were applied to the training set, while the progress of training was consistently monitored and assessed on the validation set.
- Using identical test set, performance metrics were computed for every model undergoing evaluation.
- To determine whether or not there were significant differences in performance metrics between models, statistical tests were employed, including ROC curve tests.
3. Results
3.1. Prediction Performance
3.2. ROC Curves
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Helaly, H.A.; Badawy, M.; Haikal, A.Y. Deep Learning Approach for Early Detection of Alzheimer’s Disease. Cogn. Comput. 2021, 14, 1711–1727. [Google Scholar] [CrossRef] [PubMed]
- Islam, J.; Zhang, Y. Brain MRI Analysis for Alzheimer’s Disease Diagnosis Using an Ensemble System of Deep Convolutional Neural Networks. Brain Inform. 2018, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Andrushia, A.D.; Sagayam, K.M.; Dang, H.; Pomplun, M.; Quach, L. Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis. Appl. Sci. 2021, 11, 9199. [Google Scholar] [CrossRef]
- Raju, M.; Thirupalani, M.; Vidhyabharathi, S.; Thilagavathi, S. Deep Learning Based Multilevel Classification of Alzheimer’s Disease Using MRI Scans. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1084, 012017. [Google Scholar] [CrossRef]
- Sethi, M.; Ahuja, S.; Rani, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. An Exploration: Alzheimer’s Disease Classification Based on Convolutional Neural Network. BioMed Res. Int. 2022, 2022, 8739960. [Google Scholar] [CrossRef] [PubMed]
- Pushpa, B.R.; Amal, P.S.; Kamal, N.P. Detection and stage wise classification of Alzheimer disease using deep learning methods. Int. J. Recent Technol. Eng. (IJRTE) 2019, 7, 206–212. [Google Scholar]
- Sadat, S.U.; Shomee, H.H.; Awwal, A.; Amin, S.N.; Reza, T.; Parvez, M.Z. Alzheimer’s disease detection and classification using transfer learning technique and ensemble on Convolutional Neural Networks. In Proceedings of the 2021 IEEE Interna-tional Conference on Systems, Man, and Cybernetics (SMC), Melbourne, Australia, 17–20 October 2021. [Google Scholar] [CrossRef]
- Vrahatis, A.G.; Skolariki, K.; Krokidis, M.G.; Lazaros, K.; Exarchos, T.P.; Vlamos, P. Revolutionizing the Early Detection of Alzheimer’s Disease through Non-Invasive Biomarkers: The Role of Artificial Intelligence and Deep Learning. Sensors 2023, 23, 4184. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Danker, A.; Wirgård Wiklund, J. Using Transfer Learning to Classify Different Stages of Alzheimer’s Disease; KTH Royal INSTITUTE of Technology: Stockholm, Sweden, 2021; p. 33. [Google Scholar]
- Saleem, T.J.; Zahra, S.R.; Wu, F.; Alwakeel, A.; Alwakeel, M.; Jeribi, F.; Hijji, M. Deep Learning-Based Diagnosis of Alzheimer’s Disease. J. Pers. Med. 2022, 12, 815. [Google Scholar] [CrossRef]
- Saleem, T.J.; Chishti, M.A. Deep learning for Internet of Things data analytics. Procedia Comput. Sci. 2019, 163, 381–390. [Google Scholar] [CrossRef]
- Suk, H.I.; Shen, D. Deep ensemble sparse regression network for Alzheimer’s disease diagnosis. In International Workshop on Machine Learning in Medical Imaging; Springer: Cham, Switzerland, 2016; pp. 113–121. [Google Scholar]
