Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer
Abstract
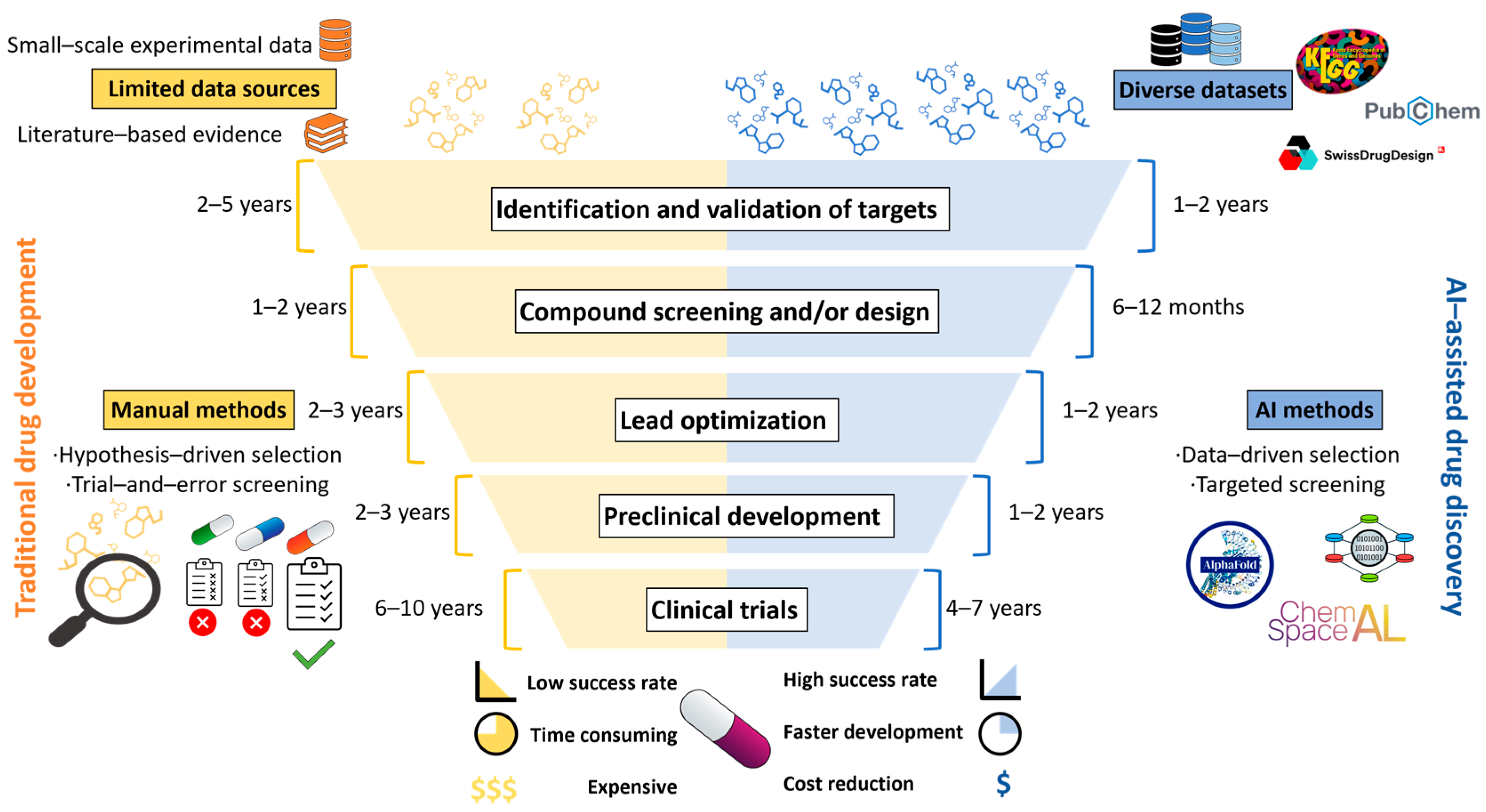
:1. Introduction
2. AI Techniques for Drug Repurposing and De Novo Drug Design
2.1. Machine and Deep Learning Techniques
2.2. Knowledge Graph-Based AI Techniques
2.3. Generative AI Models
2.4. Reinforcement Methods
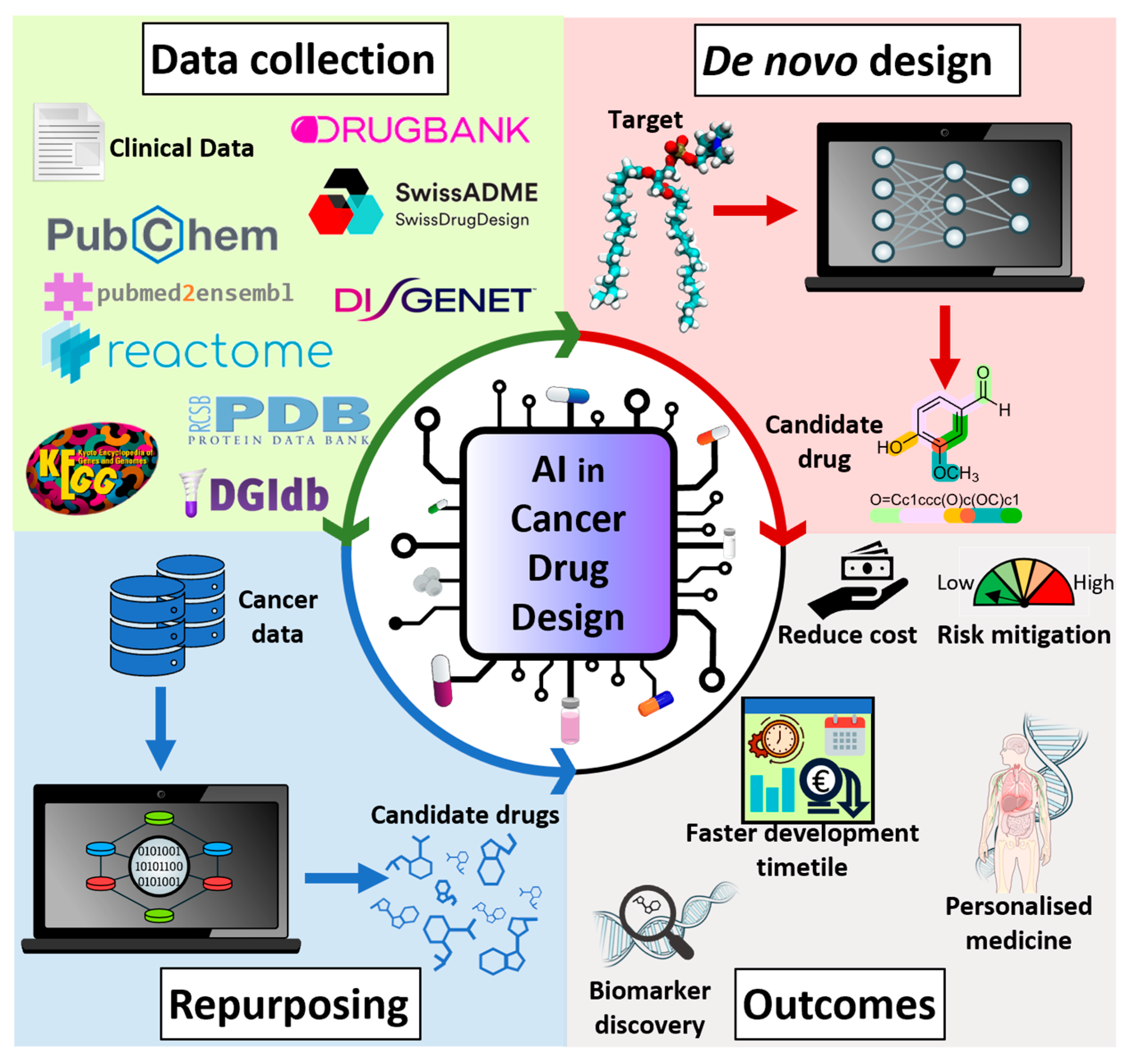
3. AI-Guided Applications in Cancer Drug Discovery
4. Integration of AI with Experimental Techniques
4.1. AI-Guided High-Throughput Screening (HTS)
4.2. AI-Assisted Drug Synthesis and Optimization
4.3. AI-Driven In Vitro and In Vivo Testing
4.4. AI for Data Integration in Preclinical Studies
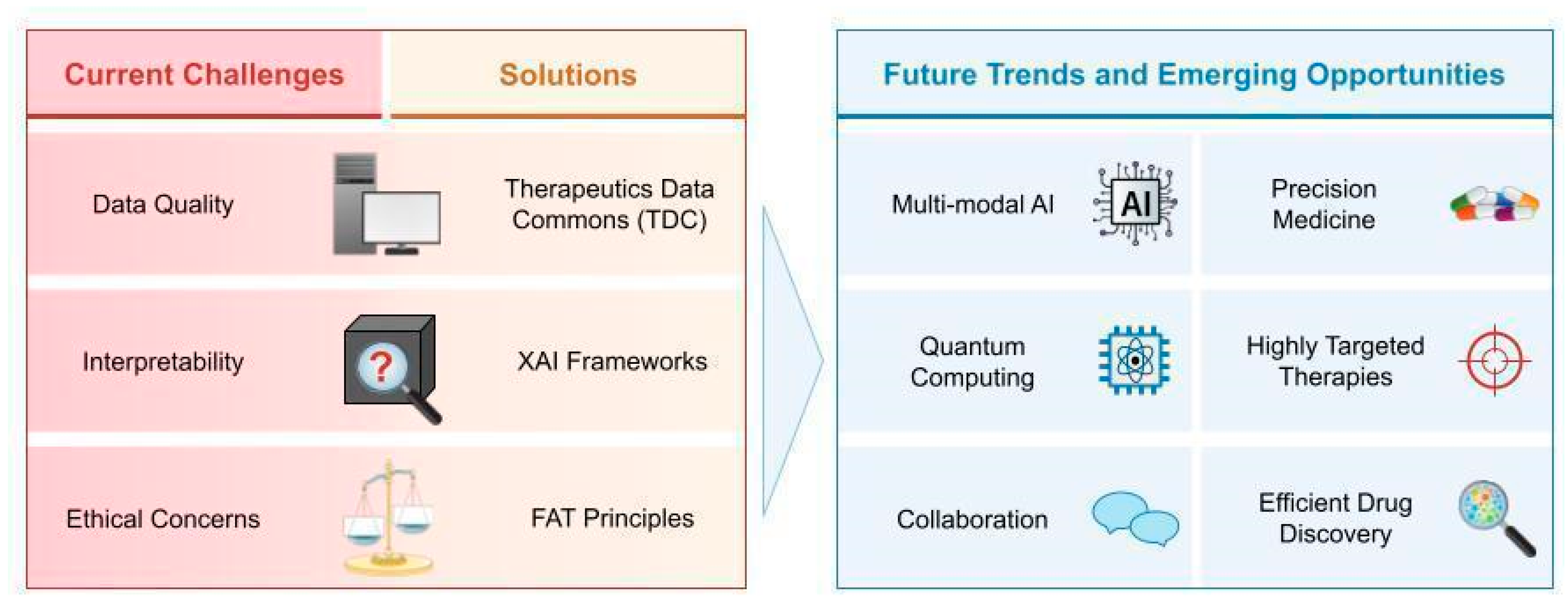
5. Challenges and Opportunities
5.1. Data Quality and Quantity
5.2. Interpretability of AI Models
5.3. Ethical Considerations
5.4. Collaboration Across Sectors, Future Trends, and Emerging Opportunities
Funding
Acknowledgments
Conflicts of Interest
References
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Garg, P.; Singhal, G.; Kulkarni, P.; Horne, D.; Salgia, R.; Singhal, S.S. Artificial Intelligence-Driven Computational Approaches in the Development of Anticancer Drugs. Cancers 2024, 16, 3884. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, Y.; Wang, H.; Zhang, X.; Wang, M.; He, J.; Li, S.; Zhang, L.; Li, K.; Cao, L. Advances of Artificial Intelligence in Anti-Cancer Drug Design: A Review of the Past Decade. Pharmaceuticals 2023, 16, 253. [Google Scholar] [CrossRef]
- Tanoli, Z.; Vaha-Koskela, M.; Aittokallio, T. Artificial intelligence, machine learning, and drug repurposing in cancer. Expert. Opin. Drug Discov. 2021, 16, 977–989. [Google Scholar] [CrossRef]
- Cai, Z.; Poulos, R.C.; Liu, J.; Zhong, Q. Machine learning for multi-omics data integration in cancer. iScience 2022, 25, 103798. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Pan, D.; Wang, X.; Xu, Y.; Yan, J.; Wang, L.; Yang, X.; Yang, M.; Liu, G.P. Applications of multi-omics analysis in human diseases. MedComm (2020) 2023, 4, e315. [Google Scholar] [CrossRef]
