Abstract
The BioBERT Named Entity Recognition (NER) model is a high-performance model designed to identify both known and unknown entities. It surpasses previous NER models utilized by text-mining tools, such as tmTool and ezTag, in effectively discovering novel entities. In previous studies, the Biomedical Entity Recognition and Multi-Type Normalization Tool (BERN) employed this model to identify words that represent specific names, discern the type of the word, and implement it on a web page to offer NER service. However, we aimed to offer a web service that includes Relation Extraction (RE), a task determining the relation between entity pairs within a sentence. First, just like BERN, we fine-tuned the BioBERT NER model within the biomedical domain to recognize new entities. We identified two categories: diseases and genes/proteins. Additionally, we fine-tuned the BioBERT RE model to determine the presence or absence of a relation between the identified gene–disease entity pairs. The NER and RE results are displayed on a web page using the Django web framework. NER results are presented in distinct colors, and RE results are visualized as graphs in NetworkX and Cytoscape, allowing users to interact with the graphs.
1. Introduction
Recently, the topic of analyzing big data through text mining has been extensively studied. This is not a trend limited to a single field but one that appears in many. However, for professional text mining to be effective in any particular field, using universal datasets may not meet expectations. As a result, we must employ models trained on specialized datasets in the respective field. Web-based text-mining tools such as tmTool [1], ezTag [2], and PubTerm [3] have achieved good NER performance in several biomedical entities recognition fields. However, they use older NER models with lower performance compared to state-of-the-art NER models. Pre-training NER models used in web-based text-mining tools such as tmChem [4] and DNorm [5] have the disadvantage of being ineffective at discovering new entities and not taking duplicate entities into account [6]. To compensate for these shortcomings, a multi-type normalization tool known as BERN [7] was developed. The services offered by BERN at this time, however, are limited in that they only display NER results on the website—not RE. If RE is applied to a website, it will not only recognize entities with names but will also determine the relationship between the pair of entities in the sentence. Therefore, we propose a new web page that implements RE, which was lacking in the BERN described above.
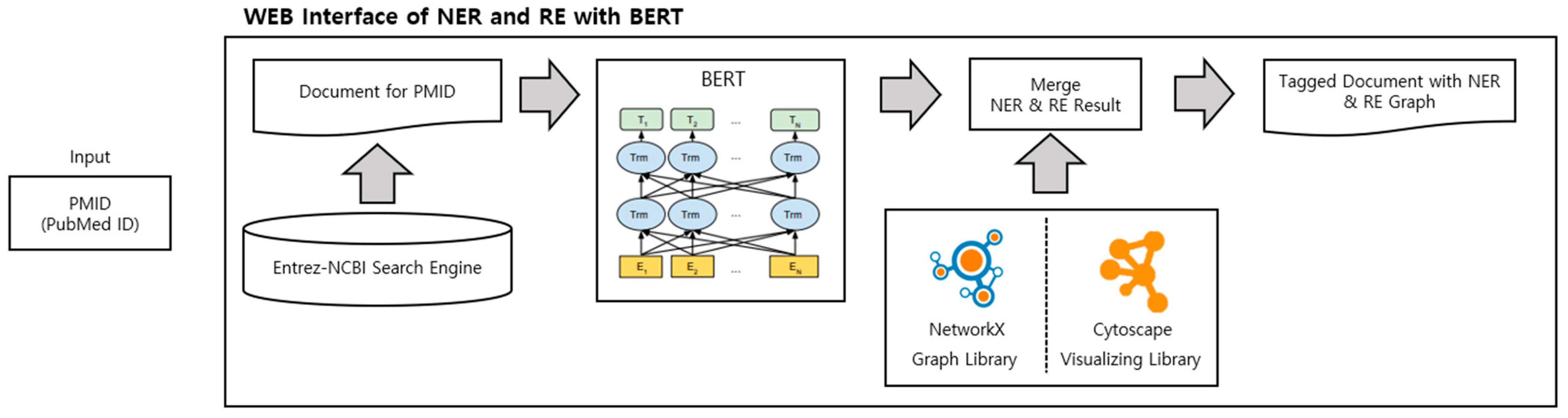
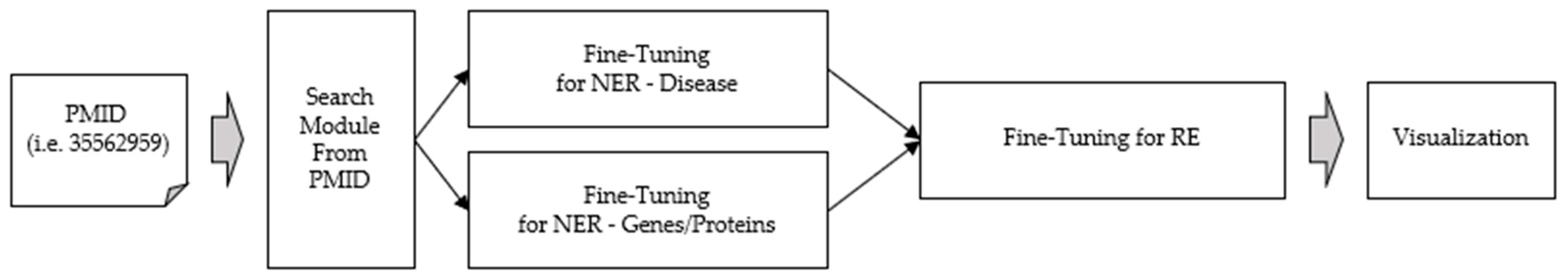
Figure 1 illustrates the structure of our website. Initially, a PubMed Identifier (PMID) is received as input. Next, the Entrez-NCBI Search Engine extracts the PubMed abstract. The extracted abstract undergoes normalization and is divided into disease and gene/protein categories by BERT (Bidirectional Encoder Representations from Transformer) [8]. The combined results for each entity type are then displayed as the NER results. We utilized NetworkX, which can extract data relationships and represent them as graphs and nodes when data is entered, and Cytoscape, which makes models in the form of graphs and nodes available as network analysis tools on web pages. Consequently, this enabled users to employ it in various ways. Users can view the results at a glance on the web page after the process of integrating the NER and RE results to display them within one page.
