Comparative Insights into the Microbial Diversity and Community Structure of Striga hermonthica-Infested Maize Rhizosphere
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sample Collection
2.2. Analysis of the Physicochemical Properties of Soil
2.3. DNA Extraction of Soil Samples and Library Preparation
2.4. Metagenome Sequence Annotation and Statistical Analysis
3. Results
3.1. Physicochemical Properties of Striga Hermonthica-Infested Maize Rhizosphere and Bulk Soils
3.2. Metagenome Dataset of the Striga Hermonthica-Infested Soil across the Sampling Sites
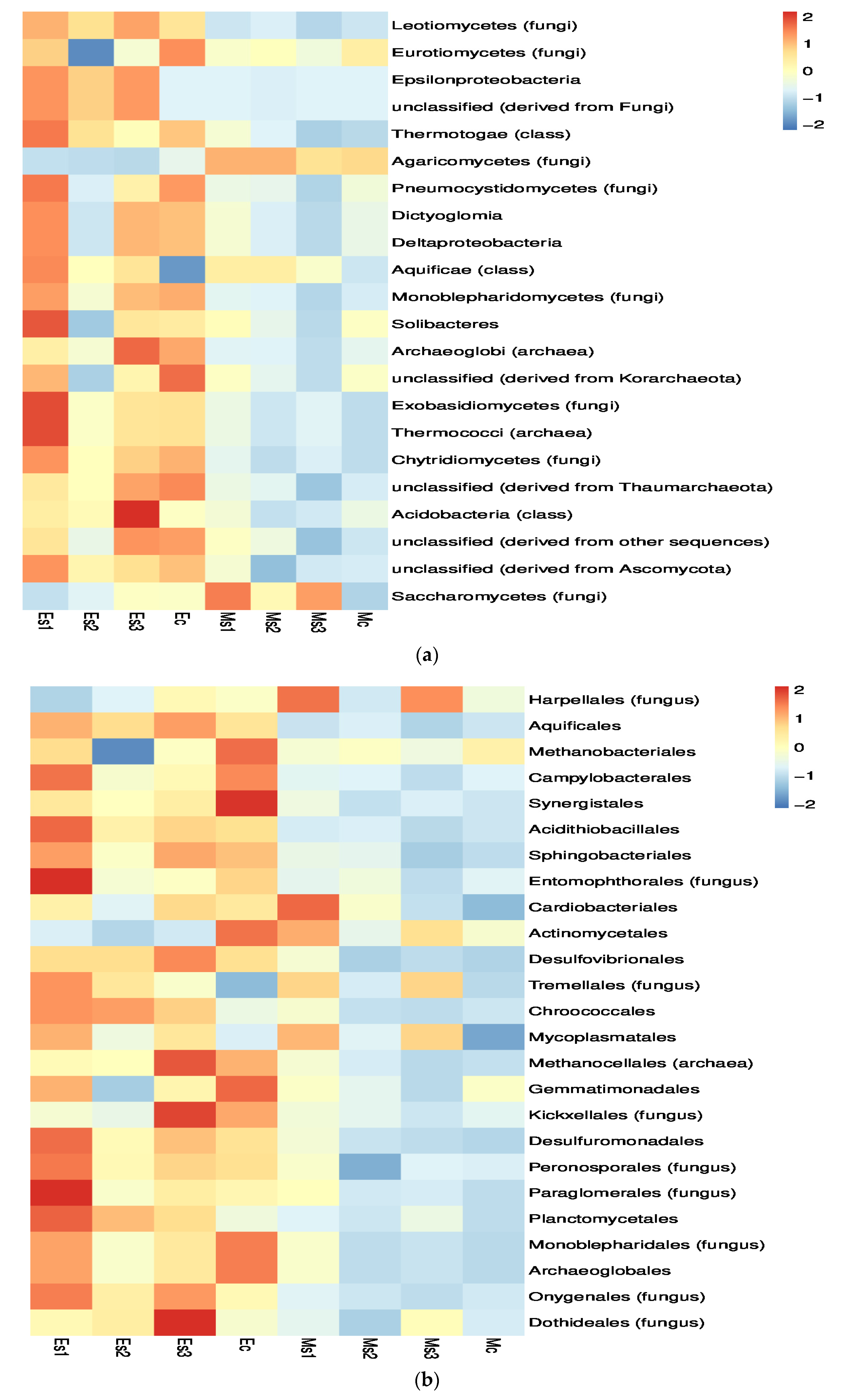
3.3. Distribution of the Major Microbiome Phyla in the Infected Soil Samples
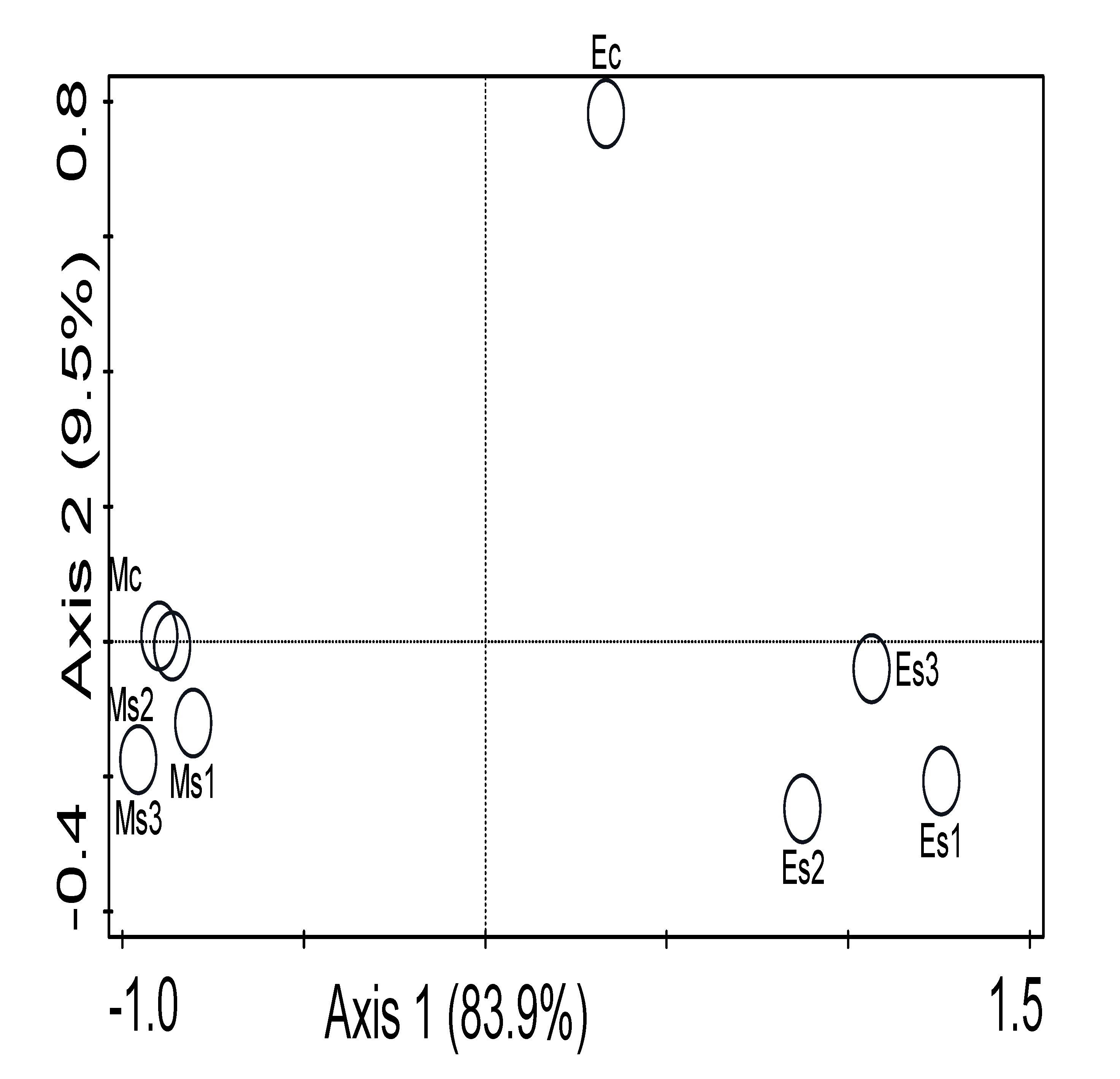
3.4. Diversity Indices of the Microbiome of the Striga-Infested Sampling Sites
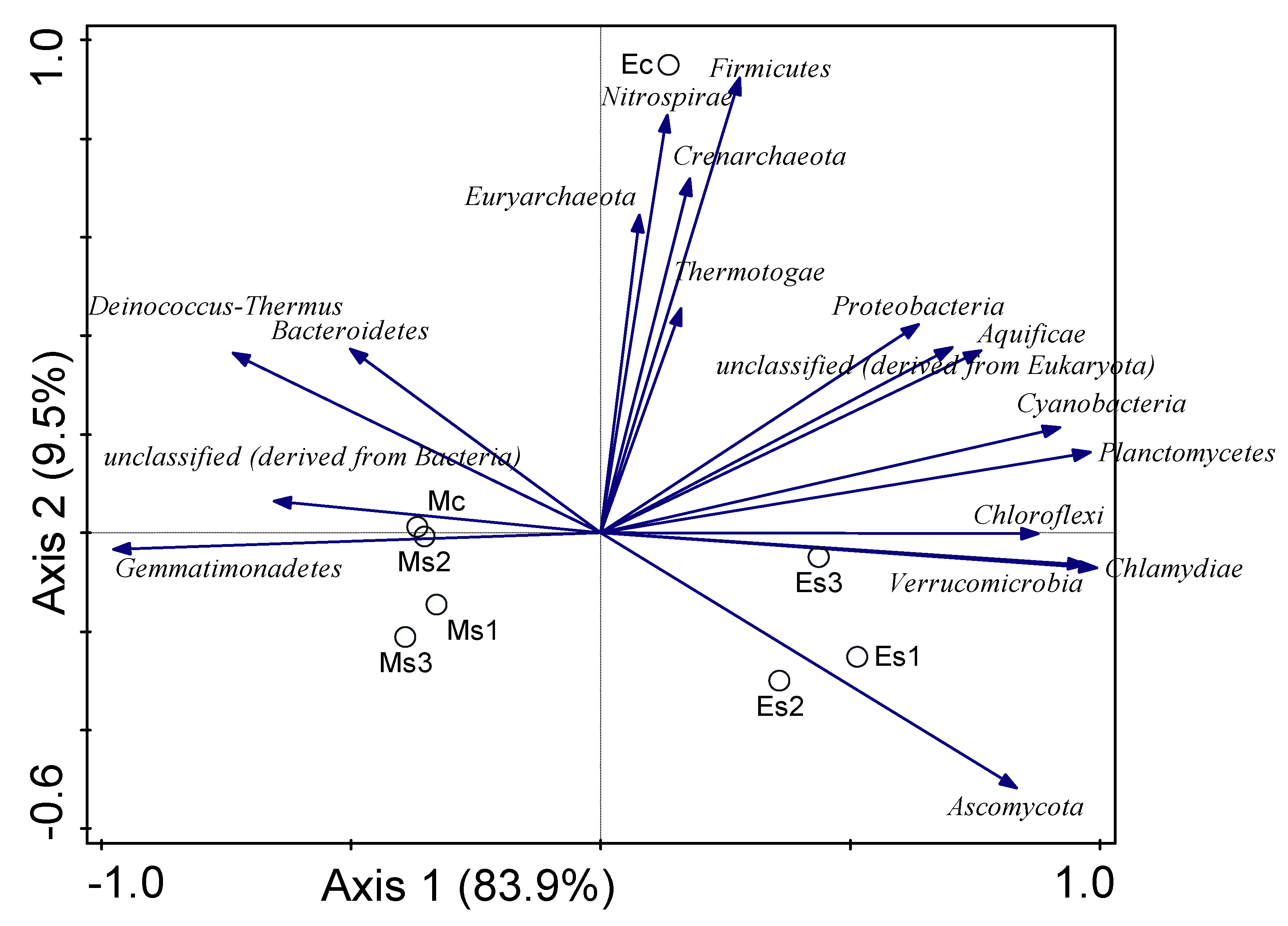
3.5. Influence of Environmental Variables on the Microbial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- David, O.G.; Ayangbenro, A.S.; Odhiambo, J.J.; Babalola, O.O. Striga hermonthica: A highly destructive pathogen in maize production. Environ. Chall. 2022, 8, 100590. [Google Scholar] [CrossRef]
- Atera, E.A.; Itoh, K.; Onyango, J.C. Evaluation of Ecologies and Severity of Striga Weed on Rice in Sub-Saharan Africa. Agric. Biol. J. N. Am. 2011, 2, 752–760. [Google Scholar] [CrossRef]
- Babalola, O.O.; Odhiambo, G.D. Effect of Inoculation with Klebsiella oxytoca ‘10 mkr 7’on Striga Suicidal Germination in Zea mays. World Appl. Sci. J. 2008, 3, 57–62. [Google Scholar]
- Mounde, L.G.; Anteyi, W.O.; Rasche, F. Tripartite interaction between Striga spp., cereals, and plant root-associated microorganisms: A review. CAB Rev. 2020, 15, 1–17. [Google Scholar]
- Seck, P.A.; Diagne, A.; Mohanty, S.; Wopereis, M. Crops that feed the world 7: Rice. Food Secur. 2012, 4, 7–24. [Google Scholar] [CrossRef]