- Xu, X.; Yue, L.; Li, B.; Liu, Y.; Wang, Y.; Zhang, W.; Wang, L. DSGAT: Predicting frequencies of drug side effects by graph attention networks. Brief. Bioinform. 2022, 23, bbab586. [Google Scholar] [CrossRef]
- Tatonetti, N.P.; Liu, T.; Altman, R.B. Predicting drug side-effects by chemical systems biology. Genome Biol. 2009, 10, 238. [Google Scholar] [CrossRef]
- Cichonska, A.; Ravikumar, B.; Rahman, R. AI for targeted polypharmacology: The next frontier in drug discovery. Curr. Opin. Struct. Biol. 2024, 84, 102771. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baião, A.R.; Cai, Z.; Poulos, R.C.; Robinson, P.J.; Reddel, R.R.; Zhong, Q.; Vinga, S.; Gonçalves, E. A technical review of multi-omics data integration methods: From classical statistical to deep generative approaches. arXiv 2025, arXiv:2501.17729. [Google Scholar]
- He, X.; Liu, X.; Zuo, F.; Shi, H.; Jing, J. Artificial intelligence-based multi-omics analysis fuels cancer precision medicine. Semin. Cancer Biol. 2023, 88, 187–200. [Google Scholar] [CrossRef]
- Ozturk, H.; Ozgur, A.; Ozkirimli, E. DeepDTA: Deep drug-target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Ekundayo, N.F. Reinforcement learning in treatment pathway optimization: A case study in oncology. Int. J. Sci. Res. Arch. 2024, 13, 2187–2205. [Google Scholar] [CrossRef]
- Duan, F.; Duan, C.; Xu, H.; Zhao, X.; Sukhbaatar, O.; Gao, J.; Zhang, M.; Zhang, W.; Gu, Y. AI-driven drug discovery from natural products. Adv. Agrochem 2024, 3, 185–187. [Google Scholar] [CrossRef]
- Ahmed, F.; Soomro, A.M.; Chethikkattuveli Salih, A.R.; Samantasinghar, A.; Asif, A.; Kang, I.S.; Choi, K.H. A comprehensive review of artificial intelligence and network based approaches to drug repurposing in Covid-19. Biomed. Pharmacother. 2022, 153, 113350. [Google Scholar] [CrossRef]
