Phytochemical Exploration of Ceruchinol in Moss: A Multidisciplinary Study on Biotechnological Cultivation of Physcomitrium patens (Hedw.) Mitt.
Abstract
:1. Introduction
2. Materials and Methods
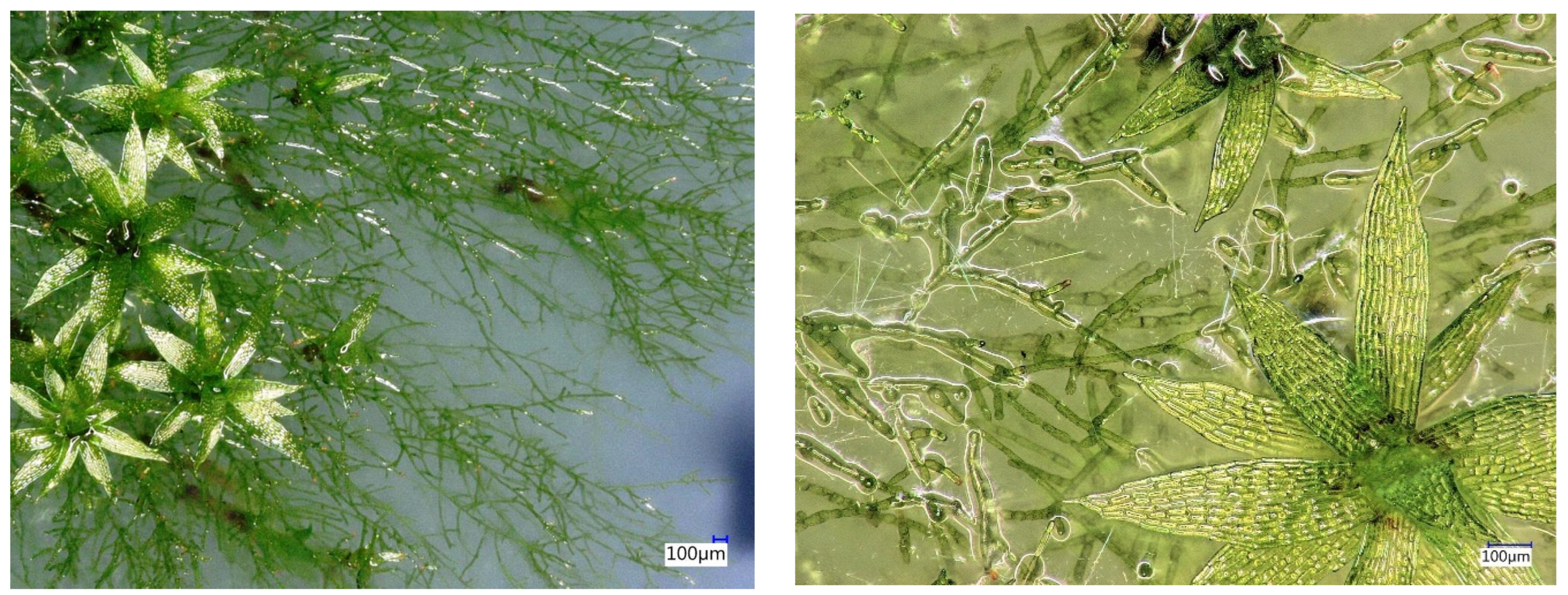
2.1. Plant Material and Generation of P. patens Biomass
2.2. Centrifugal Partition Chromatography (CPC)
2.3. NMR Analyses and Metabolite Identification
2.4. Supercritical CO2 Extraction
2.5. GC-MS Analyses
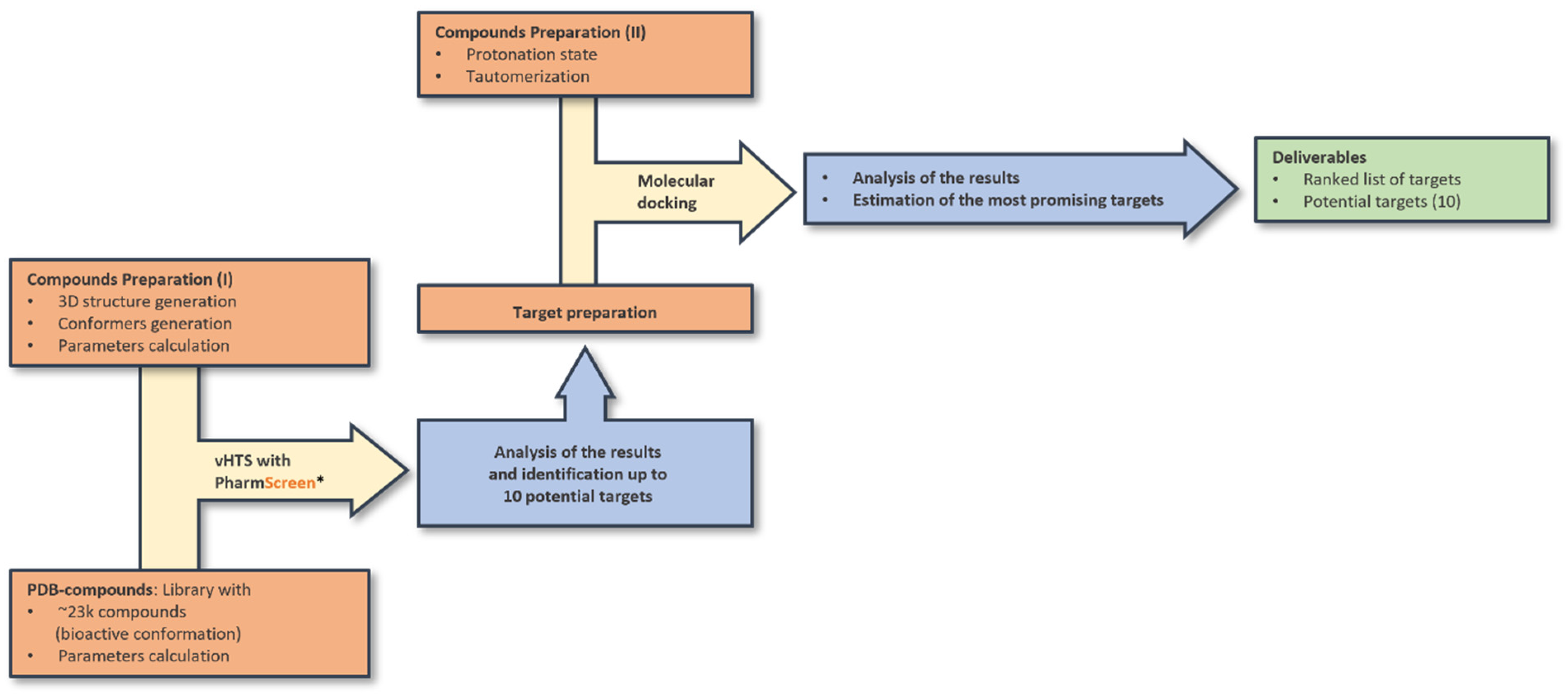
2.6. Ligand-Based and Docking Screening
- Calculation of 3D conformers and selection of more representative ones in terms of energy and root mean squared deviation (RMSD).
- Calculation of molecular charges (Gasteiger method) and LogP (atom-type method).
2.7. Preparation of Co-Crystallized Compounds Extracted from Protein Data Bank (PDB) Database (DB)
2.8. Ligand-Based Virtual Screening with PS
2.9. Docking of Ceruchinol against Selected Targets
- The selected target was prepared for the docking protocol (missing side-chains, duplicated amino acids, pH).
- For some of the targets, different PDB structures were found in the PDB database. Structures with the best resolution were selected. In the case of flexible regions, the most representative structure was chosen.
- Protonation state and tautomerization were calculated.
- Co-crystallized compounds were docked against their own target to validate the docking protocol, evaluating its capacity to retrieve the same pose available in the PDB.
- If multiple cavities were found, compounds were docked in each of them.
- The interaction energy (EI), which is given by the sum of the van der Waals and electrostatic interaction energies.
- The total energy (Etot), which is calculated by combining (i) the intermolecular interaction energy calculated as the sum of the van der Waals and electrostatic potentials between the protein–ligand atom pairs, (ii) the intramolecular interaction energy calculated as the sum of the van der Waals and electrostatic potentials between the 1–4 atom pairs, and (iii) the torsional term of the ligand.
- The affinity prediction or docking score (S), which is evaluated using an empirical function that considers the Total Energy of the top-ranked pose.
- The root-mean-square deviation (RMSD), which is calculated by comparing the atomic coordinates of each binding mode with those of a reference ligand and measures the average distance between the atoms of the two molecules.