- Gressel, J.; Meir, S.; Herschkovitz, Y.; Al-Ahmad, H.; Babalola, O.; Amsellem, Z. Transgenic biocontrol agents to overcome evolutionary barriers. In Integrating New Technologies for Striga Control: Towards Ending the Witch-Hunt; World Scientific: Singapore, 2007; pp. 313–323. [Google Scholar]
- Ejeta, G. Breeding for Striga resistance in sorghum: Exploitation of an intricate host–parasite biology. Crop Sci. 2007, 47, S-216–S-227. [Google Scholar] [CrossRef]
- Scholes, J.D.; Press, M.C. Striga infestation of cereal crops—An unsolved problem in resource limited agriculture. Curr. Opin. Plant Biol. 2008, 11, 180–186. [Google Scholar] [CrossRef]
- Massenssini, A.; Bonduki, V.; Melo, C.; Tótola, M.; Ferreira, F.; Costa, M. Soil microorganisms and their role in the interactions between weeds and crops. Planta Daninha 2014, 32, 873–884. [Google Scholar] [CrossRef] [Green Version]
- Babalola, O.O.; Sanni, A.I.; Odhiambo, G.D.; Torto, B. Plant growth-promoting rhizobacteria do not pose any deleterious effect on cowpea and detectable amounts of ethylene are produced. World J. Microbiol. Biotechnol. 2007, 23, 747–752. [Google Scholar] [CrossRef]
- Babalola, O.O.; Osir, E.O.; Sanni, A.I. Characterization of potential ethylene-producing rhizosphere bacteria of Striga-infested maize and sorghum. Afr. J. Biotechnol. 2002, 1, 67–69. [Google Scholar]
- Crowther, T.W.; Maynard, D.S.; Leff, J.W.; Oldfield, E.E.; McCulley, R.L.; Fierer, N.; Bradford, M.A. Predicting the responsiveness of soil biodiversity to deforestation: A cross-biome study. Glob. Chang. Biol. 2014, 20, 2983–2994. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.W.; Curtis, P.S. Effects of forest management on soil C and N storage: Meta analysis. For. Ecol. Manag. 2001, 140, 227–238. [Google Scholar] [CrossRef]
- Yang, T.; Ala, M.; Zhang, Y.; Wu, J.; Wang, A.; Guan, D. Characteristics of soil moisture under different vegetation coverage in Horqin Sandy Land, northern China. PLoS ONE 2018, 13, e0198805. [Google Scholar] [CrossRef]