- Bhatia, A.S.; Saggi, M.K.; Kais, S. Quantum Machine Learning Predicting ADME-Tox Properties in Drug Discovery. J. Chem. Inf. Model. 2023, 63, 6476–6486. [Google Scholar] [CrossRef]
- Qiu, Y.; Cheng, F. Artificial intelligence for drug discovery and development in Alzheimer’s disease. Curr. Opin. Struct. Biol. 2024, 85, 102776. [Google Scholar] [CrossRef]
- Kumar, V.; Krishna, S.; Siddiqi, M.I. Virtual screening strategies: Recent advances in the identification and design of anti-cancer agents. Methods 2015, 71, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Khan, F. 3D-QSAR studies on Maslinic acid analogs for Anticancer activity against Breast Cancer cell line MCF-7. Sci. Rep. 2017, 7, 6019. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef] [PubMed]
- Issa, N.T.; Stathias, V.; Schurer, S.; Dakshanamurthy, S. Machine and deep learning approaches for cancer drug repurposing. Semin. Cancer Biol. 2021, 68, 132–142. [Google Scholar] [CrossRef]
- Somolinos, F.J.; León, C.; Guerrero-Aspizua, S. Drug Repurposing Using Biological Networks. Processes 2021, 9, 1057. [Google Scholar] [CrossRef]
- Zeng, X.; Zhu, S.; Liu, X.; Zhou, Y.; Nussinov, R.; Cheng, F. deepDR: A network-based deep learning approach to in silico drug repositioning. Bioinformatics 2019, 35, 5191–5198. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mao, C.; Hou, Y.; Luo, Y.; Binder, J.L.; Zhou, Y.; Bekris, L.M.; Shin, J.; Hu, M.; Wang, F.; et al. Interpretable deep learning translation of GWAS and multi-omics findings to identify pathobiology and drug repurposing in Alzheimer’s disease. Cell Rep. 2022, 41, 111717. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Pal, S.; Islam, M.A. Generative AI in drug discovery and development: The next revolution of drug discovery and development would be directed by generative AI. Ann. Med. Surg. 2024, 86, 6340–6343. [Google Scholar] [CrossRef]