3. Results
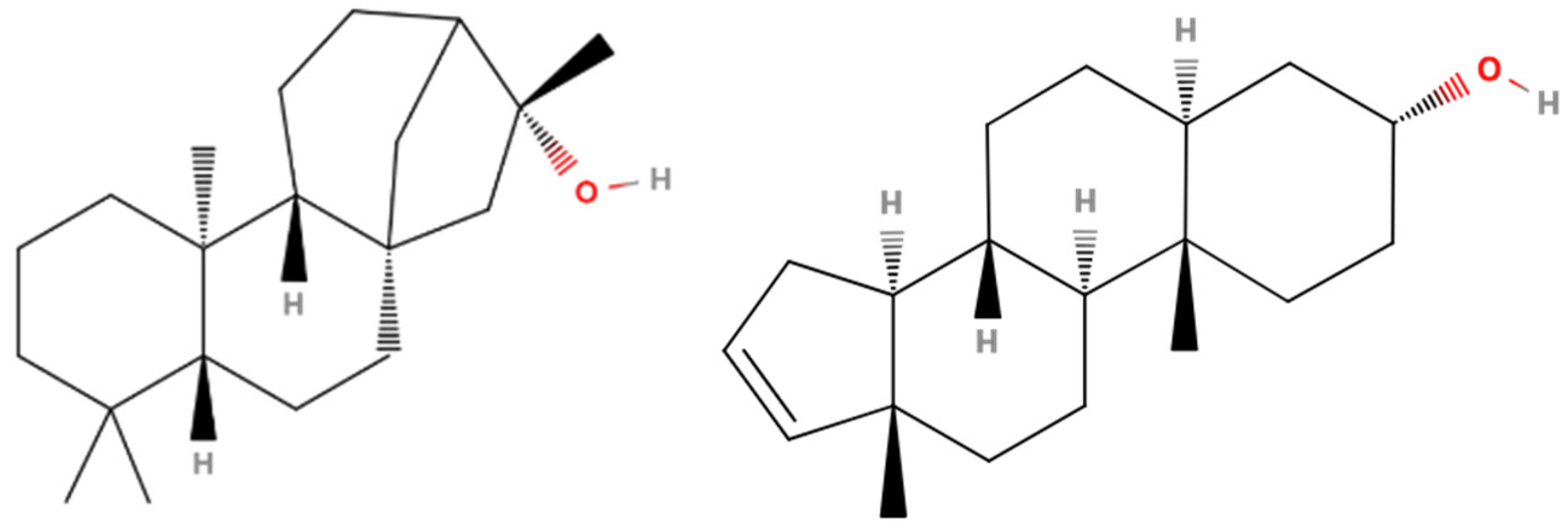
3.1. Chemical Profiling of the Hydroalcoholic Extract
3.2. GC-MS Profiling of the Supercritical CO2 Extract
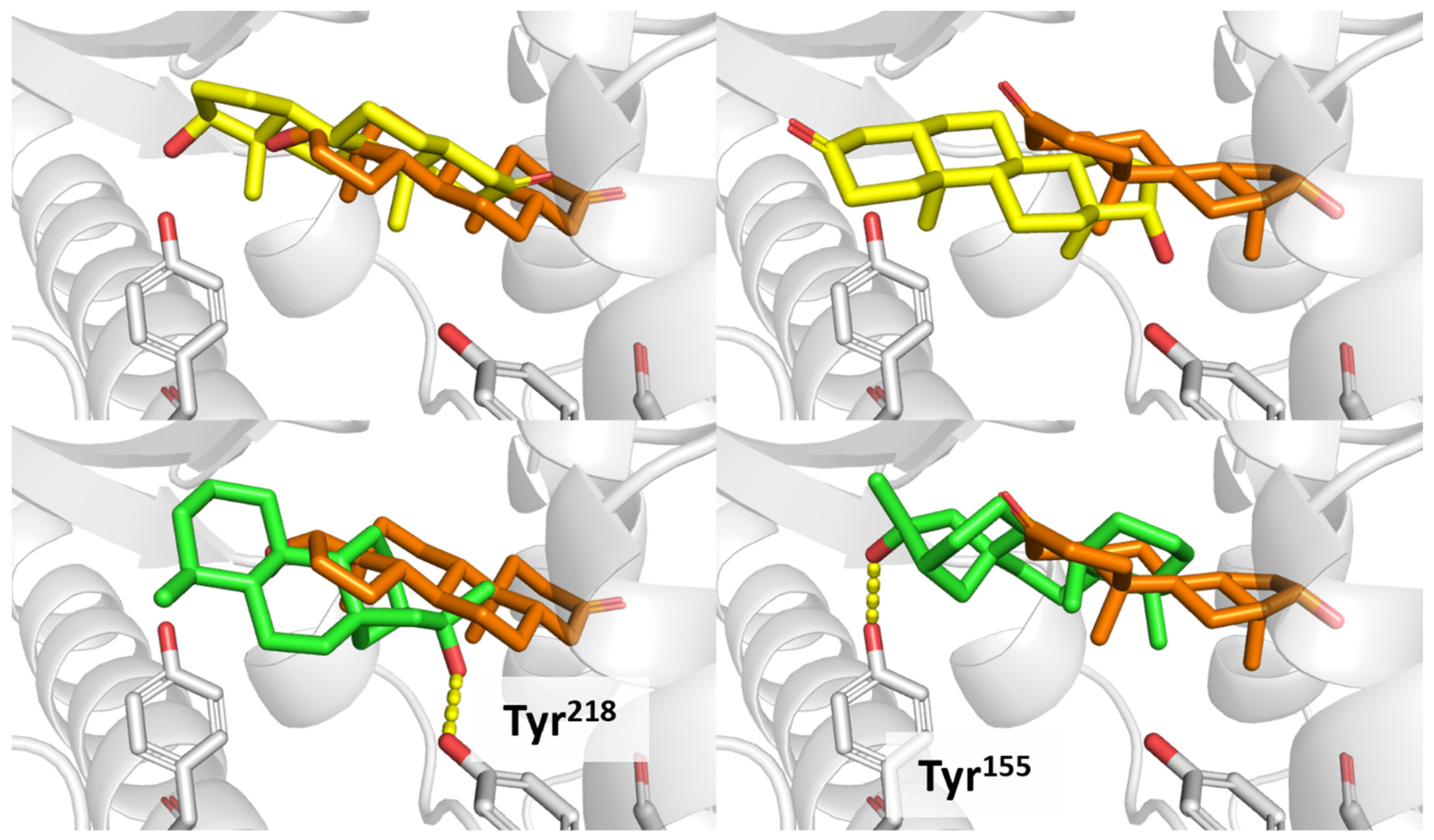
3.3. Docking Evaluation
- CAR protein (Androstan Receptors)
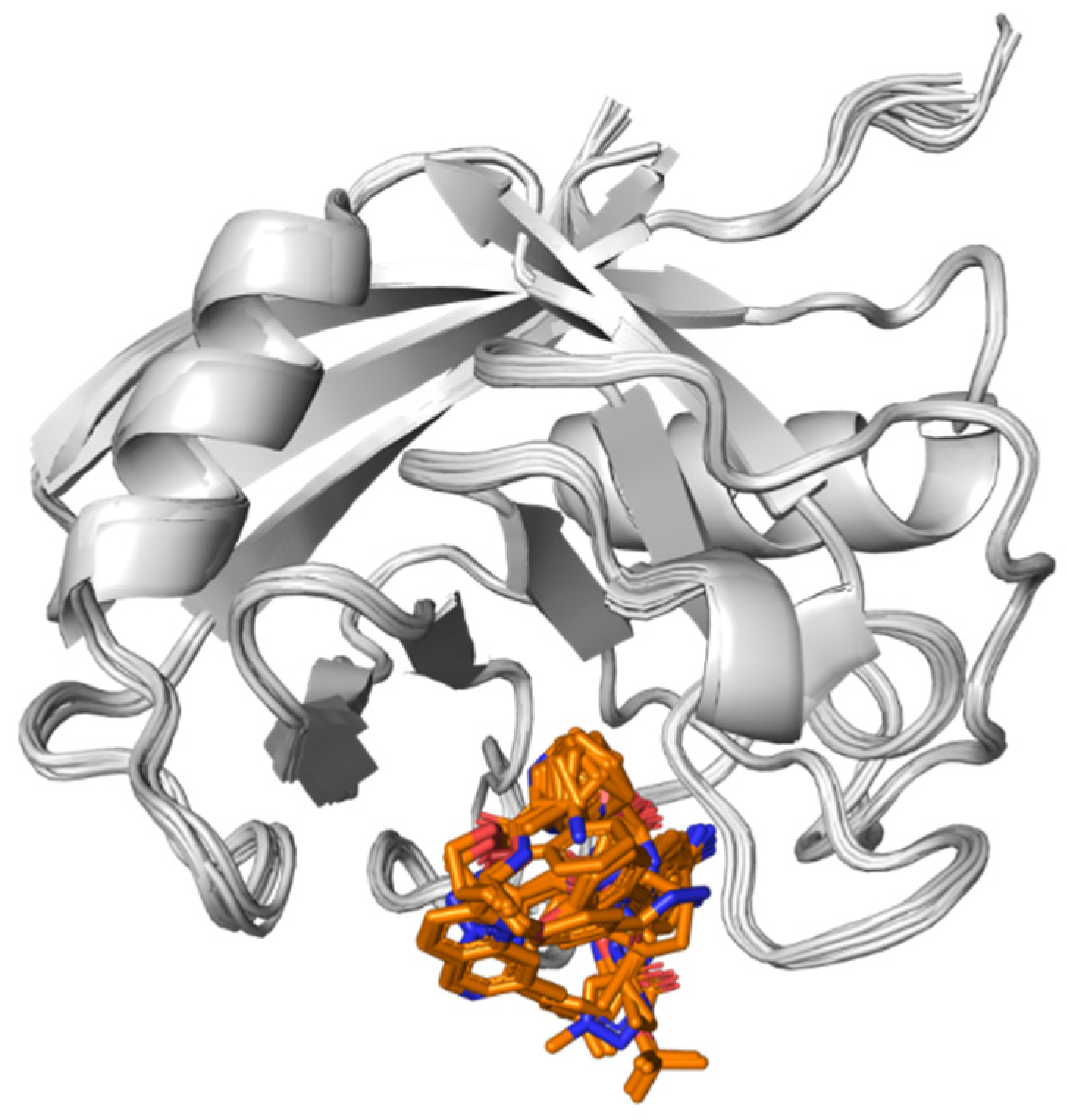
- AKR1D1 (Aldo-keto reductase)
- 17β-HSD1 (17β-Hydroxysteroid dehydrogenase)
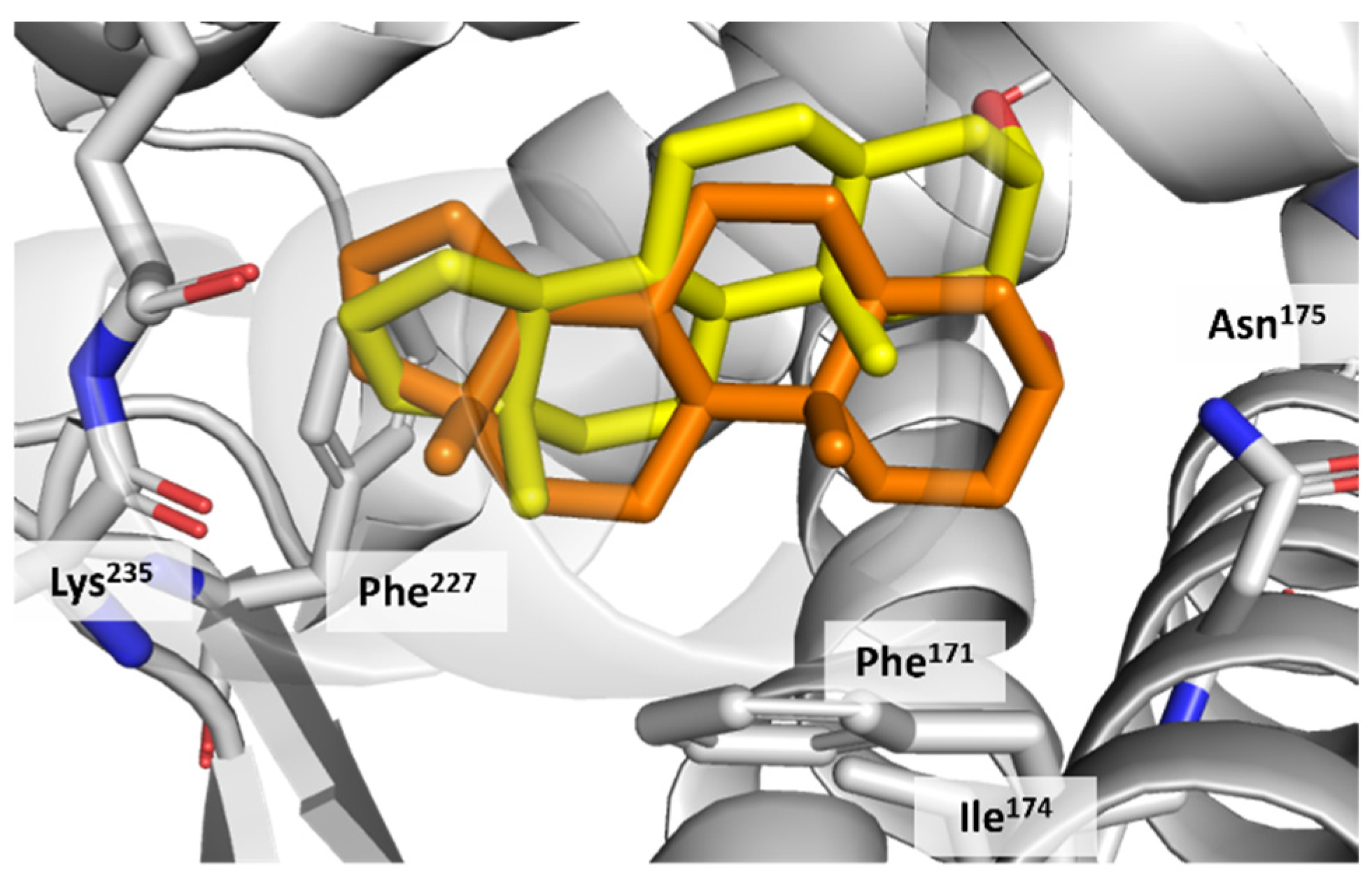
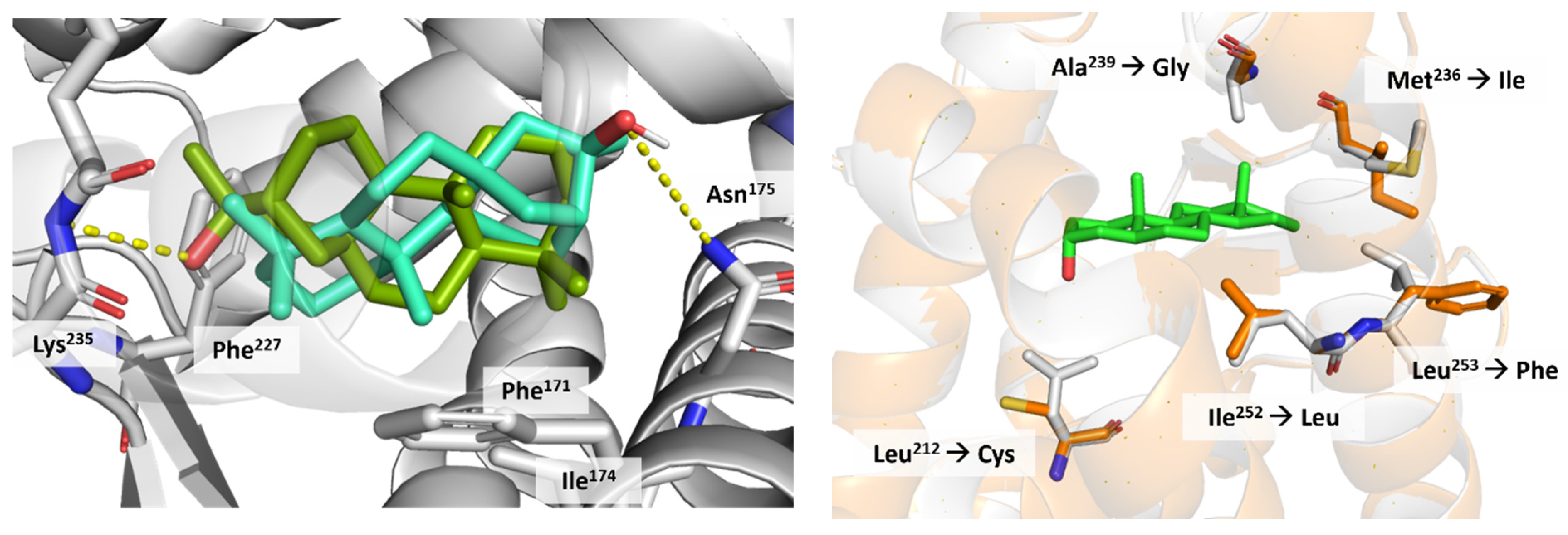
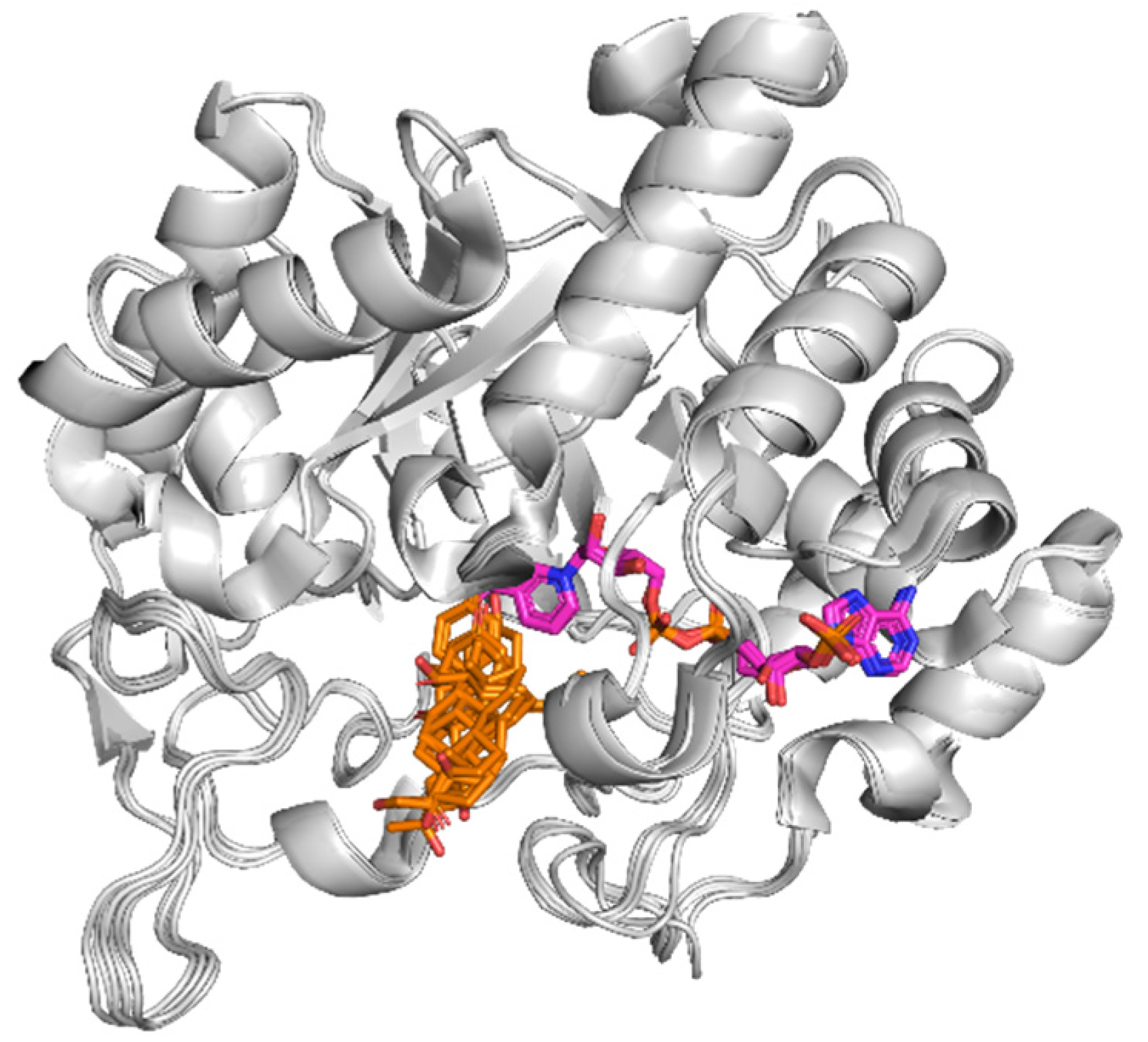
3.4. CAR Protein (Androstane Receptor)
3.5. Docking on Human Constitutive Androstane Receptor
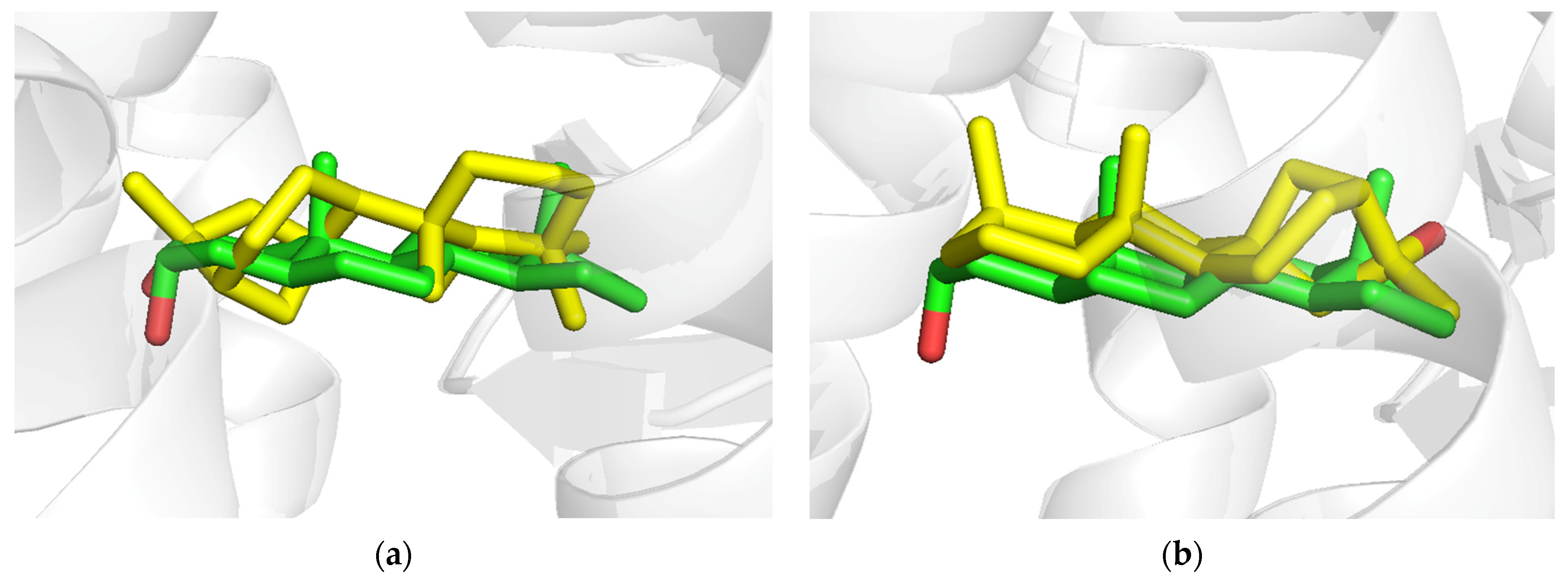
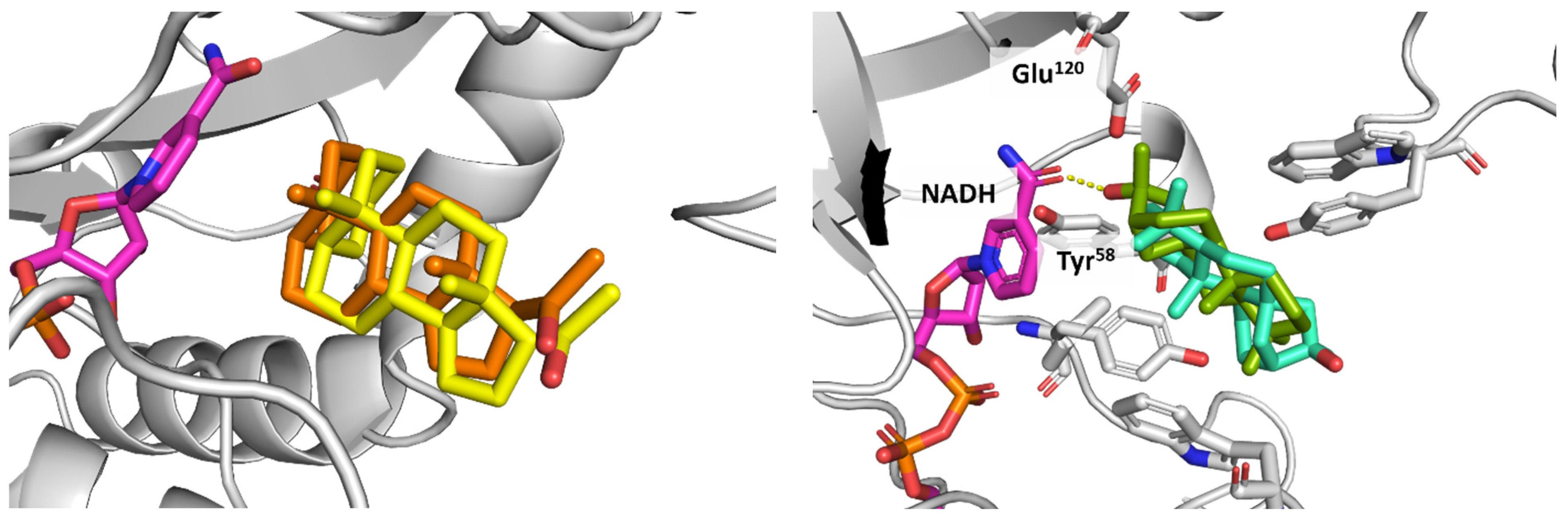
3.6. AKR1D1 (Aldo-Keto Reductase)
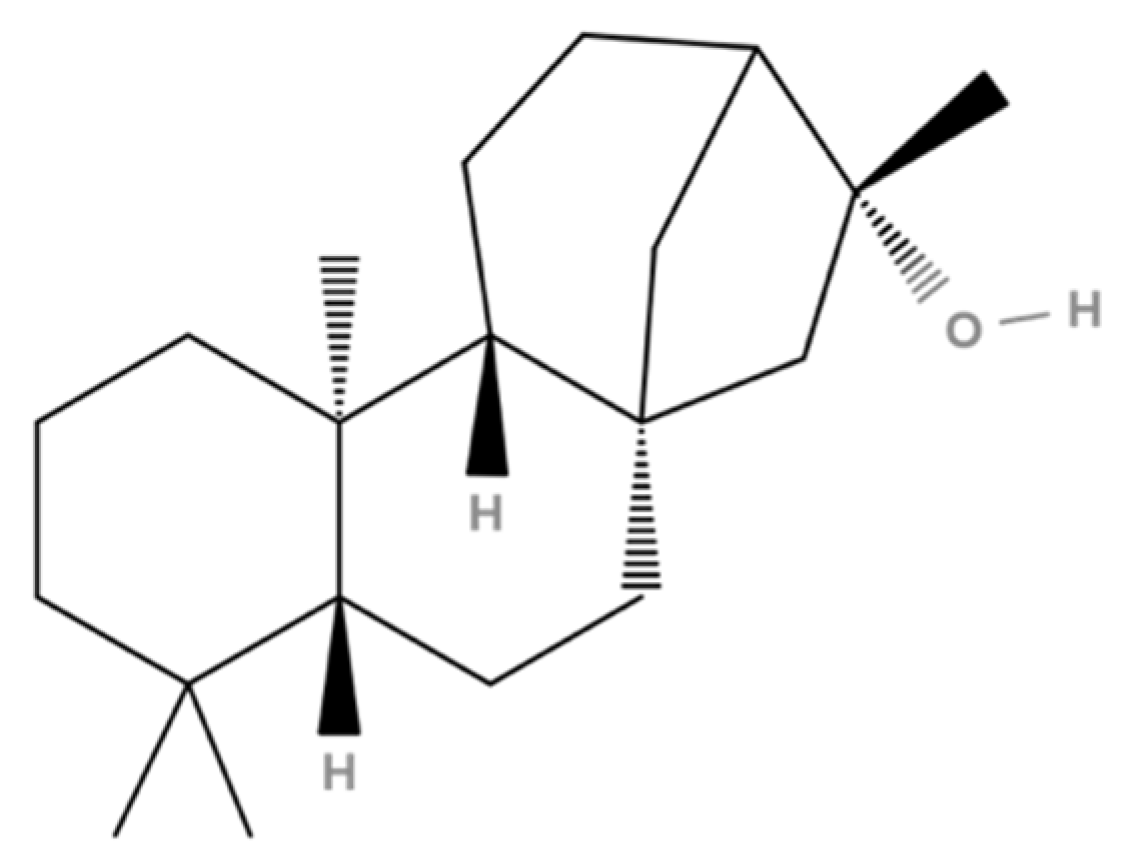
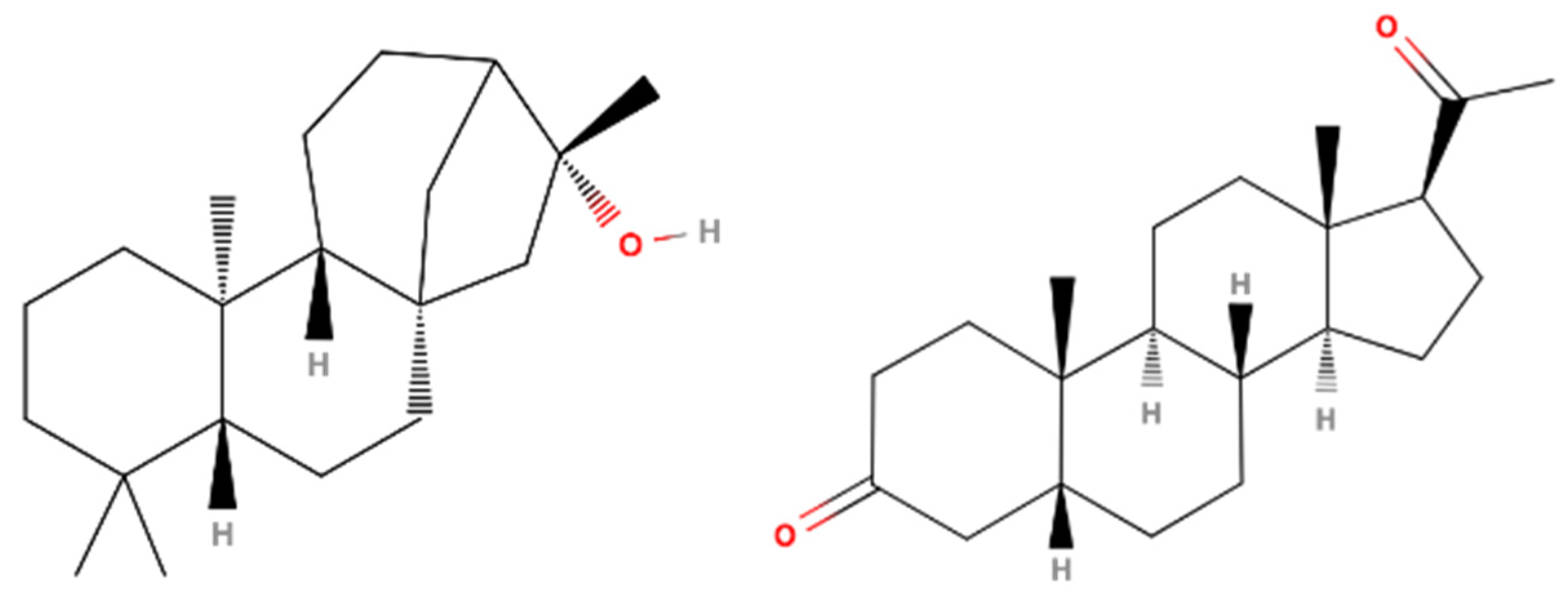
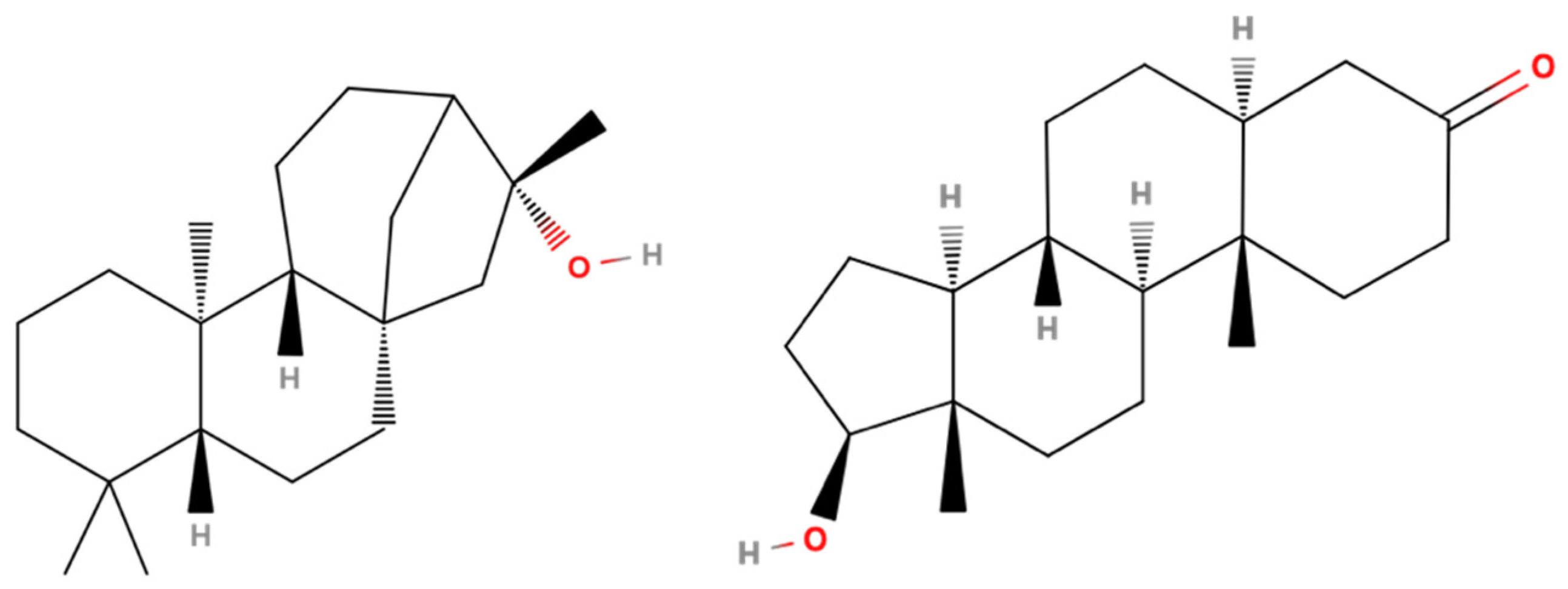
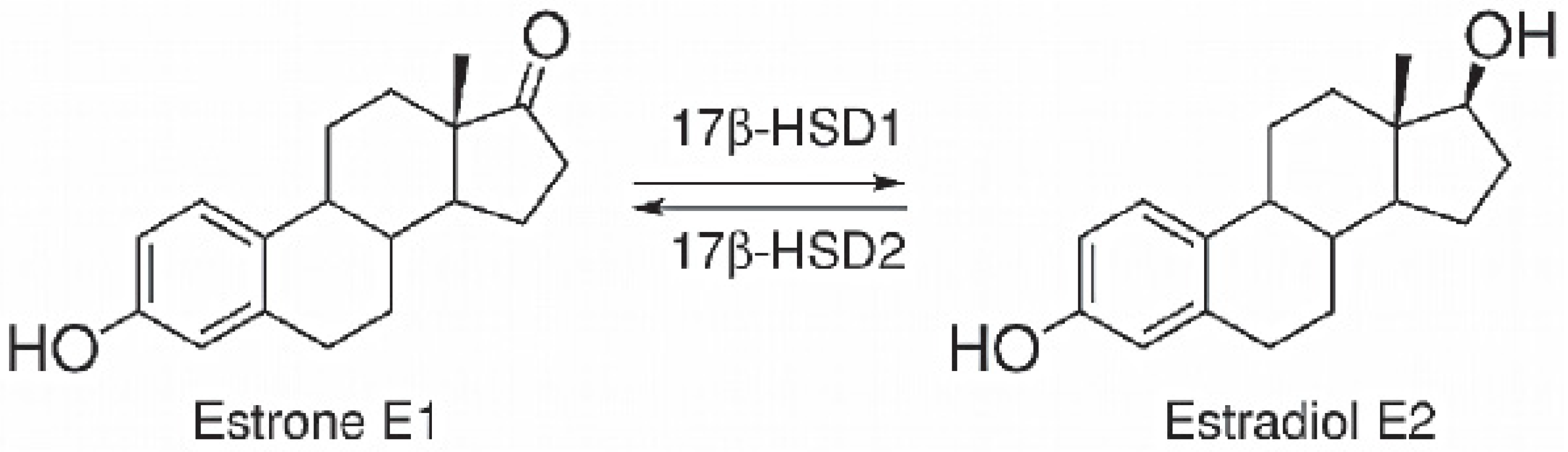
3.7. 17β-HSD1 (17β-Hydroxysteroid Dehydrogenase)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozgunova, E.; Yoshida, M.W.; Reski, R.; Goshima, G. Spindle motility skews division site determination during asymmetric cell division in Physcomitrella. Nat. Commun. 2022, 13, 2488. [Google Scholar] [CrossRef] [PubMed]
- Chater, C.C.; Caine, R.S.; Tomek, M.; Wallace, S.; Kamisugi, Y.; Cuming, A.C.; Lang, D.; MacAlister, C.A.; Casson, S.; Bergmann, D.C.; et al. Origin and function of stomata in the moss Physcomitrella patens. Nat. Plants 2016, 2, 16179. [Google Scholar] [CrossRef] [PubMed]
- Horst, N.A.; Katz, A.; Pereman, I.; Decker, E.L.; Ohad, N.; Reski, R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants 2016, 2, 15209. [Google Scholar] [CrossRef]
- Lueth, V.M.; Reski, R. Mosses. Curr. Biol. 2023, 33, R1163–R1185. [Google Scholar] [CrossRef]
- Lang, D.; van Gessel, N.; Ullrich, K.K.; Reski, R. The genome of the model moss Physcomitrella patens. Adv. Bot. Res. 2016, 78, 97–140. [Google Scholar]
- Reski, R. Quantitative moss cell biology. Curr. Opin. Plant Biol. 2018, 46, 39–47. [Google Scholar] [CrossRef]
- Reski, R.; Bae, H.; Simonsen, H.T. Physcomitrella patens, a versatile synthetic biology chassis. Plant Cell Rep. 2018, 37, 1409–1417. [Google Scholar] [CrossRef]
- Decker, E.L.; Alder, A.; Hunn, S.; Ferguson, J.; Lehtonen, M.T.; Scheler, B.; Kerres, K.L.; Wiedemann, G.; Safavi-Rizi, V.; Nor-dzieke, S.; et al. Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 2017, 216, 455–468. [Google Scholar] [CrossRef]
- Koselski, M.; Hoernstein, S.N.W.; Wasko, P.; Reski, R.; Trebacz, K. Long-distance electrical and calcium signals evoked by hydrogen peroxide in Physcomitrella. Plant Cell Physiol. 2023, 64, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Resemann, H.C.; Herrfurth, C.; Feussner, K.; Hornung, E.; Ostendorf, A.K.; Gömann, J.; Mittag, J.; van Gessel, N.; de Vries, J.; Ludwig-Müller, J.; et al. Convergence of sphingolipid desaturation across over 500 million years of plant evolution. Nat. Plants 2021, 7, 219–232. [Google Scholar] [CrossRef]
