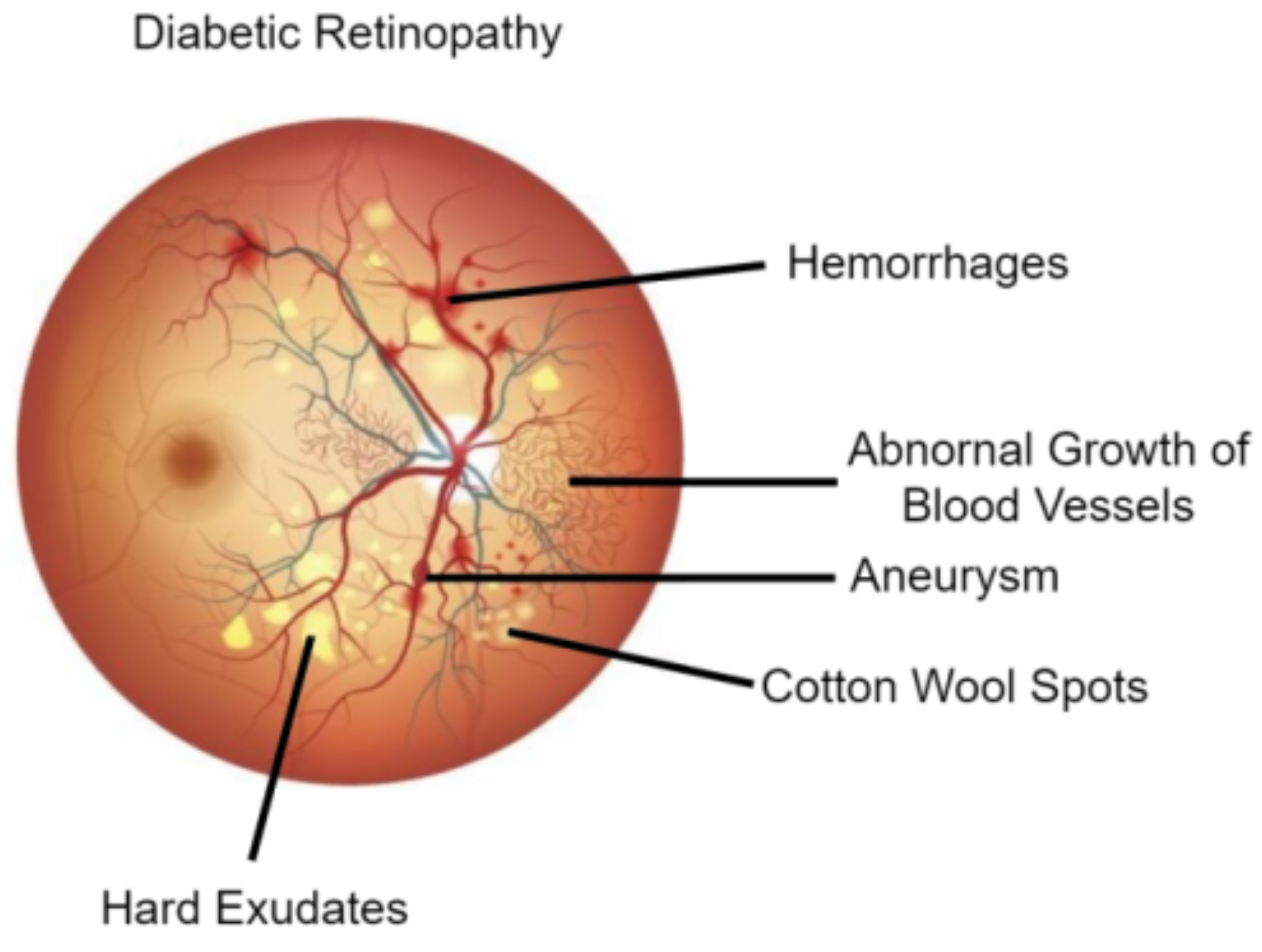
The DR studies reviewed in this article have been classified into two categories which include traditional-based approaches and DL-based studies.
