The effectiveness and superiority of the 2DOF IMC-PID controller is verified by three simulations. Firstly, the advantages of the 2DOF IMC-PID controller are explained by its output responses. Secondly, the realization of the 2DOF IMC-PID controller based on the CRNs in DSD response networks is evidenced. Thirdly, the 2DOF IMC-PID division gate control system is built by using CRNs. This ensures that the output of the control system is still correct even if leak reactions occur in the division gate.
3.1. Performance of the 2DOF IMC-PID Biomolecular Controller
In this part, the input signals are split into two situations for simulation. In Case 1, there are no disturbance inputs to the control systems (
) and the reference value input is
. In Case 2, the disturbance input of the control system is
and the reference value input is
. The two simulations shown in this section are the output response curves of the three control systems
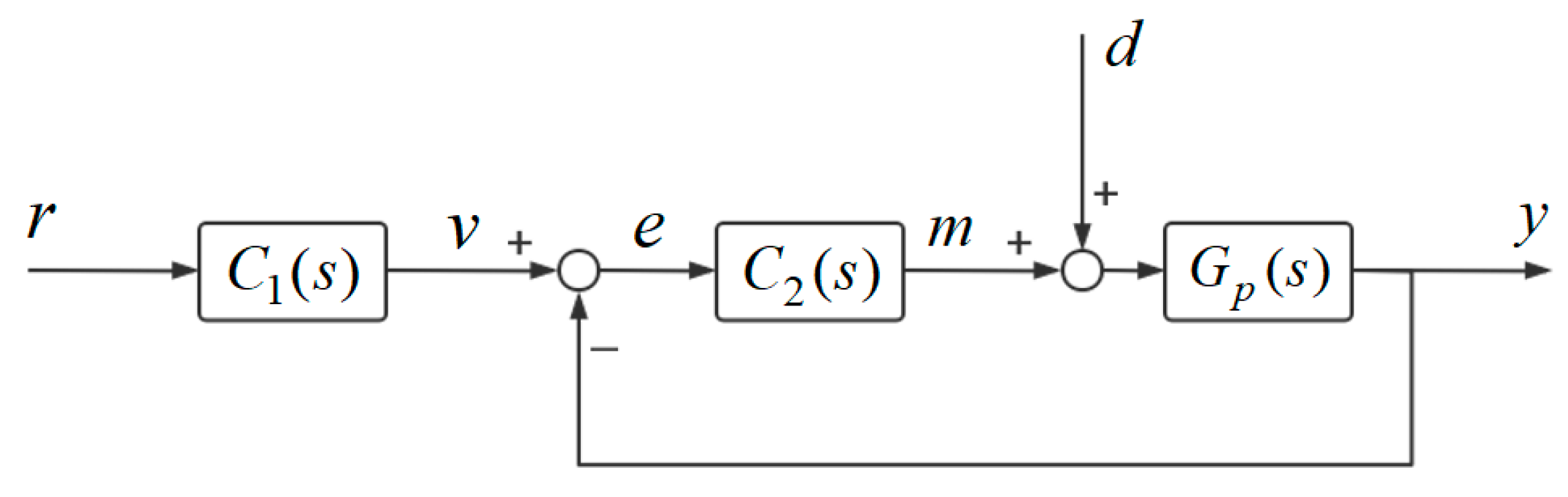
for the same input conditions. The system block diagram for the 1DOF PID control system
and the 2DOF PID control system
are given in
Figure 6 and
Figure 7.
Remark 3. The transfer functions of the controllers shown in Figure 6 and Figure 7 are given as follows: The 1DOF PID controller
based on CRNs is established as
The feedforward controller
is built by CRNs in the following form
The 2DOF PID controller shown in
Figure 7 is modeled based on CRNs through Equations (45)–(57). The parameters of the control systems
and
have been tuned in [
29,
30]. The parameters of the controlled object
and the control system
are given in
Table 1. In
Figure 8a, the rise time and settling time of the control system
are shorter than the control systems
and
. However, the control system
generates a large overshoot when the disturbance signal is
. The overshoot gets reduced by the control systems
and
, respectively. In
Figure 8b, the rise time of the control system
is longer than the control systems
and
.However, the performance of the control system
is better than the control systems
and
for the overshoot and settling time.
Table 2 and
Table 3 are given to show the performance of the three control systems
. In addition, there are only two filtering parameters (
and
) to be adjusted in the 2DOF IMC-PID controller; the complexity of the controller parameter tuning is reduced.
Remark 4. Figure 9 is given to show the output response curves of the 2DOF IMC-PID controller for different values of the two filtering parameters and .
In the simulation shown in Figure 9, the step function is chosen as the disturbance input to the 2DOF IMC-PID control system .
The parameter
is adjusted while keeping the parameter
constant in
Figure 9a. As can be seen, the set-point following characteristic of the controller can be improved by reducing the filtering parameter
. Similarly, the parameter
is adjusted while keeping the parameter
constant. In
Figure 9b, the disturbance rejection performance of the 2DOF IMC-PID controller gets improved by increasing the filtering parameter
. The performance of the controller for the different filtering parameters
and
are shown in
Table 4 and
Table 5.