- Lin, G.; He, Y.; Lu, J.; Chen, H.; Feng, J. Seasonal variations in soil physicochemical properties and microbial community structure influenced by Spartina alterniflora invasion and Kandelia obovata restoration. Sci. Total Environ. 2021, 797, 149213. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Zhang, J.; Xu, W.; Mou, Z. Microbial Community Structure in the Sediments and Its Relation to Environmental Factors in Eutrophicated Sancha Lake. Int. J. Environ. Res. Public Health 2019, 16, 1931. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Li, M.-H.; Ni, H. Land use change effects on diversity of soil bacterial, Acidobacterial and fungal communities in wetlands of the Sanjiang Plain, northeastern China. Sci. Rep. 2019, 9, 18535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Cartabia, A.; Lalaymia, I.; Declerck, S. Arbuscular mycorrhizal fungi and production of secondary metabolites in medicinal plants. Mycorrhiza 2022, 32, 221–256. [Google Scholar] [CrossRef]
- Babalola, O.; Sanni, A.; Odhiambo, G. Isolation of rhizobacteria associated with maize and assessment of their potential for use in Striga hermonthica (Del.) Benth. suicidal germination. J. Trop. Microbiol 2004, 3, 64–70. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Q.; Wang, B.; Wang, X.; Franks, A.E.; Teng, Y.; Li, Z.; Luo, Y. Changes in the abundance and structure of bacterial communities under long-term fertilization treatments in a peanut monocropping system. Plant Soil 2015, 395, 415–427. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, G.; Chen, H.; Chen, C.; Wang, J.; Ai, S.; Wei, D.; Li, D.; Ma, B.; Tang, C. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhu, N.; Zhou, G.; Dang, P.; Yang, X.; Qiu, L.; Huang, M.; Gong, Y.; Zhao, S.; Chen, J. Response of soil microbial community to plant composition changes in broad-leaved forests of the karst area in Mid-Subtropical China. PeerJ 2022, 10, e12739. [Google Scholar] [CrossRef] [PubMed]