- Mesko, B. The ChatGPT (Generative Artificial Intelligence) Revolution Has Made Artificial Intelligence Approachable for Medical Professionals. J. Med. Internet Res. 2023, 25, e48392. [Google Scholar] [CrossRef]
- Kyro, G.W.; Morgunov, A.; Brent, R.I.; Batista, V.S. ChemSpaceAL: An Efficient Active Learning Methodology Applied to Protein-Specific Molecular Generation. arXiv 2023, arXiv:2309.05853. [Google Scholar]
- Lu, H.; Wei, Z.; Wang, X.; Zhang, K.; Liu, H. GraphGPT: A Graph Enhanced Generative Pretrained Transformer for Conditioned Molecular Generation. Int. J. Mol. Sci. 2023, 24, 16761. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, C.; Han, P.; Li, X.; Chen, W.; Rodriguez Paton, A.; Wang, S.; Zheng, P. PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning. Int. J. Mol. Sci. 2023, 24, 1146. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, R.; Zhang, L.; Xie, P. DrugChat: Towards Enabling ChatGPT-Like Capabilities on Drug Molecule Graphs. arXiv 2023, arXiv:2309.03907. [Google Scholar]
- Popova, M.; Isayev, O.; Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018, 4, eaap7885. [Google Scholar] [CrossRef] [PubMed]
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in de Novo Drug Design: From Conventional to Machine Learning Methods. Int. J. Mol. Sci. 2021, 22, 1676. [Google Scholar] [CrossRef] [PubMed]
- Olivecrona, M.; Blaschke, T.; Engkvist, O.; Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminform 2017, 9, 48. [Google Scholar] [CrossRef]
- Li, J.; Shi, M.; Chen, Z.; Pan, Y. DeepPurpose-based drug discovery in chondrosarcoma. Chin. J. Plast. Reconstr. Surg. 2022, 4, 158–165. [Google Scholar] [CrossRef]
- Esteban-Medina, M.; de la Oliva Roque, V.M.; Herraiz-Gil, S.; Pena-Chilet, M.; Dopazo, J.; Loucera, C. drexml: A command line tool and Python package for drug repurposing. Comput. Struct. Biotechnol. J. 2024, 23, 1129–1143. [Google Scholar] [CrossRef]