Several traditional methods have been used for EX detection like thresholding, morphology, edge detection, mixture modeling, and region growing. Grayscale morphology was used along with active contour-based segmentation and region-wise classification by the authors of [
7] for automatic EX detection. The authors used a blend of region growing and edge detection in [
8]. They used the Luv color space to get the color difference image of objects. A mean squared Wiener filter in two dimensions was used to remove noise which was trailed by edge detection and an enhancement of the region growing method. Gray level thresholding was used by the authors in [
9] since EX have high local and gray levels. The local properties of EX were exploited and a blend of global as well as local histogram thresholding was used by them. Sanchez et al. [
10] used an EX detection method built using mixture models for obtaining image histograms. This helped them get a dynamic threshold for every image. Some machine learning approaches have also been used in the literature. Niemeijer et al. [
11] used the k-nearest neighbor classifier for detecting red lesions from fundus images. A ranking SVM-boosted convolutional neural network (CNN) was utilized for EX detection by the authors of [
12]. Damaged or falsely detected OD and HM were also detected by them.
2.1. Diabetic Retinopathy Lesion Detection
The studies that were devoted to the detection of DR lesions like MA, HM, and EX fall under this category. Another group of studies detected multiple lesions as well as OD. A set of proposed methods are discussed in this section as well as a summarization of them in terms of techniques and datasets used in
Table 1 summarizes.
Some studies reviewed, focused on detecting EX. Raja and Balaji [
16] used adaptive histogram equalization for preprocessing followed by blood vessel segmentation using CNN. After this, EX segmentation was performed using fuzzy c-means clustering. Finally, DR classification was achieved using SVM. Adem [
17], combined a circular Hough transform with CNNs to detect EX. The circular Hough transform helps to ignore OD. EX detection was performed after segmentation of EX. SVM based on the ranking was implemented along with CNN architecture to accurately segment fundus images by the authors in [
12]. They also detected EX after segmentation. HE detection with deep CNN and multi-feature joint representation was performed by Wang et al. [
18] using a new methodology. They utilized combined features that included deep features as well as handcrafted features. HE detection using SVM blended with a speedier region-based CNN (RCNN) was performed by the authors of [
19]. Wang et al. [
20] performed EX classification. They used a CNN to investigate if the classification performance improves if more importance was given to the image characteristic inside an EX. The outcomes showed that the performance improved. Recurrent neural network (RNN) based semantic features were used for EX segmentation by the authors of [
21] for detecting DR. Huang et al. [
22] used a super-pixel segmentation algorithm, followed by multi-feature extraction and patch generation. Finally, CNN was used for HE detection. RCNN with MobileNet was utilized as a feature extractor by the authors of [
23] for EX detection. Mateen et al. [
24] used pre-trained Inception-v3, ResNet-50, and VGG-19 to perform feature extraction before EX classification using the softmax classifier. Another study by the authors of [
25] used three different models including SqueezeNet, ResNet50, and GoogLeNet for EX detection. Among these three, ResNet50 was found to provide the best results. A CNN with residual skip connection was used for EX detection by the authors in [
26]. The architecture used was called RTC-net.
Few studies focused on MA detection. Multi-scale residual network (MSRNet) was used for segmentation by the authors of [
27] to detect tiny MA. It was based on U-Net architecture. After this, classification was achieved using MS-EfficientNet (multi-scale efficient network). Qiao et al. [
28] used the prognosis of MA for DR grading into three, which includes early NPDR, moderate NPDR, and severe NPDR. Deep CNN was used to perform semantic segmentation of fundus images. Gaussian answer Laplacian (LOG) and maximum matching filter response (MFR) filters were used for lesion identification. Feature extraction using VGG and Inception V3 backbones, alongside transfer learning was utilized for MA detection by the authors of [
29]. After this, several machine learning classifiers were used for classifying input images. Qomariah et al. [
30] modified the U-Net model and used it for MA segmentation. Residual units with modified identity mapping were used in this new model known as MResUNet. Subhasree et al. [
31] made use of three different transfer learning-based CNN models for MA detection. These consisted of VGG-16, Inception V3, and ResNet50. They also performed the same task by making use of a random forest approach.