- Reski, R. Enabling the water-to-land transition. Nat. Plants 2018, 4, 67–68. [Google Scholar] [CrossRef]
- Moreira, A.S.; Braz-Filho, R.; Mussi-Dias, V.; Vieira, I.J. Chemistry and Biological Activity of Ramalina Lichenized Fungi. Molecules 2015, 20, 8952–8987. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.E. The Lichen Genus Ramalina in the Gulf South Region of the United States. Master’s Thesis, Graduate School, Louisiana State University, Baton Rouge, LA, USA, 1973. [Google Scholar]
- Horst, N.A.; Reski, R. Alternation of generations—Unravelling the underlying molecular mechanism of a 165-year-old botanical observation. Plant Biol. 2016, 18, 549–551. [Google Scholar] [CrossRef] [PubMed]
- Lüth, V.M.; Rempfer, C.; van Gessel, N.; Herzog, O.; Hanser, M.; Braun, M.; Decker, E.L.; Reski, R. A Physcomitrella PIN protein acts in spermatogenesis and sporophyte retention. New Phytol. 2023, 237, 2118–2135. [Google Scholar] [CrossRef] [PubMed]
- Marttinen, E.M.; Decker, E.L.; Heinonen, P.; Reski, R.; Valkonen, J.P. Putative NAD(P)-binding Rossmann fold protein is involved in chitosan-induced peroxidase activity and lipoxygenase expression in Physcomitrium patens. Mol. Plant-Microbe Interact. 2023, 36, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Marttinen, E.M.; Lehtonen, M.T.; van Gessel, N.; Reski, R.; Valkonen, J.P.T. Viral suppressor of RNA silencing in vascular plants also interferes with the development of the bryophyte Physcomitrella patens. Plant Cell Environ. 2021, 45, 220–235. [Google Scholar] [CrossRef]
- Glime, J.M. Water Relations: Leaf Strategies—Cuticles and Waxes. In Bryophyte Ecology. Volume 1. Physiological Ecology; Glime, J.M., Ed.; Michigan Technological University: Houghton, MI, USA, 2007; Chapter 7-4b. [Google Scholar]
- Renault, H.; Alber, A.; Horst, N.A.; Lopes, A.B.; Fich, E.A.; Kriegshauser, L.; Wiedemann, G.; Ullmann, P.; Herrgott, L.; Erhardt, M.; et al. A phenol-enriched cuticle is ancestral to lignin evolution in land plants. Nat. Commun. 2017, 8, 14713. [Google Scholar] [CrossRef] [PubMed]
- Knosp, S.; Kriegshauser, L.; Tatsumi, K.; Malherbe, L.; Wiedemann, G.; Bakan, B.; Kohchi, T.; Reski, R.; Renault, H. An ancient role for the CYP73 gene family in t-cinnamic acid 4-hydroxylation, phenylpropanoid biosynthesis and embryophyte development. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kriegshauser, L.; Knosp, S.; Grienenberger, E.; Tatsumi, K.; Gütle, D.D.; Sørensen, I.; Herrgott, L.; Zumsteg, J.; Rose, J.K.; Reski, R.; et al. Function of the Hydroxycinnamoyl-CoA:Shikimate hydroxycinnamoyl Transferase is evolutionarily conserved in embryophytes. Plant Cell 2021, 33, 1472–1491. [Google Scholar] [CrossRef]
- Resemann, H.C.; Lewandowska, M.; Gömann, J.; Feussner, I. Membrane lipids, waxes and oxylipins in the moss model organism Physcomitrella patens. Plant Cell Physiol. 2018, 60, 1166–1175. [Google Scholar] [CrossRef]
