Vacuum Ultraviolet Single-Photon Postionization of Amino Acids
Abstract
:1. Introduction
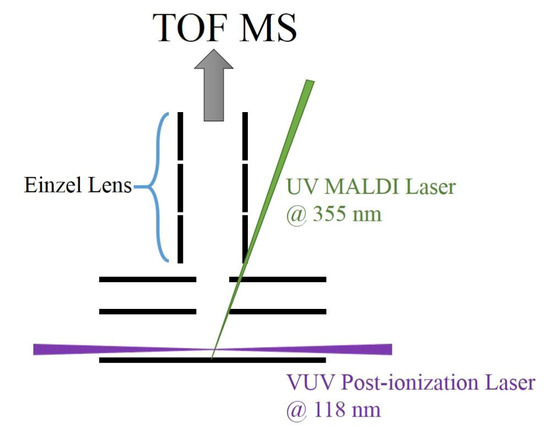
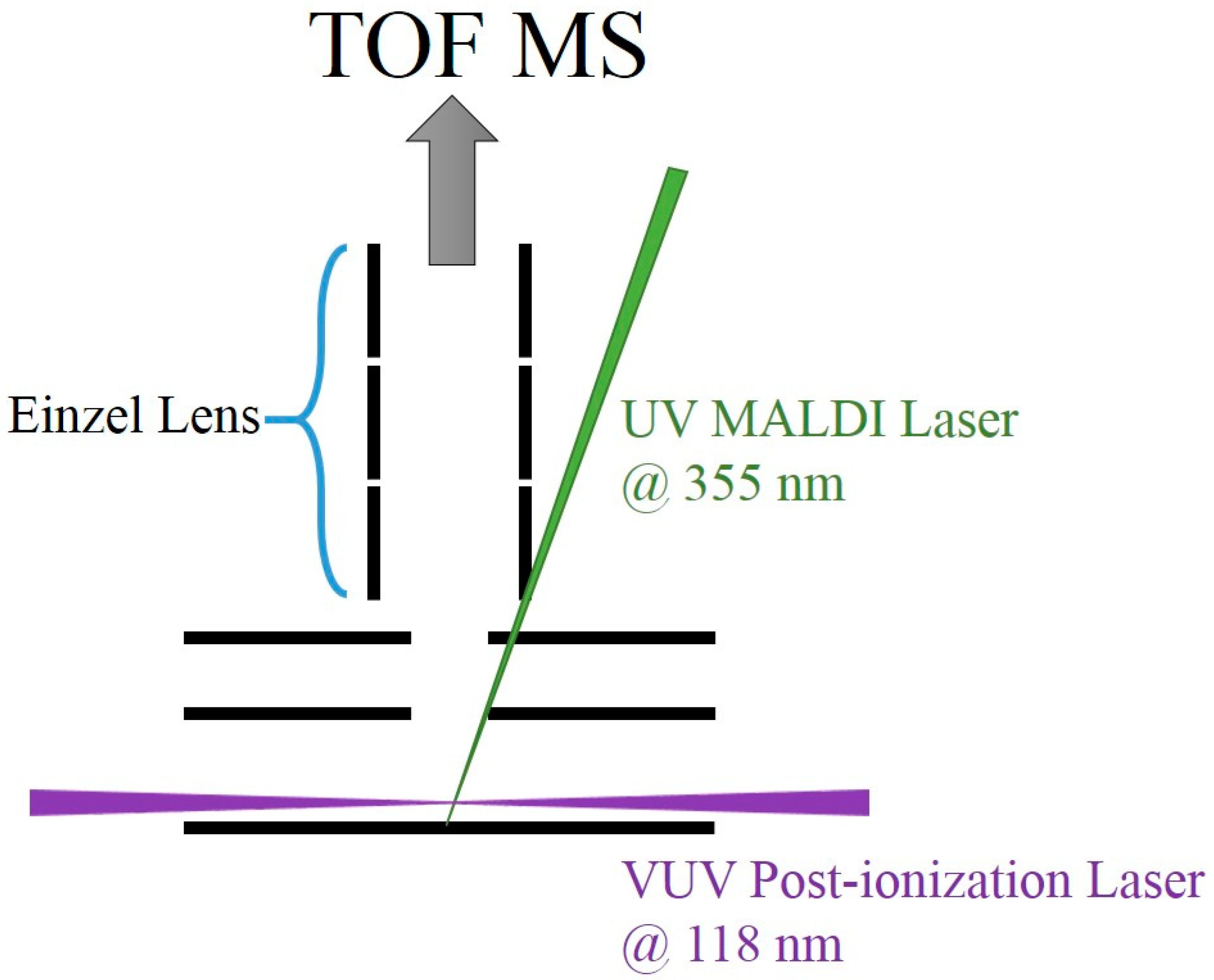
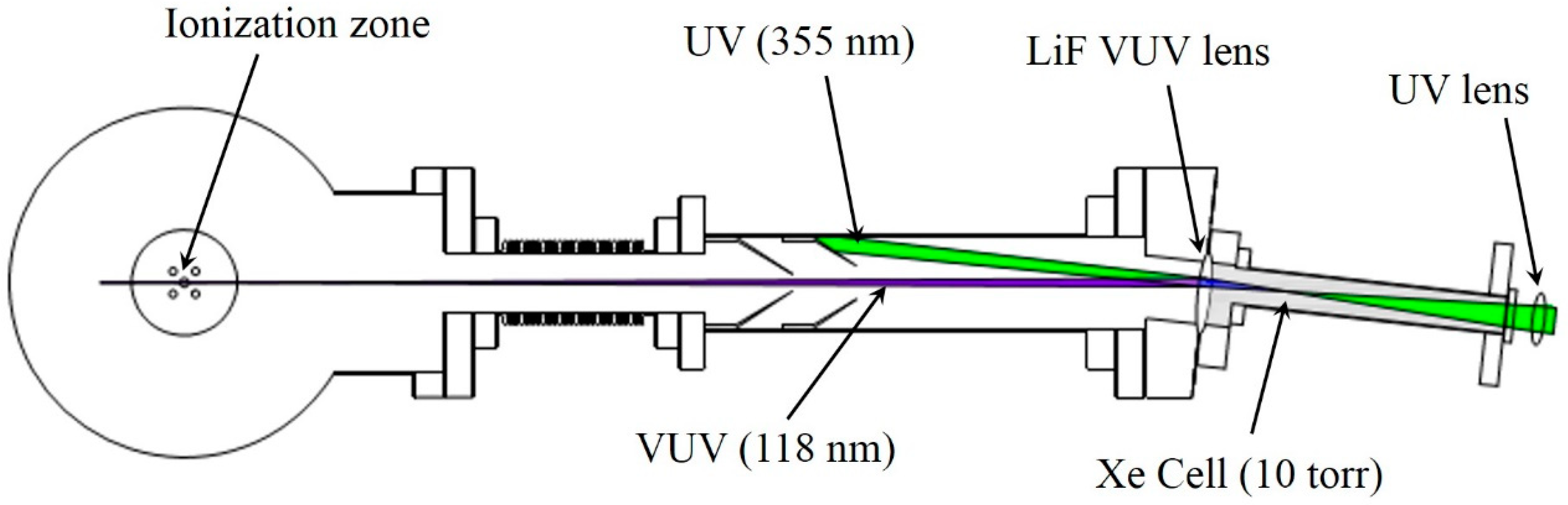
2. Experiment
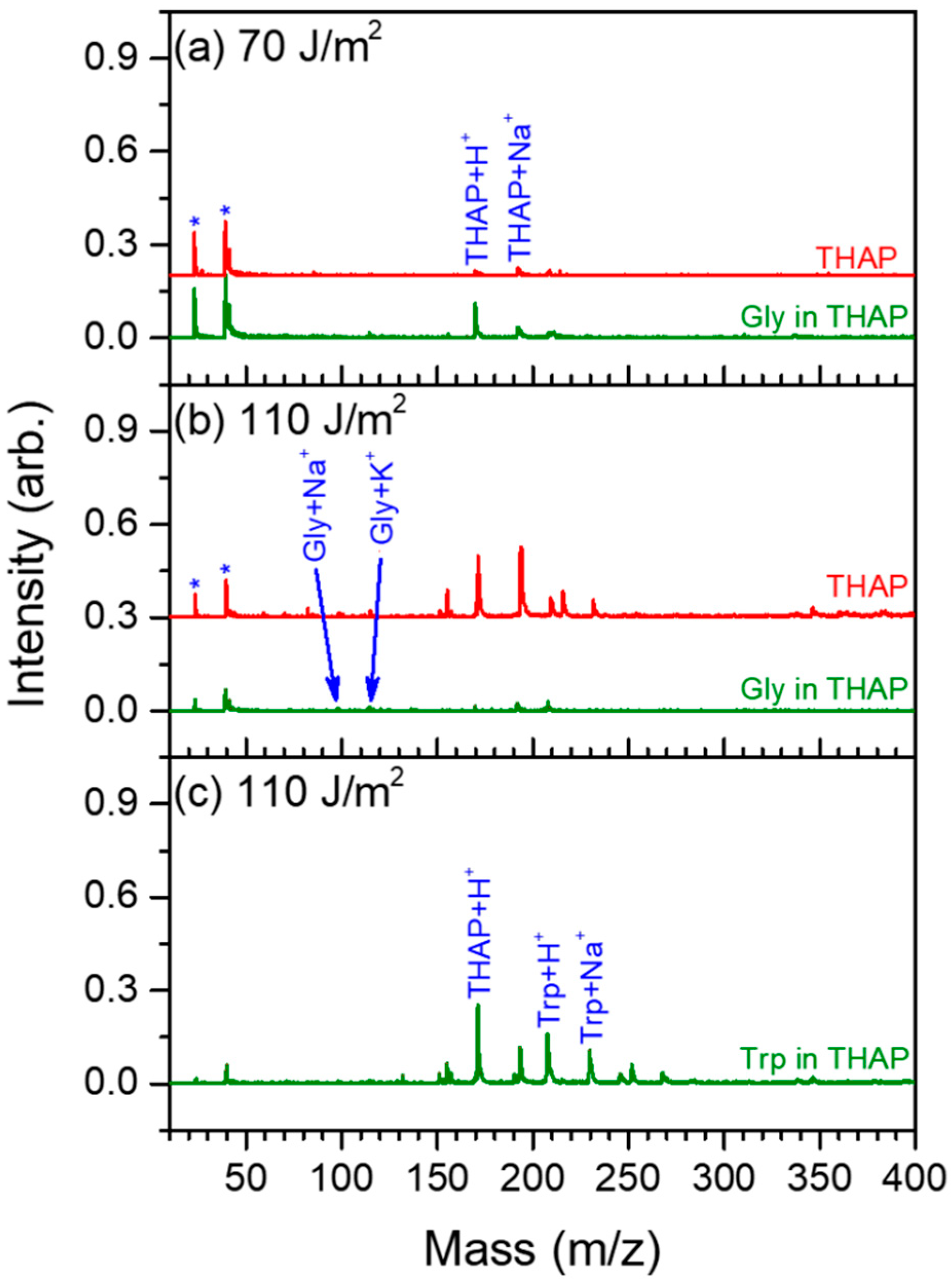
3. Results
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Karas, M.; Bachmann, D.; Bahr, U.; Hillenkamp, F. Matrix-assisted ultraviolet-laser desorption of nonvolatile compounds. Int. J. Mass Spectrom. 1987, 78, 53–68. [Google Scholar] [CrossRef]
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Huang, L.C.L.; Wang, Y.S.; Peng, W.P.; Chang, H.C.; Hsu, N.Y.; Yang, W.B.; Chen, C.H. Matrix-assisted laser desorption/ionization (MALDI) mechanism revisited. Anal. Chim. Acta 2007, 582, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Taranenko, N.I.; Golovlev, V.V.; Puretzky, A.A.; Allman, S.L.; Chen, C.H. Direct monitoring of laser-absorption of maldi matrices by fast piezoelectric transducer. Chem. Phys. Lett. 1995, 234, 165–171. [Google Scholar] [CrossRef]
- Gluckmann, M.; Pfenninger, A.; Kruger, R.; Thierolf, M.; Karas, M.; Horneffer, V.; Hillenkamp, F.; Strupat, K. Mechanisms in MALDI analysis: Surface interaction or incorporation of analytes? Int. J. Mass Spectrom. 2001, 210, 121–132. [Google Scholar] [CrossRef]
- Horneffer, V.; Dreisewerd, K.; Ludemann, H.C.; Hillenkamp, F.; Lage, M.; Strupat, K. Is the incorporation of analytes into matrix crystals a prerequisite for matrix-assisted laser desorption/ionization mass spectrometry? A study of five positional isomers of dihydroxybenzoic acid. Int. J. Mass Spectrom. 1999, 185, 859–870. [Google Scholar] [CrossRef]
- Horneffer, V.; Reichelt, R.; Strupat, K. Protein incorporation into MALDI-matrix crystals investigated by high resolution field emission scanning electron microscopy. Int. J. Mass Spectrom. 2003, 226, 117–131. [Google Scholar] [CrossRef]
- Strupat, K.; Kampmeier, J.; Horneffer, V. Investigations of 2,5-DHB and succinic acid as matrices for UV and IR MALDI. Part II: Crystallographic and mass spectrometric analysis. Int. J. Mass Spectrom. 1997, 169, 43–50. [Google Scholar] [CrossRef]