- Huang, Y.; Dong, D.; Zhang, W.; Wang, R.; Lin, Y.C.; Zuo, H.; Huang, H.Y.; Huang, H.D. DrugRepoBank: A comprehensive database and discovery platform for accelerating drug repositioning. Database 2024, 2024, baae051. [Google Scholar] [CrossRef]
- Wu, J.; Li, X.; Wang, Q.; Han, J. DRviaSPCN: A software package for drug repurposing in cancer via a subpathway crosstalk network. Bioinformatics 2022, 38, 4975–4977. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, J.; He, Y.; Huang, J.; Zhao, X.; Pan, B.; Wang, Y.; Cheng, L.; Han, J. DrugSim2DR: Systematic prediction of drug functional similarities in the context of specific disease for drug repurposing. Gigascience 2022, 12, giad104. [Google Scholar] [CrossRef]
- Ren, F.; Ding, X.; Zheng, M.; Korzinkin, M.; Cai, X.; Zhu, W.; Mantsyzov, A.; Aliper, A.; Aladinskiy, V.; Cao, Z.; et al. AlphaFold accelerates artificial intelligence powered drug discovery: Efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 2023, 14, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Born, J.; Manica, M.; Oskooei, A.; Cadow, J.; Markert, G.; Rodriguez Martinez, M. PaccMann(RL): De novo generation of hit-like anticancer molecules from transcriptomic data via reinforcement learning. iScience 2021, 24, 102269. [Google Scholar] [CrossRef] [PubMed]
- Grisoni, F.; Neuhaus, C.S.; Hishinuma, M.; Gabernet, G.; Hiss, J.A.; Kotera, M.; Schneider, G. De novo design of anticancer peptides by ensemble artificial neural networks. J. Mol. Model. 2019, 25, 112. [Google Scholar] [CrossRef]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef]
- Krenn, M.; Pollice, R.; Guo, S.Y.; Aldeghi, M.; Cervera-Lierta, A.; Friederich, P.; Dos Passos Gomes, G.; Hase, F.; Jinich, A.; Nigam, A.; et al. On scientific understanding with artificial intelligence. Nat. Rev. Phys. 2022, 4, 761–769. [Google Scholar] [CrossRef]