- Buda, G.J.; Barnes, W.J.; Fich, E.A.; Park, S.; Yeats, T.H.; Zhao, L.; Domozych, D.S.; Rose, J.K. An ATP binding cassette transporter is required for cuticular wax deposition and desiccation tolerance in the moss Physcomitrella patens. Plant Cell 2013, 25, 4000–4013. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Dolan, L. Auxin promotes the transition from chloronema to caulonema in moss protonema by positively regulating PpRSL1and PpRSL2 in Physcomitrella patens. New Phytol. 2011, 192, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, S.; Nakajima, M.; Kawaide, H. Assays of protonemal growth responses in Physcomitrella patens under blue- and red-light stimuli. Methods Mol. Biol. 2019, 1924, 35–43. [Google Scholar] [PubMed]
- Lee, S.B.; Yang, S.U.; Pandey, G.; Kim, M.S.; Hyoung, S.; Choi, D.; Shin, J.S.; Suh, M.C. Occurrence of land-plant-specific glycerol-3-phosphate acyltransferases is essential for cuticle formation and gametophore development in Physcomitrella patens. New Phytol. 2020, 225, 2468–2483. [Google Scholar] [CrossRef] [PubMed]
- Bendz, G.; Santesson, J.; Wachtmeister, C.A.; Kristiansen, L.A.; Jensen, K.A. Study on the chemistry of lichens, the chemistry of the Ramalina ceruchis group. Acta Chem. Scand. 1965, 19, 1185–1187. [Google Scholar] [CrossRef]
- Erxleben, A.; Gessler, A.; Vervliet-Scheebaum, M.; Reski, R. Metabolite profiling of the moss Physcomitrella patens reveals evolutionary conservation of osmoprotective substances. Plant Cell Rep. 2012, 31, 427–436. [Google Scholar] [CrossRef]
- Huneck, S.; Vevle, O. Inhaltsstoffe der Moose VIII. (-)-16-α-Hydroxykauran aus Anthelia julacea (L.) Dum. und Anthelia juratzkana (Limpr.) Trev. Z. Naturforsch. 1970, 25, 227. [Google Scholar] [CrossRef]
- Nilsson, E. (-)-16-α-Hydroxykaurane from Saelania glaucescens (Hedw.) Broth. Acta Chem. Scand. 1971, 25, 4. [Google Scholar] [CrossRef]
- Lehn, J.-M.; Huneck, S.Z. Die erstmalige Isolierung des Diterpens (-)-16-a-Hydroxykauran aus einer Flechte. Z. Naturforsch. 1965, 20, 1013. [Google Scholar] [CrossRef]
- Fujita, E.; Johne, S.; Kasai, R.; Node, M.; Tanaka, O.; Fujita, E.; Node, M. Diterpenoids of Rabdosia species. Progr. Chem. Org. Nat. Prod. 1984, 46, 77–157. [Google Scholar]
- Hayashi, K.-I.; Kawaide, H.; Notomi, M.; Sakigi, Y.; Matsuo, A.; Nozaki, H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006, 580, 6175–6181. [Google Scholar] [CrossRef] [PubMed]
- von Schwartzenberg, K.; Schultze, W.; Kassner, H. The moss Physcomitrella patens releases a tetracyclic diterpene. Plant Cell Rep. 2004, 22, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Kerboua, M.; Ahmed, M.A.; Samba, N.; Aitfella-Lahlou, R.; Silva, L.; Boyero, J.F.; Raposo, C.; Rodilla, J.M.L. Phytochemical investigation of new Algerian lichen species: Physcia mediterranea Nimis. Molecules 2021, 26, 1121. [Google Scholar] [CrossRef] [PubMed]