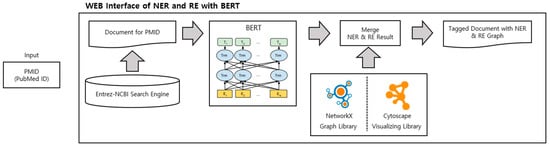
Figure 1.
Overview of web interface of NER and RE with BERT.
2. Related Work
2.1. BERT (Bidirectional Encoder Representations from Transformer)
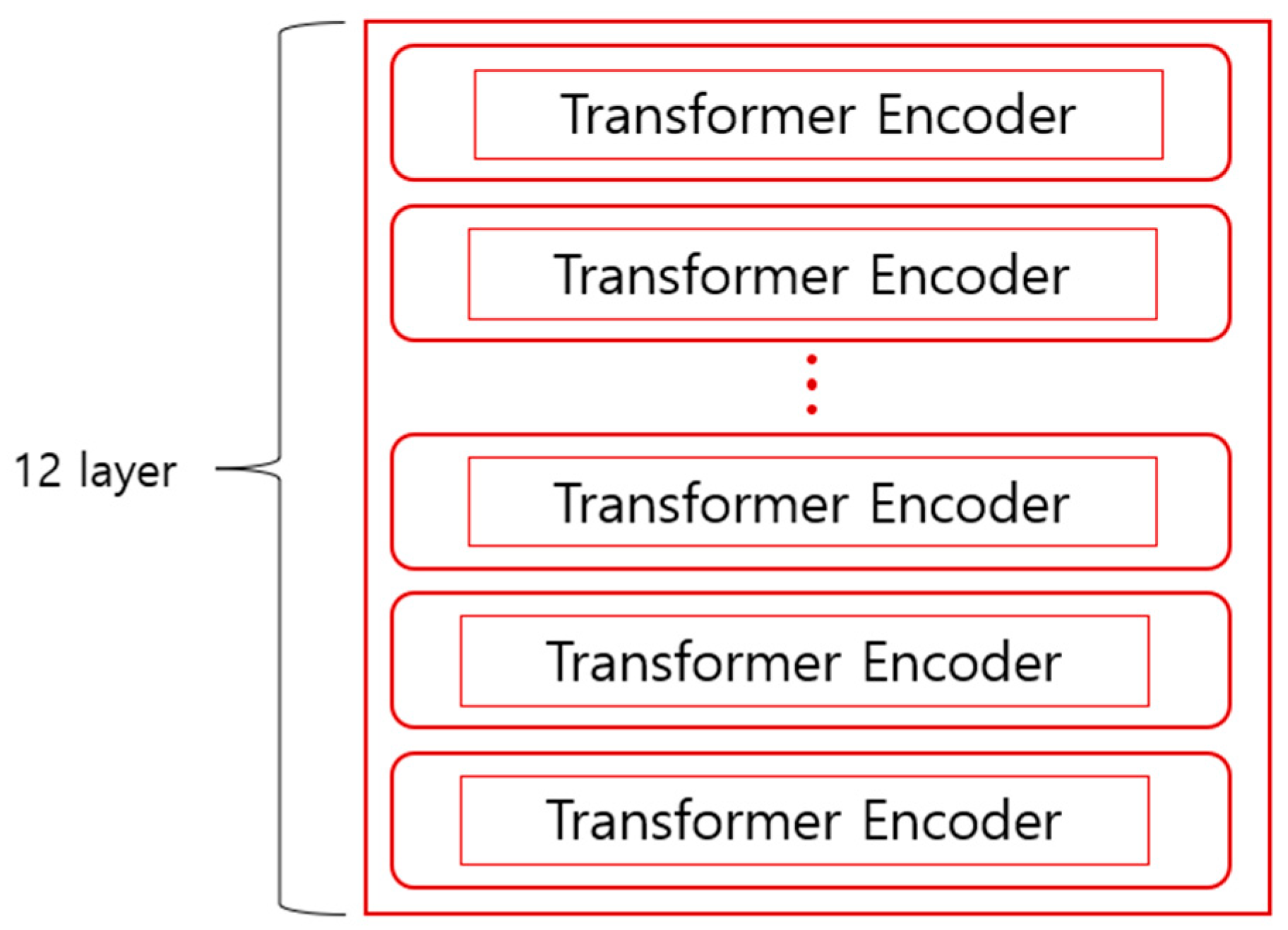
GPT, which omits the encoder part of the transformer structure and focuses on sentence generation as the basis of the language model using the decoder part, can predict sentences only from left to right. BERT was developed with the idea that GPT’s unidirectional structure limits context understanding, such as in question-and-answer tasks. In contrast to GPT, bidirectional training is possible by ignoring the decoder part of the transformer structure and using the encoder part to predict what will fill the empty space between words in the sentence. Bidirectional training examines words throughout the sentence and predicts which words will fill in the blanks, leading to a better understanding of the context compared to unidirectional training. BERT was created by overlapping 12 transformer encoders, as shown in Figure 2.
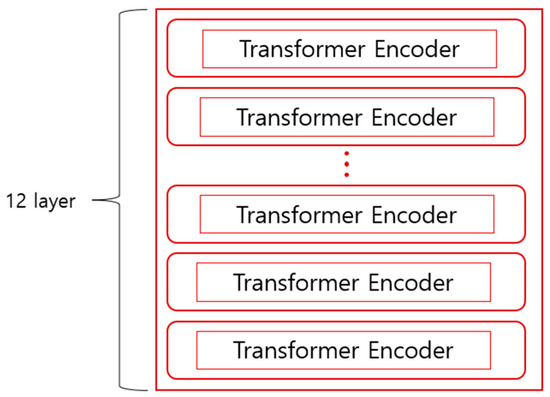
Figure 2.