- Trimpin, S.; Rader, H.J.; Mullen, K. Investigations of theoretical principles for MALDI-MS derived from solvent-free sample preparation—Part I. Preorganization. Int. J. Mass Spectrom. 2006, 253, 13–21. [Google Scholar] [CrossRef]
- Jessome, L.; Hsu, N.Y.; Wang, Y.S.; Chen, C.H. Matrix-assisted laser desorption/ionization mechanism study with dihydroxybenzoic acid isomers as matrices. Rapid Commun. Mass Spectrom. 2008, 22, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Price, D.M.; Bashir, S.; Derrick, P.R. Sublimation properties of x,y-dihydroxybenzoic acid isomers as model matrix assisted laser desorption ionisation (MALDI) matrices. Thermochim. Acta 1999, 327, 167–171. [Google Scholar] [CrossRef]
- Liang, C.W.; Lee, C.H.; Lin, Y.J.; Lee, Y.T.; Ni, C.K. MALDI mechanism of dihydroxybenzoic acid isomers: Desorption of neutral matrix and analyte. J. Phys. Chem. B 2013, 117, 5058–5064. [Google Scholar] [CrossRef] [PubMed]
- Wiegelmann, M.; Soltwisch, J.; Jaskolla, T.W.; Dreisewerd, K. Matching the laser wavelength to the absorption properties of matrices increases the ion yield in UV-MALDI mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 6925–6932. [Google Scholar] [CrossRef] [PubMed]
- Dreisewerd, K.; Schurenberg, M.; Karas, M.; Hillenkamp, F. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix-assisted laser-desorption ionization with a uniform beam profile. Int. J. Mass Spectrom. 1995, 141, 127–148. [Google Scholar] [CrossRef]
- Westmacott, G.; Ens, W.; Hillenkamp, F.; Dreisewerd, K.; Schurenberg, M. The influence of laser fluence on ion yield in matrix-assisted laser desorption ionization mass spectrometry. Int. J. Mass Spectrom. 2002, 221, 67–81. [Google Scholar] [CrossRef]
- Dreisewerd, K.; Schurenberg, M.; Karas, M.; Hillenkamp, F. Matrix-assisted laser desorption/ionization with nitrogen lasers of different pulse widths. Int. J. Mass Spectrom. 1996, 154, 171–178. [Google Scholar] [CrossRef]