Some studies did a general detection of DR lesions. Conditional generative adversarial network (cGAN) was used for pixel-level lesion segmentation by combining with holistically-nested edge detection (HEDNet) by the authors in [
32], and it was found that adversarial loss advances the lesion segmentation performance. An approach known as super-pixel-based segmentation was used by the authors of [
33] to compare the performance of three different algorithms under a single unified framework in performing multi-lesion detection. Out of these, a compacted watershed algorithm was found to be faster, whereas a linear spectral clustering (LSC) algorithm generated better segmentation results. After this, a single-layer CNN was used to detect lesions. Dai et al. [
34] introduced a novel system called DeepDR comprising of three sub-networks (based on ResNet and Mask-RCNN) to grade DR using a large dataset collected between 2014 and 2017. Lesion segmentation was performed before DR grading. The Javeria segmentation (JSeg) model, a novel model with the ResNet-50 model as the backbone, was utilized for segmentation by the authors in [
35]. In their next phase, Resnet-101 was used for feature extraction built on the equilibrium optimizer (EO). After this, support vector machine (SVM) and neural network classifiers were used for grading lesions. The YOLOv3 model was used for lesion localization by the authors in [
36]. It was fused with the CNN512 model to grade DR and localize DR lesions. Wang et al. [
37] introduced a new model called adaptive multi-target weighting network (AMWNet) to segment DR lesions. It consisted of an encoder, decoder and an adaptive multi-target weighting module. After this reverse data recovery network (RRN) was used to replicate the cycle perception as part of the hierarchy. DR lesion detection using a CNN-based one-shot detector was used by the authors in [
38]. The YOLOv4 deep neural network (DNN) architecture was used along with the Darknet framework.
Many studies reviewed, detected multiple DR lesions. Pixel-wise segmentation of MA, EX, and OD were achieved by the authors of [
39] using a novel membrane system termed “dynamic membrane system” and mask R-CNN. MA and EX segmentation was performed by the authors of [
40] by using a revised U-Net architecture which was grounded on the residual network. U-Net with incremental learning was used by the authors in [
41] for EX and HM segmentation. A new incremental learning standard was introduced to excerpt the acquaintance gained by the preceding model to advance the present model. Another study that detected EX and HM was performed by the authors in [
42]. They performed a semantic segmentation with three classes and used a color space transformation along with U-Net. Ananda et al. [
43] used one individual network per one type of lesion related to DR (e.g., one network for MA, one network for HE, etc.). They found that this modification yielded better results for U-Net to classify MA, SE, and HM and detect DR. U-Net segmentation followed by classification using support vector machine was achieved by the authors of [
44] for grading DR by detecting some DR lesions. Blood vessels, EX, MA, and HM were segmented for this purpose. A nested U-Net architecture known as U-Net++ was used for red lesion segmentation (MA and HM segmentation) from fundus images by the authors in [
45]. This was followed by sub-image classification using ResNet-18 to reduce false positives. Another study that focused on red lesion detection was one by the authors of [
46]. For this purpose, they fine-tuned various pre-trained CNN models with an augmented set of image patches and found ResNet50 to be the best. They modified it with reinforced skip connections to get a new model called DR-ResNet50.
The authors in [
47] used MA, EX, and HM diagnosis to detect DR by using regions with CNNs (R-CNN). Feature extraction using the VGG19 model trialed by classification using different machine learning classifiers by the authors of [
48]. They identified and classified DR lesions into MA, SE, HE, and HM. A single-scale CNN (SS-CNN) followed by a multi-scale CNN (MS-CNN) was used by the authors of [
49] to perform pixel-level detection of MA and HM. Four DR lesions, including MA, HE, SE, and HM, were segmented by the authors of [
50] using a relation transformer network (RTNet). In this, a relation transformer block was incorporated into attention mechanisms in two levels. Another new model was used to detect these same four lesions by the authors in [
51]. It was called faster region-based CNN (RCNN) and was a combination of a regional proposal network (RPN) and fast-RCNN. Another new model known as deeply-supervised multiscale attention U-Net (Mult-Attn-U-Net) was used by the authors of [
52] for detecting MA, EX, and HM. It was based on the U-Net architecture. Dual-input attentive RefineNet (DARNet) was used by the authors in [
53] to detect four DR lesions including HE, SE, HM, and MA. DARNet included a global image encoder, local image encoder, and attention refinement decoder. The same authors used a cascade attentive RefineNet for detecting the same four DR lesions in another study [
54]. CARNet included a global image encoder, a patch image encoder, and an attention refinement decoder. A YOLOv5 model along with a genetic algorithm was used by the authors in [
55] to detect MA, SE, HE, and HM. Another study by the authors of [
56] detected MA, EX, and HM with the help of a CNN along with a SVM. Prior to this, they used a U-Net to segment OD and retinal blood vessels for feature extraction.