- Szoboszlay, M.; Dohrmann, A.B.; Poeplau, C.; Don, A.; Tebbe, C.C. Impact of land-use change and soil organic carbon quality on microbial diversity in soils across Europe. FEMS Microbiol. Ecol. 2017, 93, fix146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Organic farming enhances the diversity and community structure of endophytic archaea and fungi in maize plant: A shotgun approach. J. Soil Sci. Plant Nutr. 2020, 20, 2587–2599. [Google Scholar] [CrossRef]
- Hoogsteen, M.J.; Lantinga, E.A.; Bakker, E.J.; Groot, J.C.; Tittonell, P.A. Estimating soil organic carbon through loss on ignition: Effects of ignition conditions and structural water loss. Eur. J. Soil Sci. 2015, 66, 320–328. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Wright, A.F.; Bailey, J.S. Organic carbon, total carbon, and total nitrogen determinations in soils of variable calcium carbonate contents using a Leco CN-2000 dry combustion analyzer. Commun. Soil Sci. Plant Anal. 2001, 32, 3243–3258. [Google Scholar] [CrossRef]
- Kachurina, O.; Zhang, H.; Raun, W.; Krenzer, E. Simultaneous determination of soil aluminum, ammonium-and nitrate-nitrogen using 1M potassium chloride extraction. Commun. Soil Sci. Plant Anal. 2000, 31, 893–903. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Calvo, N.I.R.; Echeverría, H.E.; Rozas, H.S. Determination of sulfate concentration in soil: Depth of sampling. Commun. Soil Sci. Plant Anal. 2009, 40, 1624–1633. [Google Scholar] [CrossRef]
- Gammon, N.J. Determination of total potassium and sodium in sandy soils by flame photometer. Soil Sci. 1951, 71, 211–214. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [PubMed] [Green Version]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Khomtchouk, B.B.; Hennessy, J.R.; Wahlestedt, C. shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics. PLoS ONE 2017, 12, e0176334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrell, A.A.; Frank, C. Bacterial endophyte communities in the foliage of coast redwood and giant sequoia. Front. Microbiol. 2015, 6, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-Y.; Zhou, X.; Guo, D.; Zhao, J.-H.; Yan, L.; Feng, G.