- Hohe, A.; Decker, E.; Gorr, G.; Schween, G.; Reski, R. Tight control of growth and cell differentiation in photoautotrophically growing moss (Physcomitrella patens) bioreactor cultures. Plant Cell Rep. 2002, 20, 1135–1140. [Google Scholar] [CrossRef]
- Schween, G.; Gorr, G.; Hohe, A.; Reski, R. Unique tissue-specific cell cycle in Physcomitrella. Plant Biol. 2003, 5, 50–58. [Google Scholar] [CrossRef]
- Reski, R.; Abel, W.O. Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 1985, 165, 354–358. [Google Scholar] [CrossRef]
- Vázquez, J.; Deplano, A.; Herrero, A.; Ginex, T.; Gibert, E.; Rabal, O.; Oyarzabal, J.; Herrero, E.; Luque, F.J. Development and validation of molecular overlays derived from three-dimensional hydrophobic similarity with PharmScreen. J. Chem. Inf. Model. 2018, 58, 1596–1609. [Google Scholar] [CrossRef]
- Santos, K.B.; Guedes, I.A.; Karl, A.L.; Dardenne, L.E. Highly flexible ligand docking: Benchmarking of the DockThor program on the LEADS-PEP protein-peptide dataset. J. Chem. Inf. Model. 2020, 60, 667–683. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Poole, A.; Peacock, W.J.; Dennis, E.S. Arabidopsis ent-Kaurene Oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999, 119, 507–510. [Google Scholar] [CrossRef]
- Ricci, E.J. Investigating the Leaf Cuticle of the Moss Physcomitrella patens. Master’s Thesis, Rhode Island College: Providence, RI, USA, 2013. [Google Scholar]
- Guetchueng, S.T.; Nahar, L.; Ritchie, K.J.; Ismail, F.; Wansi, J.D.; Evans, A.R.; Sarker, S.D. Kaurane diterpenes from the fruits of Zanthoxylum leprieurii. Rec. Nat. Prod. 2017, 11, 304–309. [Google Scholar]
- Horn, A.; Pascal, A.; Lončarević, I.; Marques, R.V.; Lu, Y.; Miguel, S.; Bourgaud, F.; Thorsteinsdóttir, M.; Cronberg, N.; Becker, J.D.; et al. Natural products from bryophytes: From basic biology to biotechnological applications. Crit. Rev. Plant Sci. 2021, 40, 191–217. [Google Scholar] [CrossRef]
- Kawamoto, T.; Kakizaki, S.; Yoshinari, K.; Negishi, M. Estrogen activation of the nuclear orphan receptor CAR (constitutive active receptor) in induction of the mouse Cyp2b10 gene. Mol. Endocrinol. 2000, 14, 1897–1905. [Google Scholar] [CrossRef]
- Chen, F.; Ludwiczuk, A.; Wei, G.; Chen, X.; Crandall-Stotler, B.; Bowman, J.L. Terpenoid secondary metabolites in bryophytes: Chemical diversity, biosynthesis and biological functions. Crit. Rev. Plant. Sci. 2018, 37, 210–231. [Google Scholar] [CrossRef]
- Otsuka, M.; Kenmoku, H.; Ogawa, M.; Okada, K.; Mitsuhashi, W.; Sassa, T.; Kamiya, Y.; Toyomasu, T.; Yamaguchi, S. Emission of ent-Kaurene, a diterpenoid hydrocarbon precursor for gibberellins, into the headspace from plants. Plant Cell Physiol. 2004, 45, 1129–1138. [Google Scholar] [CrossRef]