- Menzel, C.; Dreisewerd, K.; Berkenkamp, S.; Hillenkamp, F. The role of the laser pulse duration in infrared matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2002, 13, 975–984. [Google Scholar] [CrossRef]
- Soltwisch, J.; Dreisewerd, K. An ultraviolet/infrared matrix-assisted laser desorption ionization sample stage integrating scanning knife-edge and slit devices for laser beam analysis. Rapid Commun. Mass Spectrom. 2011, 25, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Beavis, R.C.; Chaudhary, T.; Chait, B.T. Alpha-cyano-4-hydroxycinnamic acid as a matrix for matrix-assisted laser desorption mass-spectrometry. Org. Mass Spectrom. 1992, 27, 156–158. [Google Scholar] [CrossRef]
- Strupat, K.; Karas, M.; Hillenkamp, F. 2,5-dihydroxybenzoic acid—A new matrix for laser desorption ionization mass-spectrometry. Int. J. Mass Spectrom. 1991, 111, 89–102. [Google Scholar] [CrossRef]
- Salum, M.L.; Itovich, L.M.; Erra-Balsells, R. Z-sinapinic acid: The change of the stereochemistry of cinnamic acids as rational synthesis of a new matrix for carbohydrate MALDI-MS analysis. J. Mass Spectrom. 2013, 48, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Jaskolla, T.W.; Onischke, K.; Schiller, J. 2,5-dihydroxybenzoic acid salts for matrix-assisted laser desorption/ionization time-of-flight mass spectrometric lipid analysis: Simplified spectra interpretation and insights into gasphase fragmentation. Rapid Commun. Mass Spectrom. 2014, 28, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Stahl, B.; Steup, M.; Karas, M.; Hillenkamp, F. Analysis of neutral oligosaccharides by matrix-assisted laser desorption—Ionization mass-spectrometry. Anal. Chem. 1991, 63, 1463–1466. [Google Scholar] [CrossRef]
- Stubiger, G.; Belgacem, O. Analysis of lipids using 2,4,6-trihydroxyacetophenone as a matrix for MALDI mass spectrometry. Anal. Chem. 2007, 79, 3206–3213. [Google Scholar] [CrossRef] [PubMed]
- Stikarovska, M.; Chmelik, J. Determination of neutral oligosaccharides in vegetables by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chim. Acta 2004, 520, 47–55. [Google Scholar] [CrossRef]
