1. Introduction
Humans share a similar body structure, but each individual possesses unique characteristics, which we define as one’s body type [
1]. Various classification methods have been devised to understand and assess these body types. Body type classification has garnered the interest of researchers in related fields to evaluate human body shape and composition [
2]. One prominent method of body type classification is the Heath-Carter somatotype. Somatotyping provides a systematic and scientific approach for quantifying human body types [
3] and has been widely utilized by researchers across various disciplines for extended periods [
4]. However, because potential measurement errors depend on the skill of the measurer, somatotyping requires technical expertise [
5]. Additionally, there are limitations to its applicability in modern individuals, as many are reluctant to expose or permit contact with sensitive areas of the body [
6].
Recent research has applied artificial intelligence technology utilizing noninvasive measurement tools, such as 3D body scanners, which minimize physical contact [
7,
8]. Compared with traditional methods, 3D body scanners analyze a larger amount of data in a shorter period of time, thereby increasing the efficiency of analyzing existing body measurement data (e.g., width, length, and circumference) [
9]. Moreover, data in the form of various body measurements including height, circumference, cross-sectional area, and volume are collected [
10]. Previous studies have shown that the use of machine learning algorithms to predict obesity based on body fat percentage [
11] outperforms BMI (Body Mass Index) and BIA (Bioelectrical Impedance Analysis) in the classification of obesity. In addition, research exploring predictive models for somatotypes based on anthropometry [
12] has shown excellent performance in classifying body types. Previous studies have indicated that 3D body scanners are reliable and valid measurement tools for predicting body type.
Furthermore, data collected during 3D body scanner scans can capture not only body measurements but also three-dimensional (3D) body images. The aforementioned studies utilized body data, requiring complex procedures, such as setting markers for each segment of the body to extract data. Therefore, in terms of practicality, the resulting models may be less scalable to provide generalized functions. To compensate for these limitations, this study seeks to utilize 3D body images that were not used in previous studies. Two-dimensional (2D) images are already used to create three-dimensional (3D) body images, and a 3D body scanner is used to derive body shape values similar to actual human body values [
13,
14]. However, it is still difficult to find research applying artificial intelligence technology related to the somatotype. The technology for generating complex 3D images from simple 2D images is predicted to have a significant impact on future scientific advancements. However, implementing systems that offer practicality to many individuals requires time and expertise from researchers. Therefore, the development of practical models by simplifying 3D body images collected using 3D body scanner into 2D images is essential. Through this study, it is anticipated that various limitations such as complex procedures and expertise can be overcome from a practical standpoint. Moreover, systems that are unaffected by time and location constraints, such as identifying body shape using a mobile phone camera, can be established. Thus, the objective of this study was to develop an artificial intelligence (AI) somatotype system capable of predicting the three body types proposed by Heath–Carter’s somatotype theory using 3D body images collected using 3D body scanners. Specifically, after performing data pre-processing, such as converting the collected three-dimensional (3D) images into two-dimensional (2D) images, the model is trained and verified by segmenting the data set. After applying data augmentation techniques to solve the unbalanced data distribution, we will finally evaluate and verify the performance of the AI somatotype system by creating a body type classification model using a transfer learning model. The results of this study are intended to provide basic data for follow-up studies that predict 13 detailed body types from the 3 main body types. In addition, in the actual field, anyone with a personal phone camera can identify various body types based on the photographed images and build programs for smartphones and websites in the form of a user interface (UI) to predict obesity, diseases, etc. We hope that the results of this study will be utilized.