Structure of BERT.
2.2. BioBERT
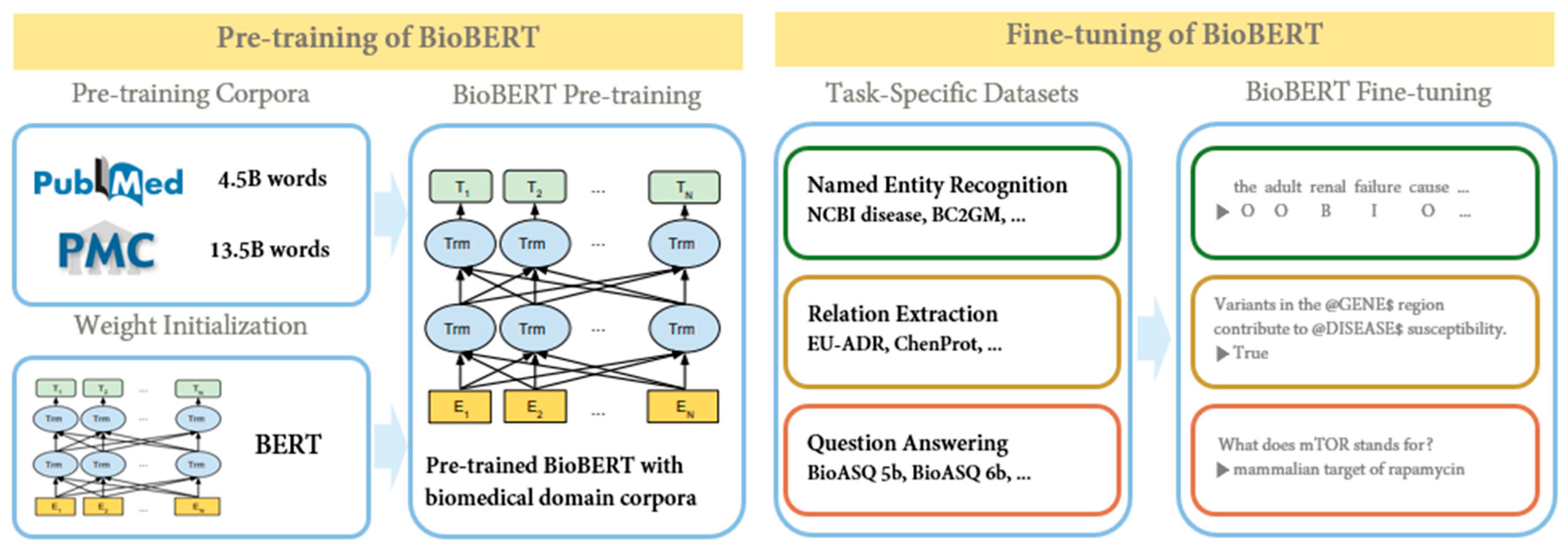
BioBERT [9] is a BERT model specialized in the biomedical domain that has been pre-trained on biomedical datasets. Biomedical terms were created using BERT, based on the assumption that the bidirectional representation employed in the existing BERT structure would be important in biomedical text mining due to the complex relationship between biomedical terms. BERT was pre-trained using English Wikipedia and BooksCorpus. However, because the biomedical domain contains a wide variety of proper names and terms, BERT models aimed at understanding general-purpose languages do not perform well in biomedical text-mining problems. To address these shortcomings, BioBERT was pre-trained not only on its datasets but also on full-text articles from PubMed and PubMed Central (PMC). BERT uses word-piece tokenization [10], an algorithm that divides new words into groups of related words. For example, to represent sub-words, “supercalifragil” can be divided into “super”, “##cali”, “##frag”, and “##gil”. Using this, BERT has the advantage of being able to predict words even when the input contains neologisms or typos, because the input is broken down into words that the deep learning model would have seen in the training phase. BioBERT also employed word-piece tokenization to capitalize on this advantage. Figure 3 is a simple schematic to illustrate the pre-training and fine-tuning of BioBERT.
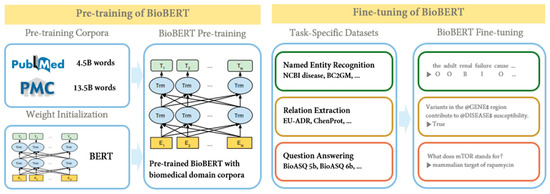
Figure 3.
BioBERT’s pre-training and fine-tuning schematic diagram (cited from [9]).
2.3. BERN (Biomedical Named Entity Recognition and Multi-Type Normalization)
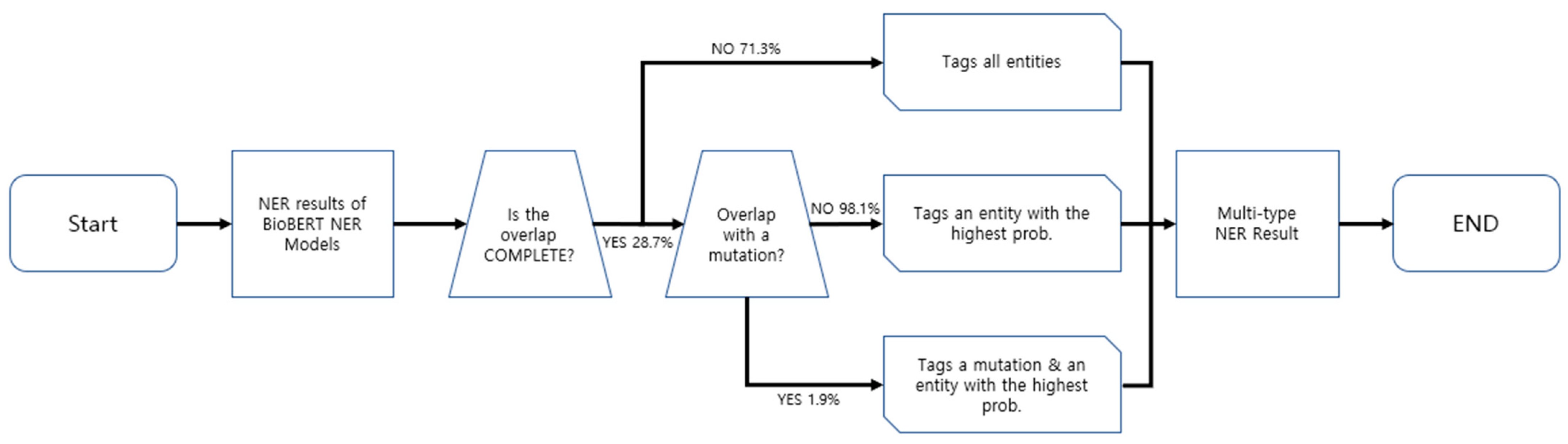
BERN stands for Biomedical Named Entity Recognition and Multi-Type Normalization, which is the basis of the web interface of NER and RE with BERT we developed. It is a biomedical text-mining tool that recognizes existing entities, identifies nested entities, and discovers new ones. The traditional input to BERN is raw text from PubMed abstracts or professional articles. There is a possibility that the text entities will overlap. Prior to BERN, other models recognized the entities in the text as they were, even when the entity overlapped. BERN, on the other hand, made progress in this area by developing a decision rule that determines which entity to select when overlapping entities are detected during named entity recognition.