-Z.; Gao, Q.; Yu, H.; Zhao, L.-P. Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann. Microbiol. 2019, 69, 1461–1473. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, R.M.; Pepper, I.L. Earth environments. In Environmental Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 57–82. [Google Scholar]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.; Han, W.; Zhang, W.; Christie, P.; Goulding, K.; Vitousek, P.; Zhang, F. Significant acidification in major Chinese croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [Green Version]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Dasgupta, D.; Brahmaprakash, G. Soil microbes are shaped by soil physico-chemical properties: A brief review of existing literature. Inter-Natl. J. Plant Soil Sci. 2021, 59–71. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Chang. Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.-Y.; Zhang, X.-Y.; Dai, X.-Q.; Fu, X.-L.; Yang, F.-T.; Liu, X.-Y.; Sun, X.-M.; Wen, X.-F.; Schaeffer, S. Changes in soil microbial community composition in response to fertilization of paddy soils in subtropical China. Appl. Soil Ecol. 2014, 84, 140–147. [Google Scholar] [CrossRef]
- Guo, Z.; Han, J.; Li, J.; Xu, Y.; Wang, X. Effects of long-term fertilization on soil organic carbon mineralization and microbial community structure. PLoS ONE 2019, 14, e0211163. [Google Scholar]
- McGowan, A.R.; Nicoloso, R.S.; Diop, H.E.; Roozeboom, K.L.; Rice, C.W. Soil organic carbon, aggregation, and microbial community structure in annual and perennial biofuel crops. Agron. J. 2019, 111, 128–142. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Adams, J.M.; Shi, Y.; Li, Y.; Song, X.; Zhao, X.; Chu, H.; Zhang, G.-L. Depth-dependent patterns of bacterial communities and assembly processes in a typical red soil critical zone. Geomicrobiol. J. 2020, 37, 201–212. [Google Scholar] [CrossRef]