Three studies among the reviewed works, focused on the segmentation of the DR lesion called HM. Skouta et al. [
57] made use of a U-Net architecture for detecting the presence of HM. Aziz et al. [
58] utilized a novel CNN model known as HE network (HemNet) for HM detection. Quality enhancement, seed points extraction, image calibration, and smart window-based adaptive thresholding were performed prior to training this model. A 3D CNN deep learning framework with feature fusion was used to perform HM detection by the authors in [
59]. A variation of the traditional CLAHE method was for pre-processing to achieve better edge detection.
Table 1.
Deep Learning based approaches for diabetic retinopathy lesion detection.
Table 1.
Deep Learning based approaches for diabetic retinopathy lesion detection.
| Type | Method | Year | Technique | Dataset |
|---|
| Exudates | Adem [17] | 2018 | Adaptive histogram equalization, CNN | DIARETDB0, DIARETDB1 |
| Raja and Balaji [16] | 2019 | CNN and fuzzy c-means clustering, SVM | DRIVE, STARE |
| Wang et al. [20] | 2019 | CNN | Self-collected |
| Wang et al. [18] | 2020 | Deep CNN, multi-feature joint representation | E-Optha, DIARETDB1 |
| Mateen et al. [24] | 2020 | Inception-v3, ResNet-50 and VGG-19 | DIARETDB1, E-ophtha |
| Ghosh and Ghosh [12] | 2021 | rSVM, CNN | STARE, DIARETDB0, DIARETDB1 |
| Kurilová et al. [19] | 2021 | SVM, RCNN, ResNet-50 | E-ophtha, DIARETDB1, MESSIDOR |
| Huang et al. [22] | 2021 | Patch-based deep CNN | DIARETDB1, E-ophtha, IDRiD |
| Bibi et al. [23] | 2021 | RCNN | DIARETDB1, E-ophtha |
| Cincan et al. [25] | 2021 | SqueezeNet, ResNet50 and GoogLeNet | - |
| Sivapriya et al. [21] | 2022 | RNN | MESSIDOR |
| Manan et al. [26] | 2022 | RTC-net | DIARETDB1, E-ophtha |
| Microaneurysms | Qiao et al. [28] | 2020 | Deep CNN | IDRiD |
| Qomariah et al. [30] | 2021 | MResUNet | DIARETDB1, IDRiD |
| Xia et al. [27] | 2021 | MSRNet, MS-EfficientNet | E-Ophtha, IDRiD, DRIVE, CHASE DB1 |
| Subhasree et al. [31] | 2022 | VGG-16, Inception V3, and ResNet50 | E-Ophtha, DIARETDB1 |
| Gupta et al. [29] | 2022 | VGG, Inception V3 | ROC |
| Multiple Lesions | Xiao et al. [32] | 2019 | cGAN, HEDNet | IDRiD |
| Ananda et al. [43] | 2019 | U-Net, SegNet | IDRiD, MESSIDOR |
| Praveena and Lavanya [33] | 2019 | Super-pixel based segmentation | DIARETDB1, DRiDB, HRF |
| Xue et al. [39] | 2019 | CNN | IDRiD, E-Optha, MESSIDOR |
| Gupta et al. [48] | 2019 | VGG-19 | IDRiD |
| Nazir et al. [51] | 2020 | RCNN | DIARETDB1, MESSIDOR |
| Sambyal et al. [40] | 2020 | Modified U-Net architecture | IDRiD, E-Optha |
| He et al. [41] | 2020 | U-Net, Incremental learning | - |
| Dai et al. [34] | 2021 | ResNet, Mask-RCNN | Self-collected |
| Alyoubi et al. [36] | 2021 | YOLOv3, CNN512 | DDR |
| Wang et al. [37] | 2021 | AMWNet | IDRiD |
| Abdelmaksoud et al. [44] | 2021 | U-Net, SVM | DRIVE, STARE, CHASEDB1, E-ophtha |
| Li et al. [49] | 2021 | SS-CNN, MS-CNN | DIARETDB1 |
| Basu and Mitra [52] | 2021 | Mult-Attn-U-Net | IDRiD |
| Santos et al. [38] | 2021 | YOLOv4 with darknet | DDR |
| Amin et al. [35] | 2022 | JSeg model | IDRiD |
| Kundu et al. [45] | 2022 | U-Net++, ResNet-18 | MESSIDOR, DIARETDB1 |
| Latchoumi et al. [47] | 2022 | R-CNN | - |
| Huang et al. [50] | 2022 | RTNet | IDRiD, DDR |
| Guo and Peng [53] | 2022 | DARNet | IDRiD, DDR, E-ophtha |
| Guo and Peng [54] | 2022 | CARNet | IDRiD, DDR, E-ophtha |
| Ashraf et al. [46] | 2022 | DR-ResNet50 | DIARETDB1, E-ophtha, IDRiD |
| Santos et al. [55] | 2022 | YOLOv5 | DDR |
| Selçuk et al. [42] | 2022 | U-Net | MESSIDOR, DIARETDB1 |
| Jena et al. [56] | 2023 | U-Net, CNN | APTOS, MESSIDOR |
| Hemorrhage | Maqsood et al. [59] | 2021 | 3D CNN with feature fusion | DIARETDB0, DIARETDB1, MESSIDOR, HRF, DRIVE, STARE |
| Skouta et al. [57] | 2022 | U-Net | IDRiD, DIARETDB1 |
| Aziz et al. [58] | 2023 | HemNet | DIARETDB0, DIARETDB1 |
2.2. Diabetic Retinopathy Retinal Blood Vessel/Optic Disc Segmentation