2. Methods
2.1. Research Subject
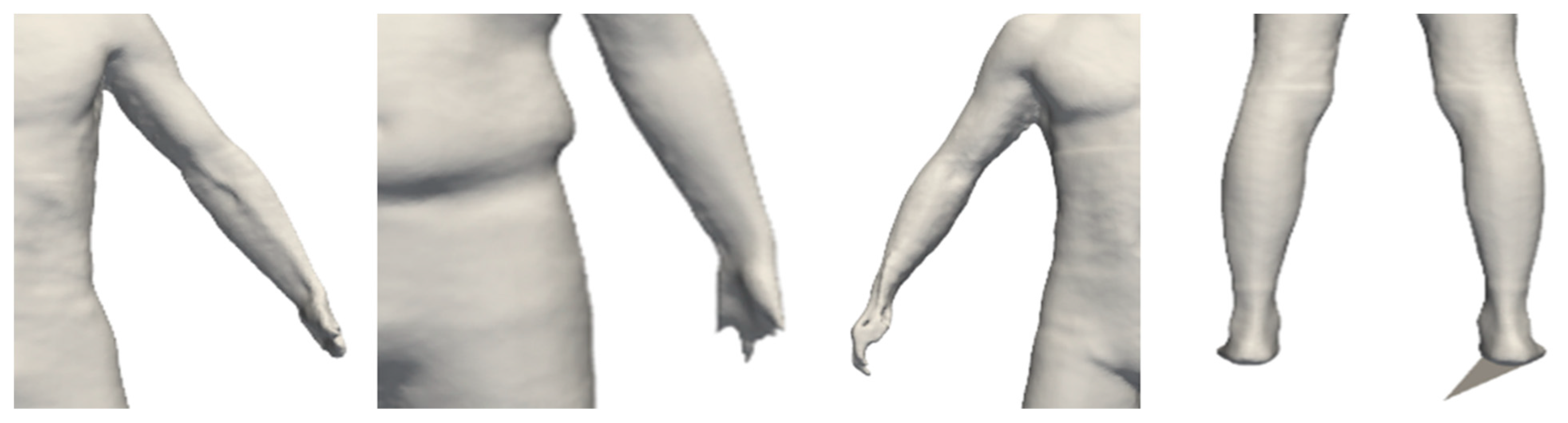
Male residents of Seoul were selected as research subjects using purposive sampling. Individuals who refused physical contact for body measurements, had mobility impairments, or did not consent to participate were excluded. Additionally, owing to various constraints, such as body exposure, menstruation, and pregnancy during body measurements, females were excluded from this study. All participants who expressed a willingness to participate provided informed consent, and the study was conducted after obtaining approval from the Korea National University of Physical Education Institutional Review Board (1263-202304-HR-009-01). Ultimately, data from 217 males were utilized after excluding data with compromised reliability, such as distortion in body parts or scanning errors, which could lead to inaccuracies in predicting body shapes. Specific details of exclusion are shown in
Figure 1.
2.2. Measurement Variable
2.2.1. Heath-Carter’s Somatotype
To classify body types, measurements were taken to determine the components of the three somatotypes (endomorphy, mesomorphy, and ectomorphy). The selection criteria were based on the guidelines of the International Society for the Advancement of Kinanthropometry (ISAK). Additionally, two experts in physical measurement assessment with extensive experience in body measurement selected and measured 10 measurement variables, including height and weight, based on Heath-Carter’s somatotype classification criteria. All participants underwent measurements of the body parts listed in
Table 1. Subsequently, the measured values were used to classify body types into the three somatotypes, as proposed by Heath-Carter.
2.2.2. Three-Dimensional Body Scanner
In this study, measurements taken during 3D body scanning may have been subject to overestimation owing to factors such as hair and clothing thickness, leading to potential errors. To minimize measurement errors, all the participants wore swim caps and thin sports tights. The piece of equipment used for the measurements was a PFS-304 model From PMTinnovation in Uiwang, Republic of Korea. The postures and attire that the participants were required to adopt during the measurements are depicted in
Figure 2.
2.3. Data
2.3.1. Three-Dimensional Image Preprocessing
In this study, a 3D body scanner was used to collect 3D images of the research subjects (participants). The collected body images are shown in
Figure 3, composed of mesh-type STL files.
Subsequently, the body regions were cropped from the 3D images in three orientations (Front, Back, Right) with the background removed, and converted into 2D images in the PNG file format. The preprocessed images used in this study are shown in
Figure 4.