- Papac, D.I.; Wong, A.; Jones, A.J.S. Analysis of acidic oligosaccharides and glycopeptides by matrix assisted laser desorption ionization time-of-flight mass spectrometry. Anal. Chem. 1996, 68, 3215–3223. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom. Rev. 1999, 18, 349–450. [Google Scholar] [CrossRef]
- Hsu, N.Y.; Yang, W.B.; Wong, C.H.; Lee, Y.C.; Lee, R.T.; Wang, Y.S.; Chen, C.H. Matrix-assisted laser desorption/ionization mass spectrometry of polysaccharides with 2′,4′,6′-trihydroxyacetophenone as matrix. Rapid Commun. Mass Spectrom. 2007, 21, 2137–2146. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.F.; Wu, H.P.; Kobayashi, T.; Solaro, R.J.; van Breemen, R.B. Enhanced ionization of phosphorylated peptides during MALDI TOF mass spectrometry. Anal. Chem. 2004, 76, 1532–1536. [Google Scholar] [CrossRef] [PubMed]
- Ueki, M.; Yamaguchi, M. Enhanced detection of sulfo-peptides as onium salts in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J.; Shaler, T.A.; Becker, C.H. Time-of-flight mass-spectrometry of underivatized single-stranded-DNA oligomers by matrix-assisted laser-desorption. Anal. Chem. 1994, 66, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J.; Steding, A.; Becker, C.H. Matrix-assisted laser desorption time-of-flight mass-spectrometry of oligonucleotides using 3-hydroxypicolinic acid as an ultraviolet-sensitive matrix. Rapid Commun. Mass Spectrom. 1993, 7, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Jaskolla, T.W.; Papasotiriou, D.G.; Karas, M. Comparison between the matrices alpha-cyano-4-hydroxycinnamic acid and 4-chloro-alpha-cyanocinnamic acid for trypsin, chymotrypsin, and pepsin digestions by MALDI-TOF mass spectrometry. J. Proteome Res. 2009, 8, 3588–3597. [Google Scholar] [CrossRef] [PubMed]