In three cases, decision rules are used. The first case occurs when the entities do not completely overlap (“Is the overlap COMPLETE?” →NO (71.3%)). In this case, BERN tags all entities. The second case happens when entities completely overlap and there is a mutation between them (“Is the overlap COMPLETE?” → YES (28.7%) × “Overlap with a mutation?” → YES (1.9%)). In this case, the mention and mutation identify the most likely entity predicted by the BioBERT NER model. The third case arises when the entities completely overlap and not all of them are mutants (“Is the overlap COMPLETE?” → YES (28.7%) × “Overlap with a mutation?” → NO (98.1%)). In this case, only the entity with the highest probability of being the actual entity is tagged. When the decision rule is applied in this manner, recall can be reduced while precision can be improved. A schematic diagram of the decision rule is shown in Figure 4.
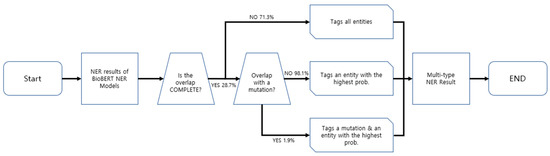
Figure 4.
Schematic diagram of BERN’s decision rule.
3. Materials and Methods
The structure of the web service proposed in this paper is depicted in Figure 5. Section 3.1 describes the user input and search modules. Section 3.2 discusses fine-tuning for named-entity recognition. Section 3.3 covers fine-tuning for relation extraction.
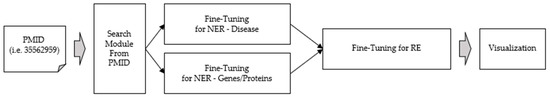
Figure 5.
Our proposed web service’s schematic diagram.
3.1. Input and Search Engine
In this section, we will explain the user input and search module. PubMed is a search engine that provides free access to biomedical information and the bioscience bibliographic database (MEDLINE), as well as summaries and references on various topics. It was created and published by the National Center for Biotechnology Information (NCBI) in 1996. PubMed contains tens of millions of abstracts and citations from biomedical and health literature, including life sciences, but does not include the full text of the article. The PMID of the literature from which the user wishes to obtain information is the input of the web service we provide. PMID stands for PubMed Identifier and PubMed Unique Identifier. This is the number assigned to the literature in PubMed to identify it. It is impossible to determine the type or quality of literature from its number. As a result, we created a module that retrieves the contents of the PubMed database using the user-supplied PMID. This module was written in Python and utilized the Bio module. The Bio module is a bioinformatics package that scientists use to conduct numerous studies in various medical fields. The package allows quick access to a variety of online services and databases. The Bio package was used for this function. Using Entrez, a package submodule, we can retrieve and import the type and abstract data of the document along with PMID information. We can check the details of the literature by sending Entrez the desired database type, PMID, and email information. Currently, the stored data type is XML (Extensible Markup Language), and data can be easily parsed using dictionary, a Python data structure form. Details include the abstract, author, explanation, and so on. Among these, we use the abstract information to perform named-entity recognition and relation extraction tasks.
3.2. Fine-Tuning for Named-Entity Recognition
In this section, we will explain the BioBERT fine-tuning task for NER. NER stands for named-entity recognition, which literally means recognizing a named entity in a document. There are two types of named entities: generic and domain-specific. The generic type refers to general entity names, such as the names of people or places, while the domain-specific type corresponds to terms in specialized fields. The NER discussed in this paper is responsible for recognizing domain-specific proper nouns in biomedical fields. We used pre-trained BioBERT-Base v1.1 with PubMed articles and PMC OA Subset articles, and we employed the BioBERT NER model construction method. This BioBERT NER model has consistently outperformed other NER models in recent studies [11,12,13,14,15,16]. Our downstream task can be defined as named-entity recognition in the genes/proteins and diseases fields. We updated the model with a fine-tuning scheme for this downstream task.