- Kadurin, A.; Nikolenko, S.; Khrabrov, K.; Aliper, A.; Zhavoronkov, A. druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico. Mol. Pharm. 2017, 14, 3098–3104. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Bonner, S.; Barrett, I.P.; Ye, C.; Swiers, R.; Engkvist, O.; Bender, A.; Hoyt, C.T.; Hamilton, W.L. A review of biomedical datasets relating to drug discovery: A knowledge graph perspective. Brief. Bioinform. 2022, 23, bbac404. [Google Scholar] [CrossRef]
- Kirboga, K.K.; Abbasi, S.; Kucuksille, E.U. Explainability and white box in drug discovery. Chem. Biol. Drug Des. 2023, 102, 217–233. [Google Scholar] [CrossRef]
- Ladbury, C.; Zarinshenas, R.; Semwal, H.; Tam, A.; Vaidehi, N.; Rodin, A.S.; Liu, A.; Glaser, S.; Salgia, R.; Amini, A. Utilization of model-agnostic explainable artificial intelligence frameworks in oncology: A narrative review. Transl. Cancer Res. 2022, 11, 3853–3868. [Google Scholar] [CrossRef] [PubMed]
- Laganà, F.; Prattico, D.; De Carlo, D.; Oliva, G.; Pullano, S.A.; Calcagno, S. Engineering Biomedical Problems to Detect Carcinomas: A Tomographic Impedance Approach. Eng 2024, 5, 1594–1614. [Google Scholar] [CrossRef]
- Salih, A.M.; Raisi-Estabragh, Z.; Galazzo, I.B.; Radeva, P.; Petersen, S.E.; Lekadir, K.; Menegaz, G. A perspective on explainable artificial intelligence methods: SHAP and LIME. Adv. Intell. Syst. 2025, 7, 2400304. [Google Scholar] [CrossRef]
- Fisher, J.L.; Jones, E.F.; Flanary, V.L.; Williams, A.S.; Ramsey, E.J.; Lasseigne, B.N. Considerations and challenges for sex-aware drug repurposing. Biol. Sex. Differ. 2022, 13, 13. [Google Scholar] [CrossRef]
- Parra-Calderon, C.L.; Sanz, F.; McIntosh, L.D. The Challenge of the Effective Implementation of FAIR Principles in Biomedical Research. Methods Inf. Med. 2020, 59, 117–118. [Google Scholar] [CrossRef]
- FAIR4Health Project. Available online: https://www.fair4health.eu/en/project (accessed on 27 February 2025).