- Jaskolla, T.; Fuchs, B.; Karas, M.; Schiller, J. The new matrix 4-chloro-alpha-cyanocinnamic acid allows the detection of phosphatidylethanolamine chloramines by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2009, 20, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Jaskolla, T.W.; Lehmann, W.D.; Karas, M. 4-chloro-alpha-cyanocinnamic acid is an advanced, rationally designed maldi matrix. Proc. Natl. Acad. Sci. USA 2008, 105, 12200–12205. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.T.; Lee, S.; Lu, I.C.; Chu, K.Y.; Liang, C.W.; Lee, C.H.; Lee, Y.T.; Ni, C.K. Ion-to-neutral ratio of 2,5-dihydroxybenzoic acid in matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 2013, 27, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Lu, I.C.; Lee, C.; Lee, Y.T.; Ni, C.K. Ionization mechanism of matrix-assisted laser desorption/ionization. Annu. Rev. Anal. Chem. 2015, 8, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Silina, Y.E.; Kochb, M.; Volmer, D.A. Impact of analyte ablation and surface acidity of Pd nanoparticles onefficiency of surface-assisted laser desorption/ionization-mass spectrometry. Int. J. Mass Spectrom. 2015, 387, 24–30. [Google Scholar] [CrossRef]
- Spencer, M.K.; Hammond, M.R.; Zare, R.N. Laser mass spectrometric detection of extraterrestrial aromatic molecules: Mini-review and examination of pulsed heating effects. Proc. Natl. Acad. Sci. USA 2008, 105, 18096–18101. [Google Scholar] [CrossRef] [PubMed]
- Kinsel, G.R.; Lindner, J.; Grotemeyer, J. Peptide sequence ions produced by postionization of neutral molecules formed during resonant 266-nm laser desorption. J. Phys. Chem. 1992, 96, 3157–3162. [Google Scholar] [CrossRef]
- Kinsel, G.R.; Lindner, J.; Grotemeyer, J. Investigations of neutral fragment formation during resonant 266-nm laser desorption. J. Phys. Chem. 1992, 96, 3162–3166. [Google Scholar] [CrossRef]
- Reilly, P.T.A.; Reilly, J.P. Laser-desorption gas-phase ionization of the amino-acid tryptophan. Rapid Commun. Mass Spectrom. 1994, 8, 731–734. [Google Scholar] [CrossRef]
- Leisner, A.; Rohlfing, A.; Berkenkamp, S.; Hillenkamp, F.; Dreisewerd, K. Infrared laser post-ionization of large biomolecules from an IR-MALDI(I) plume. J. Am. Soc. Mass Spectrom. 2004, 15, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Edirisinghe, P.D.; Moore, J.F.; Calaway, W.F.; Veryovkin, I.V.; Pellin, M.J.; Hanley, L. Vacuum ultraviolet postionization of aromatic groups covalently bound to peptides. Anal. Chem. 2006, 78, 5876–5883. [Google Scholar] [CrossRef] [PubMed]