This category includes those studies which performed retinal blood vessel segmentation as well as those which performed OD/region of interest (ROI) segmentation. Most studies fall under the first type, which helps to study the vascular tree and can thus help assess the current medical condition of the patient’s eyes. Other studies focused on optic disc (OD)/fovea segmentation and ROI segmentation. These studies can aid the feature extraction process required for developing automatic DR diagnosis systems by helping to remove unnecessary features before doing them [
60].
One of the most preferred DL methods for segmentation is to use CNNs [
61,
62,
63,
64,
65]. The presence of neovascularization was used to spot proliferative DR by the authors in [
66]. Different layers of CNNs were modeled together with VGG-16 net architecture for this. They performed blood vessel segmentation to remove the extra features before doing the classification. Chala et al. [
67] used a multi-encoder decoder architecture with two encoder units, as well as a decoder unit for retinal blood vessel segmentation. Aujih et al. [
68] studied how the U-Net model performs for the same purpose. Dropout and batch normalization with various settings were utilized for this. Batch normalization was found to make U-Net learn faster but degraded the performance after epoch thirty. They also studied how the presence/absence of retinal blood vessel segmentation affected the DR classification performance using Inception v1. Burewar et al. [
69] made use of U-Net for retinal segmentation with region merging. After this, CNN with ReLU activation function and max-pooling was utilized to perform DR grading. Yadav [
70] applied a dual-tree discrete Ridgelet transform (DT-DRT) to fundus images for feature extraction from ROIs and used a U-Net-based method for retinal blood vessel segmentation after this. The U-Net++ architecture was used to extract the retinal vasculature by the authors in [
71]. They used the extracted features to predict DR in the next stage of their experiment. A context-involved U-Net was used to perform retinal blood vessel segmentation in the study by the authors in [
72]. They made use of patch-based loss weight mapping to improve the segmentation of thin blood vessels. Another study by the authors in [
73] used an encoder enhanced atrous U-Net to extract the retinal vasculature from fundus images. They introduced two additional layers to the encoder at every stage to enhance the extraction of edge information. Jebaseeli et al. [
74] utilized contrast limited adaptive histogram equalization (CLAHE) to perform pre-processing. After this, feature generation and classification were performed. The former step was accomplished using the tandem pulse coupled neural network model and the latter was achieved using DL based support vector machine (DLBSVM). DLBSVM also extracted the blood vessel tree from the fundus images. Similar work was done by the authors of [
75]. Another study by the authors of [
76] made use of a pool-less residual segmentation network (PLRS-Net) for retinal blood vessel segmentation, which could segment smaller vessels better. They used two pool-less approaches called PLRS-Net and PLS-Net (pool-less segmentation network) which did not require any pre/post-processing. A multilevel deep neural network with a three-plane preprocessing method was used by the authors of [
77] for retinal blood vessel segmentation. The first four convolutional layers of VGG-16 were fine-tuned for blood vessel feature extraction. Jin et al. [
78] performed retinal blood vessel segmentation using a new model known as deformable U-Net (DUNet). This model combined the benefits of the U-Net architecture with those of a deformable convolutional network. Another new network followed, a network called
was used by the authors of [
79] to segment retinal blood vessels. A distinct cascaded design, along with inter-network connections made this model unique. Distinctive segmentation of thick as well as thin retinal blood vessels was performed by the authors of [