- Billones, C.D.; Demetria, O.J.L.D.; Hostallero, D.E.D.; Naval, P.C. DemNet: A convolutional neural network for the detection of Alzheimer’s disease and mild cognitive impairment. In Proceedings of the 2016 IEEE Region 10 Conference (TENCON), Singapore, 22–25 November 2016; pp. 3724–3727. [Google Scholar]
- Sarraf, S.; Tofighi, G. Classification of alzheimer’s disease using fmri data and deep learning convolutional neural networks. arXiv 2016, arXiv:1603.08631. [Google Scholar]
- Sarraf, S.; Tofighi, G. Classification of Alzheimer’s disease structural MRI data by deep learning convolutional neural networks. arXiv 2016, arXiv:1607.06583. [Google Scholar]
- Gunawardena, K.A.N.N.P.; Rajapakse, R.N.; Kodikara, N.D. Applying convolutional neural networks for pre-detection of alzheimer’s disease from structural MRI data. In Proceedings of the 2017 24th International Conference on Mechatronics and Machine Vision in Practice (M2VIP), Auckland, New Zealand, 21–23 November 2017; pp. 1–7. [Google Scholar]
- Basaia, S.; Agosta, F.; Wagner, L.; Canu, E.; Magnani, G.; Santangelo, R.; Filippi, M.; Alzheimer’s Disease Neuroimaging Initiative. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage Clin. 2019, 21, 101645. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Phillips, P.; Sui, Y.; Liu, B.; Yang, M.; Cheng, H. Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J. Med. Syst. 2018, 42, 85. [Google Scholar] [CrossRef]
- Karasawa, H.; Liu, C.-L.; Ohwada, H. Deep 3d convolutional neural network architectures for alzheimer’s disease diagnosis. In Asian Conference on Intelligent Information and Database Systems; Springer: Cham, Switzerland, 2018; pp. 287–296. [Google Scholar]
- Goceri, E. Diagnosis of Alzheimer’s disease with Sobolev gradient-based optimization and 3D convolutional neural network. Int. J. Numer. Methods Biomed. Eng. 2019, 35, e3225. [Google Scholar] [CrossRef]
- Tang, H.; Yao, E.; Tan, G.; Guo, X. A fast and accurate 3D fine-tuning convolutional neural network for Alzheimer’s disease diagnosis. In International CCF Conference on Artificial Intelligence; Springer: Singapore, 2018; pp. 115–126. [Google Scholar]
- Spasov, S.E.; Passamonti, L.; Duggento, A.; Liò, P.; Toschi, N. A multi-modal convolutional neural network framework for the prediction of Alzheimer’s disease. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1271–1274. [Google Scholar]
- Basheera, S.; Ram, M.S.S. Convolution neural network–based Alzheimer’s disease classification using hybrid enhanced independent component analysis based segmented gray matter of T2 weighted magnetic resonance imaging with clinical valuation. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2019, 5, 974–986. [Google Scholar] [CrossRef]
- Jiang, X.; Chang, L.; Zhang, Y.-D. Classification of Alzheimer’s disease via eight-layer convolutional neural network with batch normalization and dropout techniques. J. Med. Imaging Health Inform. 2020, 10, 1040–1048. [Google Scholar] [CrossRef]
- Mehmood, A.; Maqsood, M.; Bashir, M.; Shuyuan, Y. A deep siamese convolution neural network for multi-class classification of alzheimer disease. Brain Sci. 2020, 10, 84. [Google Scholar] [CrossRef]
- Raju, M.; Gopi, V.P.; Anitha, V.S.; Wahid, K.A. Multi-class diagnosis of Alzheimer’s disease using cascaded three dimensional convolutional neural network. Phys. Eng. Sci. Med. 2020, 43, 1219–1228. [Google Scholar] [CrossRef]