- Gasper, G.L.; Takahashi, L.K.; Zhou, J.; Ahmed, M.; Moore, J.F.; Hanley, L. Laser desorption postionization mass spectrometry of antibiotic-treated bacterial biofilms using tunable vacuum ultraviolet radiation. Anal. Chem. 2010, 82, 7472–7478. [Google Scholar] [CrossRef] [PubMed]
- Gasper, G.L.; Carlson, R.; Akhmetov, A.; Moore, J.F.; Hanley, L. Laser desorption 7.87 EV postionization mass spectrometry of antibiotics in staphylococcus epidermidis bacterial biofilms. Proteomics 2008, 8, 3816–3821. [Google Scholar] [CrossRef] [PubMed]
- Vanbramer, S.E.; Johnston, M.V. Tunable, coherent vacuum ultraviolet-radiation for photoionization mass-spectrometry. Appl. Spectrosc. 1992, 46, 255–261. [Google Scholar] [CrossRef]
- Edirisinghe, P.D.; Lateef, S.S.; Crot, C.A.; Hanley, L.; Pellin, M.J.; Calaway, W.F.; Moore, J.F. Derivatization of surface-bound peptides for mass spectrometric detection via threshold single photon ionization. Anal. Chem. 2004, 76, 4267–4270. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.S.; Wu, C.P.; Akhmetov, A.; Edirisinghe, P.D.; Drummond, J.L.; Hanley, L. 7.87 EV laser desorption postionization mass spectrometry of adsorbed and covalently bound bisphenol a diglycidyl methacrylate. J. Am. Soc. Mass Spectrom. 2007, 18, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Kung, A.H. 3rd-harmonic generation in a pulsed supersonic jet of xenon. Opt. Lett. 1983, 8, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Kung, A.H.; Young, J.F.; Harris, S.E. Generation of 1182-Å radiation in phase-matched mixtures of inert-gases. Appl. Phys. Lett. 1973, 22, 301–302. [Google Scholar] [CrossRef]
- Ni, C.K.; Lee, Y.T. Photodissociation of simple aromatic molecules in a molecular beam. Int. Rev. Phys. Chem. 2004, 23, 187–218. [Google Scholar] [CrossRef]