As a conclusion, the set-point following performance of the controller can be improved by keeping the parameter constant and reducing the parameter . Similarly, the excellent disturbance suppression capability of the controller is attained by keeping the parameter constant and reducing the parameter .
Remark 5. To better demonstrate the rejection performance of the 2DOF IMC-PID controller based on CRNs for different disturbance signals, the trigonometric signal is selected as the disturbance input. The suppression effect of the 2DOF IMC-PID controller based on the CRNs for the trigonometric signal with different frequencies is simulated. The CRNs of the trigonometric signal are as follows: The initial species
is selected to join Equations (58) and (59), and
is the product. In Equation (58), the frequency of the trigonometric function
is determined by the reaction rate
ω. The amplitude of the trigonometric function
is determined by the initial concentration of the reacting specie
, and the relationship between
and
is
. When the trigonometric signal
is selected as the disturbance signal, the output response curves of the 2DOF IMC-PID control system
are given in
Figure 10.
In
Figure 10, it can be seen that the trigonometric signal
can be effectively suppressed by the 2DOF IMC-PID controller. However, the suppression effects are different for the trigonometric signals
with different periods. The disturbance rejection performance of the controller is shown clearly in
Figure 10b. It can be seen that the suppression effect of the 2DOF IMC-PID controller is more obvious when the period of
is smaller. The initial concentrations and reaction rates of the trigonometric signals
with different frequencies are defined in
Table 6.
3.2. Simulation of DSD Reaction Networks
With the professional simulation software Visual DSD, the designs within the framework of CRNs can be automatically simulated to generate DSD reaction networks [
41]. The 2DOF IMC-PID controller is constructed by three basic chemical reactions (i.e., catalysis, degradation, and annihilation), and now it is necessary to map the CRNs of the 2DOF IMC-PID controller into DSD reaction networks. With deterministic simulations, the output response profiles of the control systems
in DSD response networks are depicted in
Figure 11.
The simulation results show that the 2DOF IMC-PID controller can achieve a steady-state following of the reference signal. The rise time of the control system
is shorter than the control systems
and
, but the settling time of the control system
is shorter than the control systems
and
. In addition, the overshoot generated by the 2DOF IMC-PID control system is smaller than the 1DOF PID control system and 2DOF PID control system. The performance of the three control systems
is shown in
Table 7. The DSD reactions for constructing the 2DOF IMC-PID controller can be obtained in Visual DSD. (The relevant details are given in the
Supplementary Materials.)
In this part, the effectiveness of the 2DOF IMC-PID control strategy in the DSD reaction networks is verified. The simulation results show that oscillations around the output signal become suppressed and overshoots become reduced with the 2DOF IMC-PID controller.
3.3. Building the Division Gate Control System to Restrain Leak Reactions
It has been verified that the design of the 2DOF IMC-PID controller is implementable within the framework of CRNs, and, next, the controller will be used to construct a division gate control system to suppress leak reactions. The division gate is established by combining the three operations of addition, subtraction, and multiplication, which achieves the ratio operation of the two biomolecular signals
and
[
22].
The division gate in
Figure 12 is built based on CRNs by Equations (60)–(63).
Equations (60) and (61) represent the error signal
and set
. Equation (62) produces the calculation result
, and the reaction rates
and
. Equation (63) offers the multiplication result of the output signal
with the input signal
and sets
. For example, the initial concentrations of the species are set to
and
. When the initial specie
is affected by the different extent of the leak reactions, it makes
and leads to the calculation of the division gate to a smaller
. Similarly, when the initial specie
is affected by the different extent of the leak reactions, it makes
and leads to the calculation of the division gate to a bigger
. The CRNs of the leak reactions are given as follows.
In Equations (64) and (65), the leak parameters
and
reflect the extent to which leak reactions occur.
Figure 13 presents the effect of various levels of leak reactions on the computations.
As the leak reaction occurs more severely, the error of the calculation result of the divisor gate gets larger. The leak at the dividend leads to small calculations and the leak at the divisor leads to large results. The 2DOF IMC-PID division gate control system in
Figure 14 is established to solve this problem; the calculation results are accurate even when leak responses occur in the division gate.
Different initial reaction concentrations and leak parameters are defined in
Table 8. The first case is a leak in the initial specie
, and the other is a leak in the initial specie
. As
Figure 15 shows, the calculation results of the 2DOF IMC-PID control system are acquired according to the parameters defined in
Table 8.
Remark 6. Another case is discussed for when the initial species and leak simultaneously, and how the calculation result of the division gate changes. The calculation of the division gate is correct when the initial species and leak to the same level, but this is not the ideal result. Once the two species leak to different extents, the calculation result of the division gate will be wrong. However, the problem is solved by the 2DOF IMC-PID division gate control system. Figure 16 is given to show the calculations of the 2DOF IMC-PID division gate control system when the initial species and leak at the same time. As can be observed, the 2DOF IMC-PID division gate control system still obtains correct calculation results when there is a leak at both the divisor and dividend. The concentrations of the initial reaction species and the leak parameters are defined in Table 9. The 2DOF IMC-PID division gate control system is designed to suppress leak reactions, and we discuss different cases of leak reactions of the initial species and in this part. The simulations show that the effectiveness of the 2DOF IMC-PID controller to restrain the leak reaction is demonstrated.