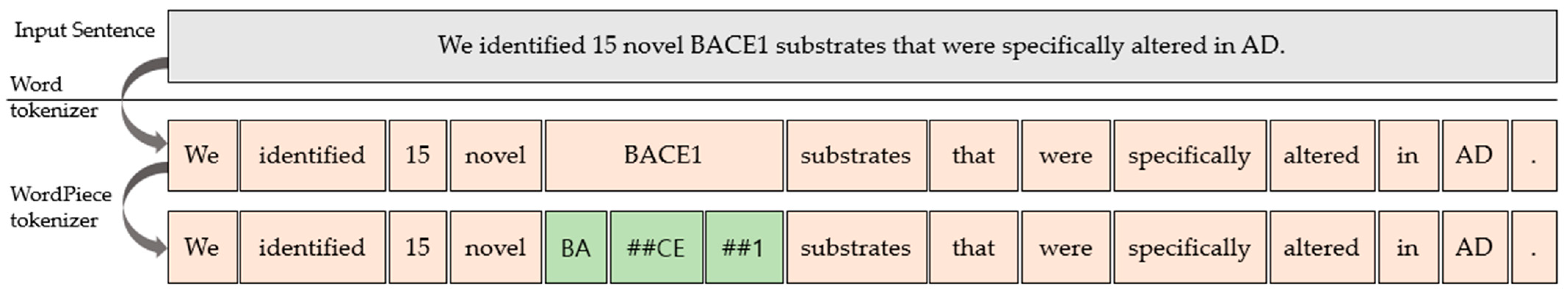
In the pre-training and fine-tuning process, BioBERT discovers new entities by directly learning word-piece embedding in addition to existing word embedding (Figure 6). In the case of a lack of vocabulary, if the word in the original text is not in the embedding’s vocabulary, providing multiple representations for the word becomes more challenging than if the word is in the embedding’s vocabulary.

Figure 6.
Word tokenizer and word-piece tokenizer.
The model results show that the input sentence is word-tokenized to produce BIO and “X”, “[CLS]”, “[SEP]”, and “PAD” tags for each word. In the NER task, after segmenting a sentence into token units, each token is tagged to determine whether or not it is a named entity. However, there may be cases where a single token is not a single entity, but several tokens are. Therefore, we used the BIO tag, one of the various tagging systems. The BIO tag is a fundamental tagging technique for representing individuals identified in the corpus, where “B” stands for “Begin”, “I” stands for “Inside”, and “O” stands for “Outside”. We identify words tagged with “B” and “I” as one entity. It considers “B” word tokens, the beginning of the object name, to consecutive “I” word tokens as an entity. To obtain a list of identified objects, the task of extracting results with a pre-trained model for each type of object was processed in parallel using thread. Furthermore, the HTML tag for the web page was added using the collected object list by locating the corresponding word in the existing abstract sentence. We processed these logics through Django’s view to generate results for NER visualization. Finally, the NER result context was passed from the view to the template, and the HTML was rendered and visualized.
3.3. Fine-Tuning for Relation Extraction
In this section, we will explain the BioBERT fine-tuning task of RE. RE is the task of determining the relationship between named entities. Identifying the relationship between words in a sentence can be highly helpful in interpreting meaning or purpose. For example, it can benefit natural-language-processing applications such as sentiment analysis, question answering, and summarization. In addition, structural triples can be extracted from sentences to summarize information and identify key components. Our RE task determines whether there is a relationship between (gene–disease) entities from different domains in a biomedical corpus sentence. Identifying entities from the NER model comes first, before performing the RE task. In Figure 6, the NER model identified “BACE1” as a genes/proteins entity and “AD” as a diseases entity in the input sentence. To prevent model overfitting, genes’/proteins’ entities are replaced with the “@GENE$” mask and diseases’ entities with the “@DISEASE$” mask in that sentence. The actual input sentence for RE in that sentence is “We identified 15 novel @GENE$ substrates that we specifically altered in @DISEASE$”. Given input sentences as text, the model tokenizes them and breaks them into words. Each word is pre-trained by BERT and mapped to a defined word ID. Through this process, token embeddings of the sentence are created, and semantic information of each token is extracted. Then, based on the object recognition results, we can extract the object’s embedding through the object’s surrounding context. In the process of extracting relationships between entities, additional models are constructed using the [CLS] tokens and their embeddings from the output of the BERT model to further understand the contextual information between entities. A single output layer is then used to output the highest probabilities of possible relationship types in the input sentence. We performed RE in this way.
With a sentence sequence of extracted named entities as input, the BioBERT RE model outputs whether the two genes/proteins and diseases objects are related in each sense. To visualize RE results, we use NetworkX. It is a Python package that makes it simple to create and analyze complex networks [17]. This allows the result data to be handled and manipulated in a graph format, as well as output in various formats such as json and txt. A named entity extracted from the NER model and found to be related to other types of objects in the RE model is added as a node in the graph. Each node’s attributes were set to the recognized types of each entity, genes/proteins, and diseases. Graph types include directed and undirected graphs. Because we do not extract directed results between entities, our RE results are represented using undirected graphs. After all of the graph’s nodes have been initialized, the undirected graph is completed by adding edges between nodes and saving the results in json format. This saved result is fed into Cytoscape for visualization. Cytoscape is a bioinformatics package for visualizing biological networks [18]. We loaded the previously saved json data into the javascript language and used the Cytoscape library to visualize the graph so that we could interact with the user.
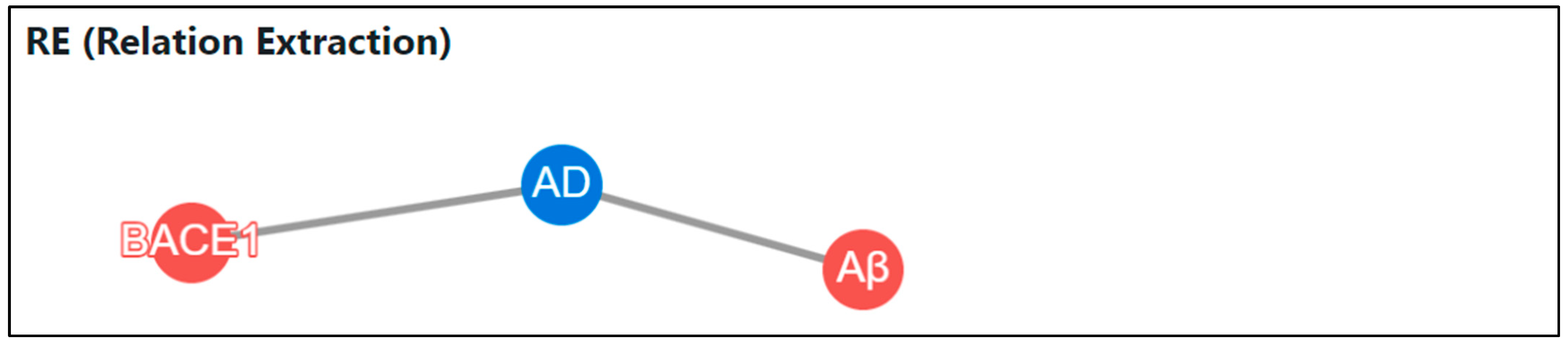
The process of extracting and analyzing relationships between entities is as follows. First, we extract sentences from the entire abstract that contain one or more disease and gene category entities. “However, despite numerous BACE1 inhibitors entering clinical trials, none have successfully improved AD pathogenesis, despite effectively lowering Aβ concentrations.” is an example of one of the extracted sentences.