2.3.2. Data Segmentation
To generalize the performance of the deep learning model in this study, the dataset was divided into three parts (training, validation, and test sets). The training and validation data were used for training, and the test data were excluded to validate the performance of the developed model. To evaluate the performance of the proposed model, approximately 10% of the data, which amounted to 21 individuals out of the 217 research subjects, were used as the test data, as shown in
Figure 5. The training and validation datasets consisted of 196 individuals divided into an 8:2 ratio, allowing for training with new datasets at each epoch. The final dataset used in this study is presented in
Table 2.
2.3.3. Data Augmentation
In order to achieve satisfactory performance in deep learning and computer vision, a large number of images are required [
15]. However, collecting body images of subjects in actual research situations is challenging, expensive and costly [
16]. Image augmentation can be useful for improving the performance of models in image classification and prediction, as well as preventing overfitting when the amount of training data is limited [
17]. The most common method of image augmentation involves image transformation through rotation, translation, resizing, and flipping.
Upon examining
Table 3, it is evident that the distribution of data in the existing training set was imbalanced. A significantly lower amount of data was collected for ectomorphy than for endomorphy and mesomorphy. Because this study utilized body images, there is a concern that past transformations, such as rotation, scaling, and resizing, may actually decrease the model’s performance. Therefore, to address this issue, while maintaining relatively balanced data for endomorphy and mesomorphy, the severely lacking ectomorphy data were augmented. Specifically, considering potential body distortions, each image was rotated by 2 degrees, resulting in three augmented images per original image.
2.4. CNN and Transfer Learning Model
Among the various deep learning techniques, convolutional neural networks (CNN) have been optimized for image data learning [
18]. However, the implementation of CNN-based deep-learning models requires a large amount of data. Excessive image augmentation can lead to overfitting, making further augmentation difficult in this study, which aimed to classify body images. Transfer learning was used to address this issue. Transfer learning is a method for resolving learning problems using a small number of labeled samples in the target field by transferring the knowledge learned from existing large-scale datasets to a new domain [
19]. Before training the somatotype model in this study, three models (MobileNetV2, VGG16, and ResNet50) that have been useful as transfer learning models in various studies [
20,
21] were tested. Subsequently, the early stopping technique was applied to determine the epoch for each model, and performance metrics, including accuracy (ACC) and loss, were evaluated. The results are shown in
Figure 6.
Through testing, it was observed that MobileNetV2 achieved the highest classification accuracy of approximately 76%, followed by VGG16, with approximately 62%, and ResNet50, with approximately 57%. These results indicate that MobileNetV2 performed the best in predicting new data among the transfer learning models tested. This observation suggests that simpler structured models with faster computational speeds perform better at identifying the features of body images than more complex models. Therefore, MobileNetV2 was selected as the final training model. MobileNetV2, proposed by Howard et al. [
22], was designed to apply CNN models to small-scale image processing tasks. This offers the advantage of reducing the computational load while maintaining accuracy, leading to shorter training times. Additionally, as a specialized model for mobile devices, it operates efficiently, even on devices with limited computational capabilities [
23]. Hence, it was deemed the most suitable for the future scalability of the system.
2.5. Data Processing and Analysis Procedure
2.5.1. Data Processing
In this study, all analyses, including preprocessing of three-dimensional (3D) images, data augmentation, data splitting, and CNN-based transfer learning, were conducted using Python 3.9. Specifically, the ‘pyvista’ library was utilized to convert 3D images to two-dimensional (2D) images and perform operations such as cropping and background removal from each direction. Subsequently, the ‘Pillow’ library was used to adjust the data of each somatotype group to the same ratio as other groups. The libraries provided by TensorFlow were used for data splitting and model training. Data splitting was performed using the ImageData Generator to divide the data into training and validation datasets. The ‘MobileNetV2’ model was employed for transfer learning, and early stopping was applied to determine the optimal epoch for the model.