- You, Y.; Wang, J.; Huang, X.; Tang, Z.; Liu, S.; Sun, O.J. Relating microbial community structure to functioning in forest soil organic carbon transformation and turnover. Ecol. Evol. 2014, 4, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Liu, J.; Yu, Z.; Li, Y.; Jin, J.; Liu, X.; Wang, G. Changes of bacterial community compositions after three years of biochar application in a black soil of northeast China. Appl. Soil Ecol. 2017, 113, 11–21. [Google Scholar] [CrossRef]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Alotaibi, M.O.; Mohammed, A.E.; Eltom, K.H. Metagenomic analysis of bacterial communities of Wadi Namar Lake, Riyadh, Saudi Arabia. Saudi J. Biol. Sci. 2022, 29, 3749–3758. [Google Scholar] [CrossRef]
- Kim, M.; Kim, W.-S.; Tripathi, B.M.; Adams, J. Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microb. Ecol. 2014, 67, 837–848. [Google Scholar] [CrossRef]
- Akinola, S.A.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic insight into the community structure of maize-rhizosphere bacteria as predicted by different environmental factors and their functioning within plant proximity. Microorganisms 2021, 9, 1419. [Google Scholar] [CrossRef]
- Chukwuneme, C.F.; Ayangbenro, A.S.; Babalola, O.O. Impacts of land-use and management histories of maize fields on the structure, composition, and metabolic potentials of microbial communities. Curr. Plant Biol. 2021, 28, 100228. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic profiling of the community structure, diversity, and nutrient pathways of bacterial endophytes in maize plant. Antonie Van Leeuwenhoek 2020, 113, 1559–1571. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, A.M.; Al-Khatri, B.; Nasehi, A.; Al-Shihi, M.; Al-Mahmooli, I.H.; N Maharachchikumbura, S.S. High Fungal Diversity and Dominance by Ascomycota in Dam Reservoir Soils of Arid Climates. Int. J. Agric. Biol. 2017, 19, 682–688. [Google Scholar] [CrossRef]