If there is more than one entity in each category, we perform the task of finding a number of sentences in all cases where a gene–disease link can be found. In the example sentence, two entities, “BACE1” and “Aβ”, were extracted as genes, and “AD” was extracted as a disease. As shown in Table 1, the RE input sample is divided into two related statements: (“BACE1”, “AD”) and (“Aβ”, “AD”). In this way, we graphically visualize the output statements in which the relationship between the two objects exists in the statements listed as inputs.

Table 1.
RE input sample.
4. Results
Section 4.1 describes the implementation system environment, and Section 4.2 describes web services that display BERT-based NER and RE results.
4.1. Environments
We tested the web services proposed in Table 2’s environment, as well as the NER and RE model learning required to provide this web service.

Table 2.
System environments.
The learning rate was 0.00005, and batch size was 32, using python TensorFlow library for NER and RE fine-tuning. Table 3 shows the number of dataset annotations by category that we used to train the NER model.

Table 3.
Statistics of NER datasets.
The NCBI Disease dataset was used to fine-tune models in the disease category, and the BC2GM dataset was used to fine-tune models in the gene/protein category. The NCBI disease dataset is a collection of 793 annotated PubMed abstracts at the mention and concept level, made available as a research resource in the biomedical field. The BC2GM dataset is a corpus of identified genes and is provided for use in bioinformatics research in the biomedical field. The datasets we used have been employed for research in many biomedical fields [21,22,23,24,25], and based on them, we used datasets that are not ethically problematic to build services.
Table 4 shows the number of relations in the dataset by category that we used to train the RE model. The categories of objects corresponding to RE learning correspond to genes and disease recognized by NER. In response, a GAD dataset representing the relationship between gene and disease was used.

Table 4.
Statistics of RE datasets.
Table 5 shows the precision, F1 score, and recall values of the model. We used the highest performing BioBERT model in the gene/protein domain and disease domain, and we show that the BioBERT model we used yielded significantly better performance than other NER models. Since there is no other text-mining tool showing the results for RE, comparisons for RE cannot be made.

Table 5.
Performance comparison of models for entity types (genes/proteins, diseases).
4.2. Implementation
The proposed web service was built with Django, which supports the Python programming language.
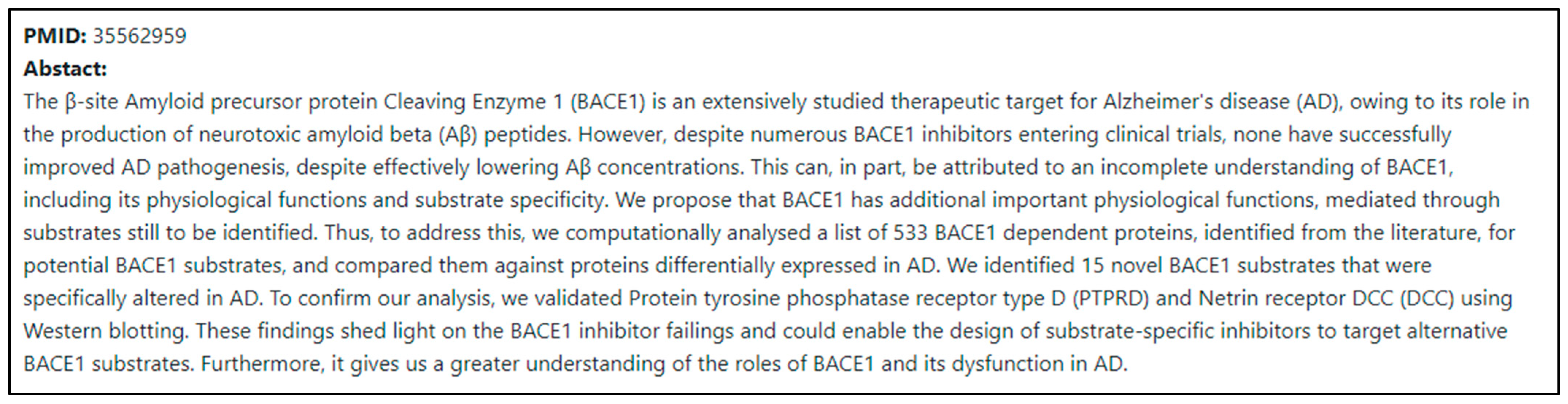
The first screen seen when accessing the implemented web page is shown in Figure 7. When the user enters the PMID and presses “Submit”, the abstract is extracted using Entrez, an NCBI search engine.

Figure 7.
First screen on our website.