- Lai, J.-J.; Chang, P.; Lai, K.-P.; Chen, L.; Chang, C. The role of androgen and androgen receptor in skin-related disorders. Arch. Dermatol. Res. 2012, 304, 499–510. [Google Scholar] [CrossRef]
- Grymowicz, M.; Rudnicka, E.; Podfigurna, A.; Napierala, P.; Smolarczyk, R.; Smolarczyk, K.; Meczekalski, B. Hormonal effects on hair follicles. Int. J. Mol. Sci. 2020, 28, 5342. [Google Scholar] [CrossRef]
- Wandrey, F.; Henes, B.; Zülli, F.; Reski, R. Biotechnologically produced moss active improves skin resilience. SOFW J. 2018, 144, 34–37. [Google Scholar]
- Imfeld, D. Inhibition of cutaneous cortisol activation: A novel approach to protect skin from stress induced damage and aging. In Proceedings of the 30th IFSCC Congress, Munich, Germany, 18–21 September 2018; Poster IFSCC Munich. pp. 1–501. [Google Scholar]
- Gambineri, A.; Forlani, G.; Munarini, A.; Tomassoni, F.; Cognigni, G.E.; Ciampaglia, W.; Pagotto, U.; Walker, B.R.; Pasquali, R. Increased clearance of cortisol by 5beta-reductase in a subgroup of women with adrenal hyperandrogenism in polycystic ovary syndrome. J. Endocrinol. Investig. 2009, 32, 210–218. [Google Scholar] [CrossRef]
- Dahlgren, A.; Kecklund, G.; Theorell, T.; Åkerstedt, T. Day-to-day variation in saliva cortisol--relation with sleep, stress and self-rated health. Biol. Psychol. 2009, 82, 149–155. [Google Scholar] [CrossRef]
- Dismukes, A.; Shirtcliff, E.; Jones, C.W.; Zeanah, C.; Theall, K.; Drury, S. The development of the cortisol response to dyadic stressors in Black and White infants. Dev. Psychopathol. 2018, 30, 1995–2008. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, N.; Gathercole, L.L.; Marchand, L.; Althari, S.; Dempster, N.J.; Green, C.J.; Van De Bunt, M.; McNeil, C.; Arvaniti, A.; Hughes, B.A.; et al. AKR1D1 is a novel regulator of metabolic phenotype in human hepatocytes and is dysregulated in non-alcoholic fatty liver disease. Metabolism 2019, 99, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, N.; Arvaniti, A.; Appanna, N.; Sharp, A.; Hughes, B.A.; Digweed, D.; Whitaker, M.J.; Ross, R.; Arlt, W.; Penning, T.M.; et al. Glucocorticoids regulate AKR1D1 activity in human liver in vitro and in vivo. J. Endocrinol. 2020, 245, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H. Signaling control of the constitutive androstane receptor (CAR). Protein Cell 2014, 5, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Oliviero, F.; Lukowicz, C.; Boussadia, B.; Forner-Piquer, I.; Pascussi, J.M.; Marchi, N.; Mselli-Lakhal, L. Constitutive Androstane Receptor: A peripheral and a neurovascular stress or environmental sensor. Cells 2020, 9, 2426. [Google Scholar] [CrossRef] [PubMed]
- Küblbeck, J.; Niskanen, J.; Honkakoski, P. Metabolism-disrupting chemicals and the Constitutive Androstane Receptor CAR. Cells 2020, 9, 2306. [Google Scholar] [CrossRef]