- Ni, C.K.; Tseng, C.M.; Lin, M.F.; Dyakov, Y. Photodissociation dynamics of small aromatic molecules studied by multimass ion imaging. J. Phys. Chem. B 2007, 111, 12631–12642. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Guan, J.W.; Bernstein, E.R. Mass-selected IR-VUV (118 nm) spectroscopic studies of radicals, aliphatic molecules, and their clusters. Mass Spectrom. Rev. 2013, 32, 484–501. [Google Scholar] [CrossRef] [PubMed]
- Lockyer, N.P.; Vickerman, J.C. Single photon ionisation mass spectrometry using laser-generated vacuum ultraviolet photons. Laser Chem. 1997, 17, 139–159. [Google Scholar] [CrossRef]
- Shin, J.-W.; Bernstein, E.R. Vacuum ultraviolet photoionization of carbohydrates and nucleotides. J. Chem. Phys. 2014, 140, 044330. [Google Scholar] [CrossRef] [PubMed]
- Hunter, E.P.L.; Lias, S.G. Evaluated gas phase basicities and proton affinities of molecules: An update. J. Phys. Chem. Ref. Data 1998, 27, 413–656. [Google Scholar] [CrossRef]
- Ni, C.K.; Huang, J.D.; Chen, Y.T.; Kung, A.H.; Jackson, W.M. Photodissociation of propyne and allene at 193 nm with VUV detection of the products. J. Chem. Phys. 1999, 110, 3320–3325. [Google Scholar] [CrossRef]
- Tsai, S.T.; Lin, C.K.; Lee, Y.T.; Ni, C.K. Multimass Ion Imaging Detection: Application to Photodissociation. Rev. Sci. Instrum. 2001, 72, 1963–1969. [Google Scholar] [CrossRef]
- Hsu, H.C.; Tsai, M.T.; Dyakov, Y.D.; Ni, C.K. Energy transfer of highly vibrationally excited molecules studied by crossed molecular beam/time-sliced velocity map ion imaging. Int. Rev. Phys. Chem. 2012, 31, 201–233. [Google Scholar] [CrossRef]
- Liang, S.P.; Lu, I.C.; Tsai, S.T.; Chen, J.L.; Lee, Y.T.; Ni, C.K. Laser pulse width dependence and ionization mechanism of matrix-assisted laser desorption/ionization. J. Am. Soc. Mass. Spectrom. 2017, 28, 2235–2245. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.Y.C.; Spicer, W.E. Photoemission and optical studies of the electronic structure of palladium. Phys. Rev. 1968, 169, 497–507. [Google Scholar] [CrossRef]
- NIST. Available online: http://webbook.nist.gov/chemistry (accessed on 1 July 2017).