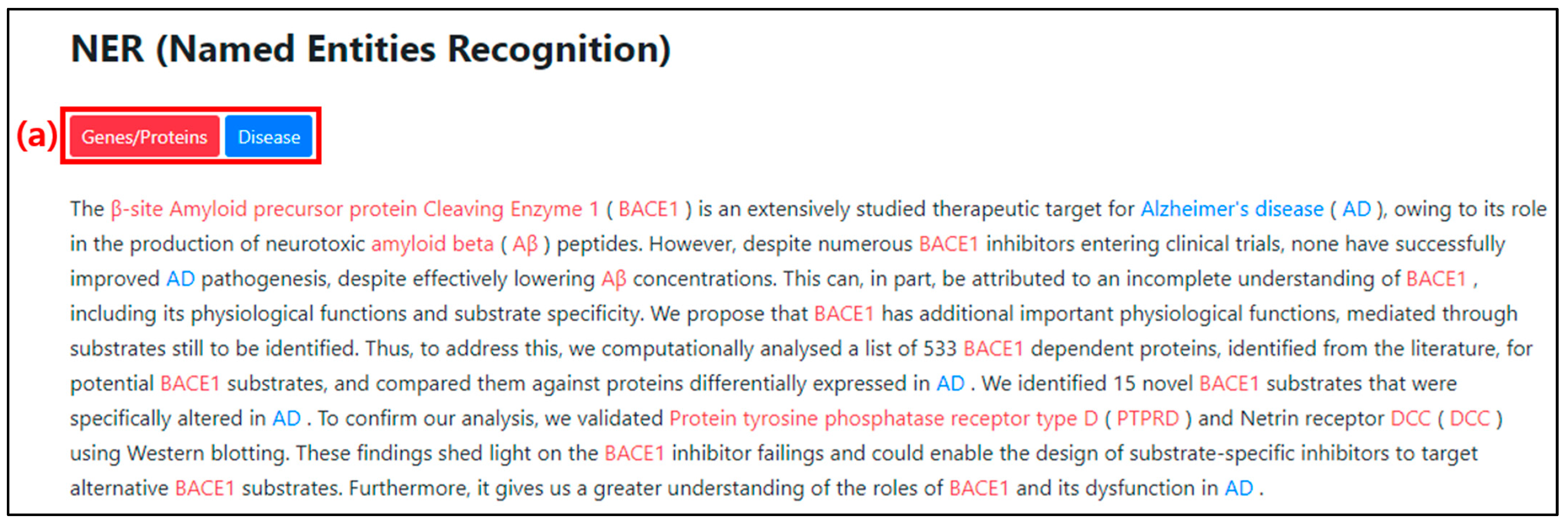
When the PMID is entered, the abstract of the article appears at the top of the web page, as shown in Figure 8. The lower section displays the NER results, as shown in Figure 9. The genes/proteins and disease categories are color-coded as shown in Figure 9(a). The named entities classified by the NER model are represented in their respective color and displayed in the abstract section so that they can be identified within the text. The entity classified as genes/proteins is colored red, while the entity classified as disease is colored blue.
Figure 8.
Abstract result screen on web page after PMID input.
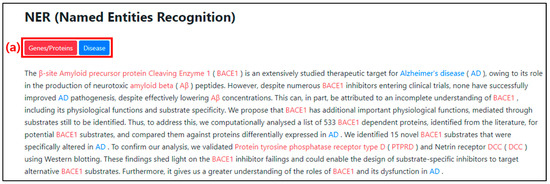
Figure 9.
NER result screen on web page after PMID input. (a) It is an interface to indicate the type and color of the result category from the NER.
At this point, we implemented RE, which classifies and correlates the relationship of named entities based on entity type, rather than just performing NER. We used NetworkX, a graph library that analyzes data and graphically expresses relationships within data, to visualize RE, and Cytoscape, a visualizing library that allows the model created in this manner to be more flexible on a web page, to implement RE. Figure 10 illustrates the correlation between words in a text using the aforemen-tioned RE process.
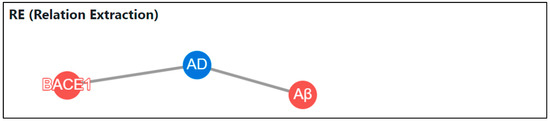
Figure 10.
RE result screen on web page after PMID input.
5. Discussion
There is a BERN web service that extracts and displays new entities for biological domain text. In contrast, this paper enables users to see the relationship between new objects at a glance by performing RE tasks in addition to simply NER. Because we perform sentence-level relationship extraction several times, our RE work requires a relatively large amount of computation when compared to performing a single task. As a result, sufficient time is required to output the result. However, if we conduct a study in which we perform a document-level relationship-extraction task on a single abstract, we will find improvements. Furthermore, in comparison to the existing BERN web service, it does not provide a variety of types, and sufficient research is required to regularize the types of overlapping objects. If these issues are resolved, they will be able to provide better and more convenient services than currently available. A model that recognizes multiple entities in general has a good chance of over- or under-representation in relation to a specific entity if the amount of a specific entity is large or small in the training dataset. However, in the case of our service, in order to recognize only one entity in the model for entity recognition, learning is performed separately for each specific entity, and the results from each model are integrated and used. That is, since we learn each of the entity-category-specific datasets in the model rather than the entire dataset containing multiple entity categories only once in the recognition model, each overall dataset contains only one specific entity category. Therefore, it seems that our approach can be a strength of our service, in that there may be fewer problems of bias in learning data.
6. Conclusions
We propose a web service that makes the results of fine-tuning the BioBERT NER and RE models accessible to users. In the biological domains where the learning dataset exists, we chose the disease and genes/proteins categories for NER model fine-tuning. RE was fine-tuned to learn the gene–disease relationship in order to determine the relationship between the recognized entities in the proposed service. This allows users to be informed about new entities in the desired document. Furthermore, the relationship between the entities can be confirmed visually at a glance. However, our services provide limited categories in certain domains and have limitations in terms of speed. Improved research will be able to provide relatively fast and practical web services. Furthermore, if the proposed web service incorporates a system such as the tagging tool provided in other existing studies, it will be possible to use it in other aspects of the system to develop more effective curation.