2.5.2. Architectural Extension and Learning Parameters of the Model Using Transfer Learning
The transfer learning model used in this study was MobileNetV2. To develop an AI somatotype model based on body images according to the research objectives, the architecture extension and setting of learning parameters were conducted as follows. First, MobileNetV2 was loaded and initialized with weights trained on ImageNet; these weights were then frozen. Subsequently, two hidden layers were added to the model, with the ‘ReLU’ activation function applied to each hidden layer. Finally, the output layer was configured with a ‘softmax’ activation function to output the probabilities of the three classes. Additionally, four metrics, namely the training accuracy, training loss, validation accuracy, and validation loss, were monitored to evaluate the training performance of the model. For the learning parameters, the optimizer used was ‘Adam’, with a learning rate of ‘1 ×10
−4’ and a batch size of 10. To prevent overfitting, ‘Dropout’ was applied after each hidden layer, and the number of epochs was determined using early stopping, which stops training if the validation loss does not improve for five consecutive epochs. The parameter information used during model training is presented in
Table 4, and the architecture of the MobileNetV2 model selected in the preliminary test conducted for applying transfer learning is shown in
Table 5. The architecture of the model constructed in this study ultimately appears as depicted in
Figure 7.
2.5.3. Analysis Procedure
The analysis procedure of this study, summarizing the aforementioned research methods, consisted of six steps, as illustrated in
Figure 8. First, data collection. Second, preprocessing and labeling of collected images. Third, data splitting. Fourth, data augmentation. Fifth, model training. Finally, the evaluation and validation of the trained model were conducted following each step accordingly.
3. Result
3.1. AI Somatotype Model Train Result
In this study, a somatotype prediction model based on MobileNetV2 transfer learning using body images yielded the following performance metrics: accuracy = 0.9058; loss = 0.2653; validation accuracy = 0.7192; and validation loss = 0.6912. The optimal epoch was determined to be 31 using early stopping. The performance indicators of the model are presented in
Table 6, and the recorded metrics for the 31 training epochs are shown in
Figure 9.
3.2. AI Somatotype Model Test Result
In this study, the data were divided into training, validation, and test sets for the model development. The training and validation data were used to train the model, whereas the test data were used to validate the performance of the trained model. The specific validation process of the AI somatotype model involved calculating the probabilities of three somatotype elements (endomorphy, mesomorphy, and ectomorphy) and classifying them based on the highest probability. The prediction results for the 21 new data samples are presented in
Table 7, indicating classification accuracy, recall, and precision.
4. Discussion
The issues that can be discussed based on the results of this study are as follows: first, the collected data were divided into training, validation, and testing sets. Approximately 10% of the 217 individuals (21 individuals) were used as test data. When dividing the test set, we considered the possibility that new data could be easily predicted for certain body types and sampled approximately 10% of each of the three body types. The remaining 90% of subjects (196 individuals) were used for training and validation, with 78 for endomorphy, 97 for mesomorphy, and 21 for ectomorphy, resulting in an imbalance in data distribution. Previous studies, such as that of Lee et al. [
12], mentioned the difficulty of ensuring an even distribution of the three body types as a limitation of their research. Similarly, in this study, some participants were opposed to exposing their bodies to a measurer for body measurements, leading to difficulties in conducting the measurements. Chiu et al. [
5] mentioned the difficulty in recruiting ectomorphic participants, leading to an overprediction of mesomorphy. In this study, while recruiting for endomorphy and mesomorphy was relatively easy, there was a significant shortage of ectomorphy data. However, researchers have made efforts to equalize the data distribution through image augmentation, and various methods have been attempted to prevent overfitting of the augmented images during the model training process. It is necessary to obtain additional data in the future to address the limitations of data distribution. Furthermore, it is necessary to collect sufficient data from females to develop a sex-agnostic AI somatotype model.
Second, MobileNetV2 was utilized as the transfer learning model. The rationale for selecting this model was based on preliminary tests conducted on various models commonly used in transfer learning, including MobileNetV2, VGG16, and ResNet50, which were found to be useful in previous studies. The model that exhibited the best performance was selected for this study. Although various other models are available for transfer learning, the researchers aim to develop the ultimate goal of a mobile AI somatotype model through subsequent research. Therefore, it was deemed appropriate to conduct preliminary tests using simpler structured models compared to other models. Ultimately, the MobileNetV2 model, which operates efficiently even on devices with limited computational capabilities, demonstrated good performance in predicting body images. Therefore, it is necessary to explore and compare lightweight models, such as MobileNetV2, for future model scalability. This would involve research methods to enhance the performance by investigating and comparing lightweight models similar to MobileNetV2.