- Carazo, A.; Dusek, J.; Holas, O.; Skoda, J.; Hyrsova, L.; Smutny, T.; Soukup, T.; Dosedel, M.; Pávek, P. Teriflunomide is an indirect human Constitutive Androstane Receptor (CAR) Activator interacting with Epidermal Growth Factor (EGF) signaling. Front. Pharmacol. 2018, 9, 993. [Google Scholar] [CrossRef]
- Cherian, M.T.; Chai, S.C.; Chen, T. Small-molecule modulators of the constitutive androstane receptor. Expert Opin. Drug Metab. Toxicol. 2015, 11, 1099–1114. [Google Scholar] [CrossRef]
- Min, G.; Kim, H.; Bae, Y.; Petz, L.; Kemper, J.K. Inhibitory crosstalk between Estrogen Receptor (ER) and Constitutively Activated Androstane Receptor (CAR): CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J. Biol. Chem. 2002, 277, 34626–34633. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Thiboutot, D.; Zouboulis, C.C. Cutaneous androgen metabolism: Basic research and clinical perspectives. J. Investig. Dermatol. 2002, 119, 992–1007. [Google Scholar] [CrossRef] [PubMed]



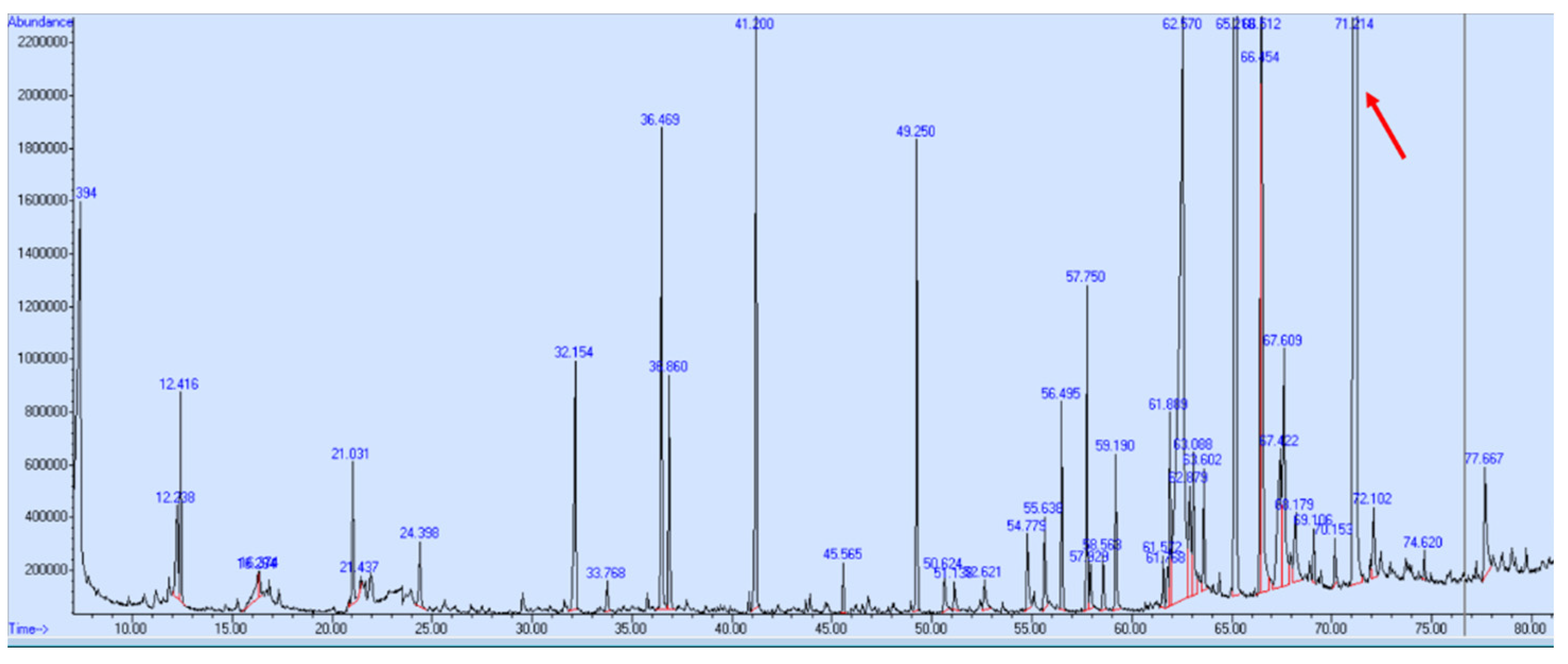

| PK | RT | Library/ID | CAS | Qual | Area | Confirmed? |
|---|---|---|---|---|---|---|
| 1 | 7.39 | Acetic acid | 64-19-7 | 86 | 1.30 × 108 | Yes |
| 2 | 12.24 | HEXANAL | 66-25-1 | 93 | 30,055,867 | Yes |
| 3 | 21.03 | 2-Octen-1-ol, (Z)- | 26001-58-1 | 93 | 25,079,272 | Yes |
| 4 | 24.4 | 9.14 Benzene acetaldehyde | 122-78-1 | 87 | 14,788,190 | Yes |
| 5 | 32.15 | 15.77 Dodecane | 112-40-3 | 95 | 56,976,919 | Yes |
| 6 | 33.77 | 2-Cyclopenten-1-one, 2-methyl- | 1120-73-6 | 89 | 5,702,674 | Yes |
| 7 | 36.47 | 19.71 Thymol | 89-83-8 | 95 | 72,963,027 | Yes |
| 8 | 36.86 | 20.14 Carvacrol | 499-75-2 | 95 | 37,180,865 | Yes |
| 9 | 41.2 | 24.61 Tetradecane (C14) | 629-59-4 | 97 | 1.14 × 108 | Yes |
| 10 | 45.57 | Butylated Hydroxytoluene | 128-37-0 | 97 | 7,402,971 | Yes |
| 11 | 49.25 | 32.89 Hexadecane (C16) | 544-76-3 | 98 | 63,206,613 | Yes |
| 12 | 50.62 | Benzene, 1-methyl-4-[(1-methylethylidene)cyclopropyl]- | 24578-28-7 | 71 | 9,609,670 | Yes |
| 13 | 51.14 | 34.73 Amyl cinnamaldehyde<Z-> | 101365-33-7 | 99 | 6,484,050 | Yes |
| 14 | 52.62 | 1-Methyl-5-nitro-1H-benzoimidazol-2-ol | 66108-85-8 | 65 | 9,853,674 | Yes |
| 15 | 54.78 | 39.42 Cinnamaldehyde<2-hexyl-(Z)-> | 364364-06-7 | 99 | 17,036,726 | Yes |
| 16 | 55.64 | 38.90 Benzyl benzoate | 120-51-4 | 95 | 17,676,783 | Yes |
| 17 | 56.49 | 40.40 Octadecane (C18) | 593-45-3 | 98 | 28,462,303 | Yes |
| 18 | 57.75 | 3,7,11,15-Tetramethyl-2-hexadecen-1-ol | 102608-53-7 | 87 | 40,608,898 | Yes |
| 19 | 57.93 | Hexa-hydro-farnesol | 6750-34-1 | 93 | 8,759,193 | Yes |
| 20 | 58.56 | 9-Eicosyne | 71899-38-2 | 81 | 11,027,985 | Yes |
| 21 | 59.19 | BENZYL SALICYLATE (ALLERGENE) | 118-58-1 | 87 | 28,689,063 | Yes |
| 22 | 61.57 | 44.98 Beyerene | 2359-73-1 | 99 | 9,040,448 | Yes |
| 23 | 61.77 | Biformen | 5957-33-5 | 83 | 6,353,250 | Yes |
| 24 | 61.89 | 45.59 Pimaradiene | 1686-61-9 | 99 | 34,535,194 | Yes |
| 25 | 62.57 | n-Hexadecanoic acid | 57-10-3 | 99 | 4.01 × 108 | Yes |