Author Contributions
Conceptualization, Y.-J.P., M.-a.L. and G.-J.Y.; methodology, C.-B.S. and S.J.P.; software, Y.-J.P. and G.-J.Y.; validation, Y.-J.P., M.-a.L., G.-J.Y., S.J.P. and C.-B.S.; formal analysis, C.-B.S.; investigation, M.-a.L.; resources, C.-B.S.; data curation, Y.-J.P.; writing—original draft preparation, Y.-J.P. and M.-a.L.; writing—review and editing, Y.-J.P., M.-a.L., S.J.P. and C.-B.S.; visualization, Y.-J.P. and M.-a.L.; supervision, S.J.P. and C.-B.S.; project administration, Y.-J.P.; funding acquisition, S.J.P. and C.-B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Education of the Republic of Korea and the National Research Foundation of Korea (NRF-2020S1A5B8101323), The present research has been conducted by the Research Grant of Kwangwoon University in 2022 and This research was sup-ported by the National Research Foundation of Korea (NRF) grant funded by the Korea gov-ernment Ministry of Science and ICT (MSIT) (NRF-2014M3C9A3064706).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chih-Hsuan, W.; Leaman, R.; Lu, Z. Beyond accuracy: Creating interoperable and scalable text-mining web services. Bioinformatics 2016, 32, 1907–1910. [Google Scholar]
- Kwon, D.; Kim, S.; Wei, C.H.; Leaman, R.; Lu, Z. ezTag: Tagging biomedical concepts via interactive learning. Nucleic Acids Res. 2018, 46, W523–W529. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pelaez, J.; Rodriguez, D.; Medina-Molina, R.; Garcia-Rivas, G.; Jerjes-Sánchez, C.; Trevino, V. PubTerm: A web tool for organizing, annotating and curating genes, diseases, molecules and other concepts from PubMed records. Database 2019, 2019, bay137. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.; Wei, C.-H.; Lu, Z. tmChem: A high performance approach for chemical named entity recognition and normalization. J. Cheminformatics 2015, 7, S3. [Google Scholar]
- Robert, L.; Doğan, L.I.; Lu, Z. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013, 29, 2909–2917. [Google Scholar]
- Sohrab, M.G.; Miwa, M. Deep exhaustive model for nested named entity recognition. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018. [Google Scholar]
- Kim, D.; Lee, J.; So, C.H.; Jeon, H.; Jeong, M.; Choi, Y.; Yoon, W.; Sung, M.; Kang, J. A neural named entity recognition and multi-type normalization tool for biomedical text mining. IEEE Access 2019, 7, 73729–73740. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Wu, Y.; Schuster, M.; Chen, Z.; Le, Q.V.; Norouzi, M.; Macherey, W.; Krikun, M.; Cao, Y.; Gao, Q.; Macherey, K.; et al. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv 2016, arXiv:1609.08144. [Google Scholar]
- Wang, X.; Zhang, Y.; Ren, X.; Zhang, Y.; Zitnik, M.; Shang, J.; Langlotz, C.; Han, J. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics 2019, 35, 1745–1752. [Google Scholar] [CrossRef]
- Yoon, W.; So, C.H.; Lee, J.; Kang, J. Collabonet: Collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinform. 2019, 20, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Habibi, M.; Weber, L.; Neves, M.; Wiegandt, D.L.; Leser, U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 2017, 33, i37–i48. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.H.; Le, H.Q.; Nguyen, T.M.; Vu, S.T. D3NER: Biomedical named entity recognition using CRF-biLSTM improved with fine-tuned embeddings of various linguistic information. Bioinformatics 2018, 34, 3539–3546. [Google Scholar] [CrossRef]
- Luo, L.; Yang, Z.; Yang, P.; Zhang, Y.; Wang, L.; Lin, H.; Wang, J. An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Bioinformatics 2018, 34, 1381–1388. [Google Scholar] [CrossRef]
- Giorgi, J.M.; Bader, G.D. Transfer learning for biomedical named entity recognition with neural networks. Bioinformatics 2018, 34, 4087–4094. [Google Scholar] [CrossRef]
- Hagberg, A.; Swart, P.; SChult, D. Exploring Network Structure, Dynamics, and Function using NetworkX.; Los Alamos National Lab. (LANL): Los Alamos, NM, USA, 2008; No. LA-UR-08-05495; LA-UR-08-5495. [Google Scholar]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Doğan, R.I.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Tanabe, L.K.; Kuo, C.J.; Chung, I.; Hsu, C.N.; Lin, Y.S.; Klinger, R.; Friedrich, C.M.; Ganchev, K.; Torii, M.; et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008, 9, S2. [Google Scholar] [CrossRef]
- Weber, L.; Sänger, M.; Münchmeyer, J.; Habibi, M.; Leser, U.; Akbik, A. HunFlair: An easy-to-use tool for state-of-the-art biomedical named entity recognition. Bioinformatics 2021, 37, 2792–2794. [Google Scholar] [CrossRef]
- Veysel, K.; Talby, D. Accurate Clinical and Biomedical Named Entity Recognition at Scale. Softw. Impacts 2022, 13, 100373. [Google Scholar]
- Yuan, Z.; Liu, Y.; Tan, C.; Huang, S.; Huang, F. Improving biomedical pretrained language models with knowledge. arXiv 2021, arXiv:2104.10344. [Google Scholar]
- Rohanian, O.; Nouriborji, M.; Kouchaki, S.; Clifton, D.A. On the effectiveness of compact biomedical transformers. Bioinformatics 2023, 39, btad103. [Google Scholar] [CrossRef] [PubMed]
- Veysel, K.; Talby, D. Biomedical named entity recognition at scale. In Proceedings of the Pattern Recognition. ICPR International Workshops and Challenges, Virtual Event, 10–15 January 2021; Part I. Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar]
- Bravo, À.; Piñero, J.; Queralt-Rosinach, N.; Rautschka, M.; Furlong, L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: Implications for translational research. BMC Bioinform. 2015, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.H.; Kao, H.Y.; Lu, Z. GNormPlus: An integrative approach for tagging genes, gene families, and protein domains. BioMed Res. Int. 2015, 2015, 918710. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).