80] by using a three-stage DL model. These stages consisted of the segmentation of thick vessels, the segmentation of thin vessels, and the fusion of vessels. Another model known as multi-scale CNN with attention mechanisms (MSCNN-AM) was used by the authors in [
81] to segment retinal blood vessels using various dimensions. Atrous separable convolutions which had differing dilation rates were used to seize global and multi-scale vessel data more effectively. Li et al. [
82] used an enhanced U-Net architecture in order to segment retinal blood vessels. They made use of data from the West China Hospital of Sichuan University to perform their experiments. Three DL models consisting of SegNet, U-Net, and CNN were used by the authors of [
83] for the same task. The SegNet model was found to be better than the other two models in segmenting both thin and thick blood vessels. A back propagation neural network was utilized by the researchers in [
84] to extract the retinal vasculature from fundus images. They could obtain a reduction in operation time in addition to improved accuracy. A pre-processing AlexNet block followed by an encoder-decoder U-Net model was utilized by the authors in [
85] to extract the retinal vasculature. Their study aimed to support the diagnosis of several ophthalmological diseases including DR. Another similar study that performed extraction of the retinal vasculature was performed by the authors in [
86] and used a multi-scale residual CNN known as MSR-Net, along with a generative adversarial network (GAN). The generator of the GAN was a segmentation network, whereas the discriminator was a classification network. Another GAN called Pix2Pix GAN was used by the authors in [
87] for retinal blood vessel segmentation to help studies that detect different ophthalmological diseases. A novel model known as D-MNet with multi-scale attention and a residual mechanism, was used by the authors in [
88] for retinal blood vessel segmentation. It was used along with an enhanced version of pulse-coupled neural network to achieve this task.
Some studies performed OD segmentation. This was achieved by using an encoder-decoder with the VGG-16 backbone by the authors of [
89]. Convolutional long short term memory was merged into the encoder to improve accuracy. Kumar et al. [
90] used a mathematical morphology operation before extracting the retinal blood vessels from fundus images. OD segmentation was done with the help of watershed transform. Finally, classification was done using radial basis function neural network (RBFNN). Yeh et al. [
91] used a preprocessing technique for OD segmentation called local differential filter (LDF). Good prediction results were obtained when LDF was used for U-Net model training data to differentiate between OD and background regions. Retinal blood vessel segmentation, as well as extraction of the optic disc as well as fovea centers, were performed by the authors in [
92] using an end-to-end encoder-decoder network known as DRNet. Shankar et al. [
93] introduced a deep CNN model named Synergic DL model for retinopathy grading. They also performed histogram-based OD segmentation prior to this.
Another set of studies in this category focused on ROI segmentation. An algorithm named “adaptive-learning-rate-based” enhanced GMM was used for ROI segmentation in fundus images by Mansour [
94]. After this, they used connected component analysis (CCA) to process the input to AlexNet architecture. This was followed by feature selection and grading using a polynomial kernel-based SVM classifier. Active DL was used in a seven-layer CNN architecture (ADL-CNN) by Qureshi et al. [
95] to automatically grade the images into the five stages of DR after retinal blood vessel segmentation as well as ROI localization.
Table 2 summarizes the studies that were presented in this section.