| 26 | 62.88 | 47.09 Ethyl hexadecanoate | 628-97-7 | 94 | 32,838,290 | Yes |
| 27 | 63.09 | 47.33 Eicosane (C20) | 112-95-8 | 98 | 30,418,475 | Yes |
| 28 | 63.6 | 47.25 Kaur-15-ene | 5947-50-2 | 99 | 21,893,926 | Yes |
| 29 | 65.22 | 48.66 Kaur-16-ene | 562-28-7 | 99 | 7.40 × 108 | Yes |
| 30 | 66.45 | 50.22 Benzyl cinnamate | 103-41-3 | 95 | 74,793,795 | Yes |
| 31 | 68.18 | Octadecanoic acid | 57-11-4 | 98 | 24,803,583 | Yes |
| 32 | 69.11 | 53.54 Docosane | 629-97-0 | 93 | 7,580,271 | Yes |
| 33 | 70.15 | Kaur-15-ene, (5α,9α,10β)- | 511-85-3 | 87 | 7,534,955 | Yes |
| 34 | 71.21 | Kauran-16-ol (Ceruchinol) | 5524-17-4 | 90 | 1.44 × 109 | Yes |
| 35 | 72.1 | Arachidonic acid | 506-32-1 | 95 | 19,255,524 | Yes |
| 36 | 74.62 | 7-Methyl-Z-tetradecen-1-ol acetate | 959269-58-0 | 92 | 4,176,098 | Yes |
| 37 | 77.67 | Dipalmitin | 761-35-3 | 91 | 34,085,962 | Yes |
| 38 | 91.75 | Glycerol tricaprylate | 538-23-8 | 81 | 4.31 × 108 | Yes |
| 39 | 92.75 | Cholesta-8,24-dien-3-ol, 4-methyl-, | 7199-92-0 | 95 | 41,964,203 | Yes |
| 40 | 94.37 | Campesterol | 474-62-4 | 87 | 2.36 × 108 | Yes |
| 41 | 95.04 | Cholesta-6,22,24-triene, 4,4-dimethyl- | 1000128-66-9 | 95 | 4.29 × 108 | Yes |
| 42 | 96.01 | Caprin | 621-71-6 | 70 | 1.46 × 108 | Yes |
| Total Energy (kcal/mol) | Interaction Energy (kcal/mol) | Docking Score (kcal/mol) | |
|---|---|---|---|
| Pose 1 | 12.4 | −30.1 | −10.2 |
| Pose 2 | 15.7 | −27.0 | −10.1 |
| Total Energy (kcal/mol) | Interaction Energy (kcal/mol) | Docking Score (kcal/mol) | |
|---|---|---|---|
| Pose 1 | 29.8 | −25.6 | −9.3 |
| Pose 2 | 30.6 | −25.0 | −9.2 |
| Total Energy (kcal/mol) | Interaction Energy (kcal/mol) | Docking Score (kcal/mol) | ||
|---|---|---|---|---|
| Ligand DHT | Pose 4 | 61.8 | −23.4 | −8.7 |
| Pose 9 | 62.7 | −22.2 | −8.1 | |
| Ceruchinol | Pose 1 | 31.3 | −24.3 | −8.4 |
| Pose 3 | 33.3 | −23.4 | −8.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munoz, C.; Schröder, K.; Henes, B.; Hubert, J.; Leblond, S.; Poigny, S.; Reski, R.; Wandrey, F. Phytochemical Exploration of Ceruchinol in Moss: A Multidisciplinary Study on Biotechnological Cultivation of Physcomitrium patens (Hedw.) Mitt. Appl. Sci. 2024, 14, 1274. https://doi.org/10.3390/app14031274
Munoz C, Schröder K, Henes B, Hubert J, Leblond S, Poigny S, Reski R, Wandrey F. Phytochemical Exploration of Ceruchinol in Moss: A Multidisciplinary Study on Biotechnological Cultivation of Physcomitrium patens (Hedw.) Mitt. Applied Sciences. 2024; 14(3):1274. https://doi.org/10.3390/app14031274
Chicago/Turabian StyleMunoz, Carlos, Kirsten Schröder, Bernhard Henes, Jane Hubert, Sébastien Leblond, Stéphane Poigny, Ralf Reski, and Franziska Wandrey. 2024. "Phytochemical Exploration of Ceruchinol in Moss: A Multidisciplinary Study on Biotechnological Cultivation of Physcomitrium patens (Hedw.) Mitt." Applied Sciences 14, no. 3: 1274. https://doi.org/10.3390/app14031274