- Dube, J.P.; Valverde, A.; Steyn, J.M.; Cowan, D.A.; Van der Waals, J.E. Differences in bacterial diversity, composition and function due to long-term agriculture in soils in the eastern free State of South Africa. Diversity 2019, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Jat, S.L.; Suby, S.; Parihar, C.M.; Gambhir, G.; Kumar, N.; Rakshit, S. Microbiome for sustainable agriculture: A review with special reference to the corn production system. Arch. Microbiol. 2021, 203, 2771–2793. [Google Scholar] [CrossRef]





| pH | E.C | Potassium | Sodium | Available Phosphorus | Organic Carbon | Organic Matter | Total Nitrogen | Total Carbon | Nitrate | Ammonium | Sulfate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Es1 | 6.8 c | 14.0 g | 0.1 g | 1.0 h | 50.0 a | 11.4 c | 19.7 c | 1.3 c | 57.0 c | 0.3 f | 0.01 f | 0.0 e |
| Es2 | 7.5 a | 54.0 d | 0.4 e | 1.1 f | 33.8 d | 17.4 a | 29.10 a | 1.9 a | 87.0 a | 0.8 e | 0.02 e | 0.0 e |
| Es3 | 7.1 b | 18.0 f | 0.20 f | 1.2 e | 41.7 b | 12.6 b | 21.6 b | 1.4 b | 63.0 b | 0.3 h | 0.01 g | 0.0 e |
| Ec | 6.1 d | 11.0 h | 0.1 h | 1.1 g | 40.3 c | 6.9 d | 11.9 d | 0.8 d | 34.0 d | 0.3 g | 0.01 f | 0.0 e |
| Ms1 | 5.6 e | 195.0 a | 569.0 a | 9.6 b | 2.4 e | 1.02 h | 0.1 e | 0.1 g | 1.1 h | 56.6 a | 5.2 d | 17.8 a |
| Ms2 | 5.4 f | 71.0 c | 248.0 c | 11.9 a | 0.5 h | 2.8 e | 0.0 h | 0.2 e | 3.0 e | 42.2 b | 15.3 a | 4.4 c |
| Ms3 | 5.4 f | 101.3 b | 338.3 b | 9.1 c | 1.2 f | 1.6 f | 0.04 g | 0.1 f | 1.8 f | 40.6 c | 8.1 b | 7.9 b |
| Mc | 5.3 g | 38.0 e | 198.0 d | 5.9 d | 0.7 g | 1.1 g | 0.1 f | 0.1 h | 1.2 g | 22.9 d | 6.3 c | 1.6 d |
| Es1 | Es2 | Es3 | Ec | Ms1 | Ms2 | MS3 | Mc | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Simpson_1-D | 0.767 | 0.75 | 0.755 | 0.74 | 0.73 | 0.719 | 0.711 | 0.714 | 0.998 |
| Shannon_H | 1.936 | 1.834 | 1.88 | 1.836 | 1.77 | 1.733 | 1.694 | 1.709 | ||
| Evenness_e^H/S | 0.289 | 0.2609 | 0.273 | 0.2613 | 0.24 | 0.2357 | 0.227 | 0.23 | ||
| Class | Simpson_1-D | 0.672 | 0.658 | 0.641 | 0.657 | 0.79 | 0.77 | 0.797 | 0.783 | 0.976 |
| Shannon_H | 1.651 | 1.603 | 1.564 | 1.684 | 1.94 | 1.878 | 1.981 | 1.929 | ||
| Evenness_e^H/S | 0.2084 | 0.199 | 0.1911 | 0.215 | 0.28 | 0.262 | 0.29 | 0.275 | ||