- King, B.V.; Pellin, M.J.; Moore, J.F.; Veryovkin, I.V.; Savina, M.R.; Tripa, C.E. Estimation of useful yield in surface analysis using single photon ionisation. Appl. Surf. Sci. 2003, 203, 244–247. [Google Scholar] [CrossRef]
- Wilson, K.R.; Jimenez-Cruz, M.; Nicolas, C.; Belau, L.; Leone, S.R.; Ahmed, M. Thermal vaporization of biological nanoparticles: Fragment-free vacuum ultraviolet photoionization mass spectra of tryptophan, phenylalanine-glycine-glycine, and, beta-carotene. J. Phys. Chem. A 2006, 110, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- Close, D.M. Calculated vertical ionization energies of the common alpha-amino acids in the gas phase and in solution. J. Phys. Chem. A 2011, 115, 2900–2912. [Google Scholar] [CrossRef] [PubMed]
- Close, D.M.; Ohman, K.T. Ionization energies of the nucleotides. J. Phys. Chem. A 2008, 112, 11207–11212. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Hernandez, C.E.; Arce, R.; Ishikawa, Y.; Gorb, L.; Leszczynski, J.; Close, D.M. Ab initio ionization energy thresholds of DNA and RNA bases in gas phase and in aqueous solution. J. Phys. Chem. A 2004, 108, 6373–6377. [Google Scholar] [CrossRef]
- Hanley, L.; Kornienko, O.; Ada, E.T.; Fuoco, E.; Trevor, J.L. Surface mass spectrometry of molecular species. J. Mass Spectrom. 1999, 34, 705–723. [Google Scholar] [CrossRef]
- Boesl, U. Multiphoton excitation and mass-selective ion detection for neutral and ion spectroscopy. J. Phys. Chem. 1991, 95, 2949–2962. [Google Scholar] [CrossRef]
- Adam, T.; Zimmermann, R. Determination of single photon ionization cross sections for quantitative analysis of complex organic mixtures. Anal. Bioanal. Chem. 2007, 389, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Hanley, L.; Zimmermann, R. Light and molecular ions: The emergence of vacuum UV single-photon ionization in MS. Anal. Chem. 2009, 81, 4174–4182. [Google Scholar] [CrossRef] [PubMed]



| Amino Acids | Parent Ion (Relative Ion Abundance) a,b | Fragment Ion (Relative Ion Abundance) a,b | Amino Acids | Parent Ion (Relative Ion Abundance) | Fragment Ion (Relative Ion Abundance) |
|---|---|---|---|---|---|
| Glycine | 75 (300) | 30 (1370) | Serine | 105 d | 43, 60 (4), 74/75 (2) |
| Alanine | 89 (7) | 44 (800) | Threonine | 119 e | 57 (445), 74/75 (4200) |
| Valine | 117 (15) | 72 (160) | Phenylalanine | 165 e | 74 (1750), 91 (500), 120 (1640) |
| Proline | 115 e | 70 (2000) | Tryptophan | 204 (27) | 130 (3600) |
| Leucine | 131 (15) | 86 (530) | Glutamic acid | 147 (5) | 102 (25) |
| Isoleucine | 131 (0.4) | 75 (13), 86 (10) | Aspartic acid c | 133 e | 88 (3) |
| Tyrosine | 181 e | 107 (10) | Glutamine | 146 (0.7) | 84, 101 (30) |
| Histidine | 155 e | 82 (10) | Asparagine c | 132 e | 74, 87 (13) |
| Arginine | 174 e | 60, 74 (6) | Cysteine | 121 (23) | 74/75/76 (110) |
| Lysine | 146 (5) | 30, 44 (50), 56, 72 (180), 84, 101 (75) | Methionine | 149 (18) | 75 (155), 83, 88, 101, 104 (90), 116, 131 (100) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, H.C.; Ni, C.-K. Vacuum Ultraviolet Single-Photon Postionization of Amino Acids. Appl. Sci. 2018, 8, 699. https://doi.org/10.3390/app8050699
Hsu HC, Ni C-K. Vacuum Ultraviolet Single-Photon Postionization of Amino Acids. Applied Sciences. 2018; 8(5):699. https://doi.org/10.3390/app8050699
Chicago/Turabian StyleHsu, Hsu Chen, and Chi-Kung Ni. 2018. "Vacuum Ultraviolet Single-Photon Postionization of Amino Acids" Applied Sciences 8, no. 5: 699. https://doi.org/10.3390/app8050699
APA StyleHsu, H. C., & Ni, C.-K. (2018). Vacuum Ultraviolet Single-Photon Postionization of Amino Acids. Applied Sciences, 8(5), 699. https://doi.org/10.3390/app8050699