- Warraich, H.J.; Tazbaz, T.; Califf, R.M. FDA Perspective on the Regulation of Artificial Intelligence in Health Care and Biomedicine. JAMA 2025, 333, 241–247. [Google Scholar] [CrossRef]
- Artificial intelligence, EMA. Available online: https://www.ema.europa.eu/en/about-us/how-we-work/big-data/artificial-intelligence (accessed on 27 February 2025).
- Nene, L.; Flepisi, B.T.; Brand, S.J.; Basson, C.; Balmith, M. Evolution of Drug Development and Regulatory Affairs: The Demonstrated Power of Artificial Intelligence. Clin. Ther. 2024, 46, e6–e14. [Google Scholar] [CrossRef]
- Uema, R.; Hayashi, Y.; Kizu, T.; Igura, T.; Ogiyama, H.; Yamada, T.; Takeda, R.; Nagai, K.; Inoue, T.; Yamamoto, M.; et al. A novel artificial intelligence-based endoscopic ultrasonography diagnostic system for diagnosing the invasion depth of early gastric cancer. J. Gastroenterol. 2024, 59, 543–555. [Google Scholar] [CrossRef]
- Chang, Y.J.; Hung, K.C.; Wang, L.K.; Yu, C.H.; Chen, C.K.; Tay, H.T.; Wang, J.J.; Liu, C.F. A Real-Time Artificial Intelligence-Assisted System to Predict Weaning from Ventilator Immediately after Lung Resection Surgery. Int. J. Environ. Res. Public Health 2021, 18, 2713. [Google Scholar] [CrossRef]
- Feng, J.; Phillips, R.V.; Malenica, I.; Bishara, A.; Hubbard, A.E.; Celi, L.A.; Pirracchio, R. Clinical artificial intelligence quality improvement: Towards continual monitoring and updating of AI algorithms in healthcare. npj Digit. Med. 2022, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Biamonte, J.; Wittek, P.; Pancotti, N.; Rebentrost, P.; Wiebe, N.; Lloyd, S. Quantum machine learning. Nature 2017, 549, 195–202. [Google Scholar] [CrossRef] [PubMed]
| Case Study | Computational Approach | Model/ Algorithm | Relevant Results | |
|---|---|---|---|---|
| AI drug repurposing | Chondrosarcoma (CS) [38] | Knowledge + network-based methods 1. Genetic data of disease: pubmed2ensembl. 2. Drug–gene interaction: Drug Gene Interaction Database (DGIdb). 3. Drug–target information: DeepPurpose. | Deep learning-based algorithm | A total of 25 candidate drugs were identified. Among the listed drugs, there are drugs that have been approved for various solid tumors and have been applied to patients with CS: everolimus, paclitaxel, sirolimus, 2-methoxyestradiol, and sunitinib. |
| Familiar Melanoma [39] | Knowledge + network-based methods 1. Genetic data of disease: databases + disease knowledge. 2. Disease Mechanistic Map: HiPathia + Genotype-Tissue Expression Project. 3. Drug–target information: Drexml. | Explainable machine learning model | A total of 78 candidate drugs correspond to currently approved chemotherapeutic agents used to treat various types of cancer. Paclitaxel, docetaxel, moxetumomab, and ruxolitinib are drugs that target specific melanogenesis circuits. | |
| Liver and lung cancers [40] | Similarity-based, artificial intelligence-based, signature-based, and network-based methods 1. Integrating heterogeneous data (drugs, targets, diseases, side effects and pathways) from databases and the literature. 2. Drug–target information: DrugRepoBank. | Artificial intelligence model | AI-predicted a CYP3A4 target for sildenafil repositioning in the treatment of liver cancer. The drug candidate verteporfin may influence lung cancer by modulating the Hippo signaling pathway and insulin secretion. | |