- Sun, J.; Yan, S.; Song, C.; Han, B. Dual-functional neural network for bilateral hippocampi segmentation and diagnosis of Alzheimer’s disease. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 445–455. [Google Scholar] [CrossRef]
- Dyrba, M.; Hanzig, M.; Altenstein, S.; Bader, S.; Ballarini, T.; Brosseron, F.; Buerger, K.; Cantré, D.; Dechent, P.; Dobisch, L.; et al. Improving 3D convolutional neural network comprehensibility via interactive visualization of relevance maps: Evaluation in Alzheimer’s disease. arXiv 2020, arXiv:2012.10294. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Halm-Lutterodt, N.V.; Tang, H.; Mecum, A.; Mesregah, M.K.; Ma, Y.; Li, H.; Zhang, F.; Wu, Z.; Yao, E.; et al. Automated MRI-based deep learning model for detection of Alzheimer’s disease process. Int. J. Neural Syst. 2020, 30, 2050032. [Google Scholar] [CrossRef] [PubMed]
- Solano-Rojas, B.; Villalón-Fonseca, R. A Low-Cost Three-Dimensional DenseNet Neural Network for Alzheimer’s Disease Early Discovery. Sensors 2021, 21, 1302. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Marques, G.; Agarwal, D.; de la Torre Díez, I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020, 96, 106691. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, D.; Berbis, M.A.; Martín-Noguerol, T.; Luna, A.; Garcia, S.C.P.; de la Torre-Díez, I. End-to-end deep learning architectures using 3D neuroimaging biomarkers for early Alzheimer’s diagnosis. Mathematics 2022, 10, 2575. [Google Scholar] [CrossRef]
- Agarwal, D.; Berbís, M.Á.; Luna, A.; Lipari, V.; Ballester, J.B.; de la Torre-Díez, I. Automated Medical Diagnosis of Alzheimer’s Disease Using an Efficient Net Convolutional Neural Network. J. Med. Syst. 2023, 47, 57. [Google Scholar] [CrossRef] [PubMed]
- Luz, E.; Silva, P.L.; Silva, R.; Silva, L.; Guimarães, J.; Miozzo, G.; Moreira, G.; Menotti, D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. arXiv 2021, arXiv:2004.05717. [Google Scholar] [CrossRef]
- Zebin, T.; Rezvy, S. COVID-19 Detection and Disease Progression Visualization: Deep Learning on Chest X-rays for Classification and Coarse Localization. Appl. Intell. 2021, 51, 1010–1021. [Google Scholar] [CrossRef]
- Dubey, S. Alzheimer’s Dataset (4 Class of Images). Available online: https://www.kaggle.com/tourist55/alzheimers-dataset-4-class-of-images (accessed on 29 November 2020).
- Fu’adah, Y.N.; Wijayanto, I.; Pratiwi, N.K.C.; Taliningsih, F.F.; Rizal, S.; Pramudito, M.A. Automated Classification of Alzheimer’s Disease Based on MRI Image Processing Using Convolutional Neural Network (CNN) With AlexNet Architecture. J. Physics Conf. Ser. 2021, 1844, 012020. [Google Scholar] [CrossRef]
- Atlases—NIST. Available online: https://nist.mni.mcgill.ca/atlases/ (accessed on 20 March 2023).
- Kang, H.; Park, H.-M.; Ahn, Y.; Van Messem, A.; De Neve, W. Towards a Quantitative Analysis of Class Activation Mapping for Deep Learning-Based Computer-Aided Diagnosis. In Proceedings of the Medical Imaging 2021: Image Perception, Observer Performance, and Technology Assessment, Online, 15–19 February 2021; Samuelson, F.W., Taylor-Phillips, S., Eds.; SPIE: Bellingham, WA, USA, 2021; Volume 11599. [Google Scholar] [CrossRef]