Third, model training resulted in approximately 91% accuracy for the training set and approximately 72% accuracy for the validation set. The corresponding loss values were approximately 0.26 for the training and 0.69 for the validation set. Using the test data, the predictions for the 21 new data points showed that 18 were accurately classified, resulting in a classification accuracy of approximately 85%, whereas three predictions were incorrect. The adequacy of the model architecture extension and parameter settings could be a topic of discussion among researchers questioning whether they are truly optimal. In this study, test dataset separation was conducted for external validity verification, and image augmentation was employed to address the imbalanced data. Considering the risk of overfitting with excessive augmentation on a small dataset and the possibility of model performance deterioration with a more complex architecture, the researchers opted to use early stopping to determine the number of epochs. Additionally, as mentioned in the preliminary test results, it is anticipated that overly complex architectures may degrade the performance of the model. However, due to the limited amount of data used in this study and the need for fast computation speed to ensure model scalability, simpler architectures were employed. If an adequate amount of data is secured in the future, experimenting with slightly more complex architectures and applying various combinations of training parameters will be possible. It is anticipated that a model with superior performance than the results of this study can be developed. Moreover, because the validation loss value exceeded 0.6, indicating a higher-than-expected value, adjustments and improvements to the model’s architecture extension and training parameters are necessary in future follow-up studies. Nonetheless, this study holds significant value because it explored a transfer learning model suitable for classifying body images and identified areas of improvement, such as securing insufficient data for each body type, developing models considering gender differences, and adding test data samples. Considering these limitations, future research is anticipated to lead to the development of more advanced AI somatotype models.
5. Conclusions
The purpose of this study was to develop an artificial intelligence (AI) somatotype system capable of predicting the three body types proposed by Heath-Carter’s somatotype theory using 3D body images collected using a 3D body scanner. The following conclusions were drawn from this study. First, the AI somatotype model showed approximately 91% training accuracy and approximately 72% validation accuracy. The corresponding loss values were 0.26 for the training and 0.69 for the validation set. Second, the performance of the trained model was validated using test data, resulting in accurate predictions for 18 out of the 21 new data points, achieving approximately 85% classification accuracy, whereas three predictions were incorrect. These findings serve as foundational data for subsequent studies aiming to predict 13 detailed body types from the initial three body types. Additionally, in practical applications, anyone with a personal phone camera can identify various body types based on captured images and develop programs for smartphones and websites in the form of a user interface (UI) to predict obesity, diseases, etc. However, the 3D images used in this study had limitations due to the restricted attire of the subjects, and there are certain constraints to providing services that can be utilized by anyone. Nevertheless, despite these limitations, it is deemed significant that a model for classifying body types more conveniently than existing methods has been developed in this research. It is hoped that the results of this study will evolve to be utilized in various contexts in the future.
Author Contributions
Conceptualization, J.Y.; methodology, S.-Y.L. and J.-Y.L.; software, J.-Y.L.; validation, S.-Y.L. and J.-Y.L.; formal analysis, J.Y.; investigation, J.Y.; resources, J.Y. and J.-Y.L.; data curation, S.-Y.L.; writing original draft preparation, J.-Y.L.; writing—review and editing, S.-Y.L. and J.-Y.L.; visualization, J.Y.; supervision, J.Y.; project administration, J.Y.; funding acquisition, J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2021R1G1A1094776).