| Breast cancer [41] | Network-based method 1. Genetic data of disease: breast cancer gene expression profiles from GEO database. 2. Drug–disease interaction: DRviaSPCN. | Random walk with restart algorithm | Four of ten candidate drugs have been demonstrated to be associated with breast cancer: azacitidine, valproic acid, doxorubicin, and exemestane. | |
| Breast and lung cancers [42] | Network-based method 1. Genetic data of disease: breast cancer and lung cancer gene expression profiles from GEO database. 2. Drug–disease interaction: DrugSim2DR. | Random walk with restart algorithm | Five potential anti-breast cancer drugs were identified: fluoxymesterone, gestrinone, pyrazole, fomepizole, and medroxyprogesterone acetate. Fluoxymesterone has received approval for breast cancer treatment. Of nine candidate drugs, methotrexate and pemetrexed have been approved for the treatment of lung cancer. | |
| AI de novo drug design | Hepatocellular carcinoma [43] | Structure-based drug design of novel targets 1. Target selection: PandaOmics. 2. Determination of putative binding sites: Chemistry42. 3. Generation of novel hits targeting CDK20 inhibitor: AlphaFold. | Deep learning-based algorithm | A novel therapeutic target was identified from a pool of dark targets (without experimental structure) that were predicted using AlphaFold (v.2.3.0). ISM042-2-048 generated a compound that showed good CDK20 inhibitory activity. |
| Carcinoma and neuroblastoma [44] | Reinforcement learning approach 1.Genetic data of disease: carcinoma and neuroblastoma gene expression profiles. 2. Generation of anticancer hit molecules: PaccMannRL. | Deep learning-based algorithm | The generated compounds exhibited similar physicochemical properties to real cancer drugs. | |
| Breast and lung cancers [45] | Counter-propagation artificial neural networks (CPANNs) 1. Two peptide datasets targeting breast and lung cancer cells were assembled and curated manually from CancerPPD. 2. Training CPANN model to classify peptides according to their activity. 3. Library class generation with 1000 presumed alpha-helical peptides sequences with the amino acid distribution of alpha-helical anticancer peptides (ACPs): modlAMP. 4. Evaluation and ranking of the activity of de novo designed peptides from the library: CPANNs. 5. Selection of candidate peptides with anticancer activity to in vitro assays. | Deep learning-based algorithm | From a total of 1000 de novo designs, 6 peptides showed anticancer activity in vitro, including 5 against both MCF7 and A549 cell lines. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herráiz-Gil, S.; Nygren-Jiménez, E.; Acosta-Alonso, D.N.; León, C.; Guerrero-Aspizua, S. Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer. Appl. Sci. 2025, 15, 2798. https://doi.org/10.3390/app15052798
Herráiz-Gil S, Nygren-Jiménez E, Acosta-Alonso DN, León C, Guerrero-Aspizua S. Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer. Applied Sciences. 2025; 15(5):2798. https://doi.org/10.3390/app15052798
Chicago/Turabian StyleHerráiz-Gil, Sara, Elisa Nygren-Jiménez, Diana N. Acosta-Alonso, Carlos León, and Sara Guerrero-Aspizua. 2025. "Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer" Applied Sciences 15, no. 5: 2798. https://doi.org/10.3390/app15052798
APA StyleHerráiz-Gil, S., Nygren-Jiménez, E., Acosta-Alonso, D. N., León, C., & Guerrero-Aspizua, S. (2025). Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer. Applied Sciences, 15(5), 2798. https://doi.org/10.3390/app15052798