- Lim, B.Y.; Lai, K.W.; Haiskin, K.; Kulathilake, K.A.S.H.; Ong, Z.C.; Hum, Y.C.; Dhanalakshmi, S.; Wu, X.; Zuo, X. Deep Learning Model for Prediction of Progressive Mild Cognitive Impairment to Alzheimer’s Disease Using Structural MRI. Front. Aging Neurosci. 2022, 14, 876202. [Google Scholar] [CrossRef]
- Kabani, A.; El-Sakka, M.R. Object Detection and Localization Using Deep Convolutional Networks with Softmax Activation and Multi-class Log Loss. In Proceedings of the 2016 International Conference on Image Analysis and Recognition (ICIAR 2016), Póvoa de Varzim, Portugal, 13–15 July 2016; pp. 358–366. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Prakash, D.; Madusanka, N.; Bhattacharjee, S.; Park, H.-G.; Kim, C.-H.; Choi, H.-K. A Comparative Study of Alzheimer’s Disease Classification using Multiple Transfer Learning Models. J. Multimedia Inf. Syst. 2019, 6, 209–216. [Google Scholar] [CrossRef]
- Kandel, I.; Castelli, M. The Effect of Batch Size on the Generalizability of the Convolutional Neural Networks on a Histopathology Dataset. ICT Express 2020, 6, 312–315. [Google Scholar] [CrossRef]









| Parameters | Proposed Method (PDL) | VGG16 | ResNet50 |
|---|---|---|---|
| Number of epochs | 30 | 30 | 30 |
| Batch Size | 34 | 34 | 34 |
| Optimiser | Adam | Adam | Adam |
| Learning Rate | 0.0001 | 0.0001 | 0.0001 |
| Loss Function | Categorical cross-entropy | Categorical cross-entropy | Categorical cross-entropy |
| Steps | Operator | Resolution | Channels | Layers |
|---|---|---|---|---|
| 1 | Conv 3 × 3 | 224 × 224 | 32 | 1 |
| 2 | MBconv1, 3 × 3 | 112 × 112 | 16 | 1 |
| 3 | MBconv1, 3 × 3 | 112 × 112 | 24 | 2 |
| 4 | MBconv1, 5 × 5 | 56 × 56 | 40 | 2 |
| 5 | MBconv1, 3 × 3 | 28 × 28 | 80 | 3 |
| 6 | MBconv1, 5 × 5 | 14 × 14 | 112 | 3 |
| 7 | MBconv1, 5 × 5 | 14 × 14 | 192 | 4 |
| 8 | MBconv1, 3 × 3 | 7 × 7 | 320 | 1 |
| 9 | Conv 1 × 1 pooling | 7 × 7 | 1280 | 1 |
| Models | Class Label | Precision (%) | Recall (%) | F1-Score (%) | Average Score |
|---|---|---|---|---|---|
| Normal | 98 | 97 | 98 | 97.6% | |
| ResNet50 | Very Mild | 93 | 97 | 95 | 95% |
| Mild | 97 | 91 | 94 | 94% | |
| Moderate | 100 | 100 | 100 | 100% | |
| Normal | 98 | 99 | 99 | 98.6% | |
| VGG16 | Very Mild | 98 | 97 | 97 | 97.3% |
| Mild | 100 | 98 | 99 | 99% | |
| Moderate | 100 | 100 | 100 | 100% | |
| Normal | 99 | 99 | 98 | 98.6% | |
| Proposed Method | Very Mild | 100 | 99 | 99 | 99.3% |
| Mild | 100 | 99 | 99 | 99.3% | |
| Moderate | 100 | 100 | 100 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altwijri, O.; Alanazi, R.; Aleid, A.; Alhussaini, K.; Aloqalaa, Z.; Almijalli, M.; Saad, A. Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI. Appl. Sci. 2023, 13, 13051. https://doi.org/10.3390/app132413051
Altwijri O, Alanazi R, Aleid A, Alhussaini K, Aloqalaa Z, Almijalli M, Saad A. Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI. Applied Sciences. 2023; 13(24):13051. https://doi.org/10.3390/app132413051
Chicago/Turabian StyleAltwijri, Omar, Reem Alanazi, Adham Aleid, Khalid Alhussaini, Ziyad Aloqalaa, Mohammed Almijalli, and Ali Saad. 2023. "Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI" Applied Sciences 13, no. 24: 13051. https://doi.org/10.3390/app132413051
APA StyleAltwijri, O., Alanazi, R., Aleid, A., Alhussaini, K., Aloqalaa, Z., Almijalli, M., & Saad, A. (2023). Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI. Applied Sciences, 13(24), 13051. https://doi.org/10.3390/app132413051