Institutional Review Board Statement
This study was conducted after obtaining approval from the Korea National University of Physical Education Institutional Review Board (1263-202304-HR-009-01).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Frenzel, A.; Binder, H.; Walter, N.; Wirkner, K.; Loeffler, M.; Loeffler-Wirth, H. The aging human body shape. NPJ Aging Mech. Dis. 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Bolonchuk, W.W.; Hall, C.B.; Lukaski, H.C.; Siders, W.A. Relationship between body composition and the components of somatotype. Am. J. Hum. Biol. 1989, 1, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Heath, B.H.; Carter, J.E. A modified somatotype method. Am. J. Phys. Anthropol. 1967, 27, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Drywień, M.; Górnicki, K.; Górnicka, M. Application of artificial neural network to somatotype determination. Appl. Sci. 2021, 11, 1365. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Ciems, R.; Thelwell, M.; Bullas, A.; Choppin, S. Estimating somatotype from a single-camera 3D body scanning system. Eur. J. Sport Sci. 2022, 22, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.P.; Liu, Y.E.; Quon, B.K.; Kelly, N.N.; Wong, M.C.; Kennedy, S.F.; Chow, D.C.; Garber, A.K.; Weiss, E.J.; Heymsfield, S.B.; et al. Assessment of clinical measures of total and regional body composition from a commercial 3-dimensional optical body scanner. Clin. Nutr. 2022, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Berg, E.; Cheng, X.; Shen, W. How to best assess abdominal obesity. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Tinsley, G.M.; Moore, M.L.; Dellinger, J.R.; Adamson, B.T.; Benavides, M.L. Digital anthropometry via three-dimensional optical scanning: Evaluation of four commercially available systems. Eur. J. Clin. Nutr. 2020, 74, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.; Ng, B.K.; Kennedy, S.F.; Hwaung, P.; Liu, E.Y.; Kelly, N.N.; Pagano, I.S.; Garber, A.K.; Chow, D.C.; Heymsfield, S.B.; et al. Children and adolescents’ anthropometrics body composition from 3-D optical surface scans. Obesity 2019, 27, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, J.H.; Kim, D.G.; Lee, H.G.; Yoon, J. The classification validity of obese body shapes using 3D body data. Korean J. Phys. Educ. 2022, 61, 515–527. [Google Scholar] [CrossRef]
- Jeon, S.; Kim, M.; Yoon, J.; Lee, S.; Youm, S. Machine learning-based obesity classification considering 3D body scanner measurements. Sci. Rep. 2023, 13, 3299. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, S.Y.; Park, J.H.; Yoon, J. Exploring a 3D body images based somatotype prediction model using multi-class classification machine learning. Korean J. Meas Eval. Phys. Educ. Sport Sci. 2023, 25, 13–28. [Google Scholar]
- Kim, C.; Youm, S. Development of 3D Body Shape Creation Methodology for Obesity Information and Body Shape Management for Tracking Body Condition Check: Body Type in Their 20s and 30s. Res. Sq. 2022; Preprint. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kwon, K.; Kim, C.; Youm, S. Development of a Non-Contact Sensor System for Converting 2D Images into 3D Body Data: A Deep Learning Approach to Monitor Obesity and Body Shape in Individuals in their 20s and 30s. Sensors 2024, 24, 270. [Google Scholar] [CrossRef] [PubMed]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yoon, S.; Fuentes, A.; Park, D.S. A comprehensive survey of image augmentation techniques for deep learning. Pattern Recognit. 2023, 137, 109347. [Google Scholar] [CrossRef]
- Wong, S.C.; Gatt, A.; Stamatescu, V.; McDonnell, M.D. Understanding data augmentation for classification: When to warp? In Proceedings of the 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, QLD, Australia, 30 November–2 December 2016; IEEE Publications: Piscataway, NJ, USA, 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a Convolutional Neural Network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; IEEE Publications: Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar] [CrossRef]
- Rachburee, N.; Punlumjeak, W. Lotus species classification using transfer learning based on VGG16, ResNet152V2, and MobileNetV2. IAES Int. J. Artif. Intell. 2022, 11, 1344. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobile-nets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Xiang, Q.; Wang, X.; Li, R.; Zhang, G.; Lai, J.; Hu, Q. Fruit image classification based on Mobilenetv2 with transfer learning technique. In Proceedings of the 3rd International Conference on Computer Science and Application Engineering, Sanya, China, 22–24 October 2019; pp. 1–7. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).