| Order | Simpson_1-D | 0.793 | 0.772 | 0.777 | 0.792 | 0.74 | 0.764 | 0.732 | 0.756 | 0.998 |
| Shannon_H | 1.952 | 1.884 | 1.894 | 1.949 | 1.88 | 1.915 | 1.845 | 1.892 | ||
| Evenness_e^H/S | 0.2817 | 0.263 | 0.266 | 0.281 | 0.26 | 0.272 | 0.253 | 0.265 | ||
| Family | Simpson_1-D | 0.852 | 0.843 | 0.845 | 0.851 | 0.87 | 0.875 | 0.87 | 0.874 | 1 |
| Shannon_H | 2.256 | 2.234 | 2.237 | 2.272 | 2.38 | 2.386 | 2.378 | 2.349 | ||
| Evenness_e^H/S | 0.382 | 0.374 | 0.375 | 0.388 | 0.43 | 0.435 | 0.432 | 0.491 | ||
| Genus | Simpson_1-D | 0.815 | 0.768 | 0.76 | 0.887 | 0.85 | 0.857 | 0.861 | 0.849 | 0.991 |
| Shannon_H | 2.314 | 2.152 | 2.143 | 2.605 | 2.41 | 2.438 | 2.434 | 2.323 | ||
| Evenness_e^H/S | 0.405 | 0.344 | 0.341 | 0.541 | 0.45 | 0.458 | 0.456 | 0.408 |
| Physicochemical Parameter | Contribution % | Pseudo F | p-Value |
|---|---|---|---|
| Available Phosphorus | 86.0 | 22.0 | 0.006 |
| pH | 78.5 | 15.2 | 0.002 |
| Nitrate | 66.8 | 9.4 | 0.034 |
| Sulfate | 35.7 | 2.9 | 0.084 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olowe, O.M.; Ayangbenro, A.S.; Akanmu, A.O.; Kutu, F.R.; Odhiambo, J.J.O.; Babalola, O.O. Comparative Insights into the Microbial Diversity and Community Structure of Striga hermonthica-Infested Maize Rhizosphere. Appl. Sci. 2023, 13, 3260. https://doi.org/10.3390/app13053260
Olowe OM, Ayangbenro AS, Akanmu AO, Kutu FR, Odhiambo JJO, Babalola OO. Comparative Insights into the Microbial Diversity and Community Structure of Striga hermonthica-Infested Maize Rhizosphere. Applied Sciences. 2023; 13(5):3260. https://doi.org/10.3390/app13053260
Chicago/Turabian StyleOlowe, Olumayowa Mary, Ayansina Segun Ayangbenro, Akinlolu Olalekan Akanmu, Funso Raphael Kutu, Jude J. O. Odhiambo, and Olubukola Oluranti Babalola. 2023. "Comparative Insights into the Microbial Diversity and Community Structure of Striga hermonthica-Infested Maize Rhizosphere" Applied Sciences 13, no. 5: 3260. https://doi.org/10.3390/app13053260







