Identification of Neurodegenerative Diseases Based on Vertical Ground Reaction Force Classification Using Time–Frequency Spectrogram and Deep Learning Neural Network Features
Abstract
:1. Introduction
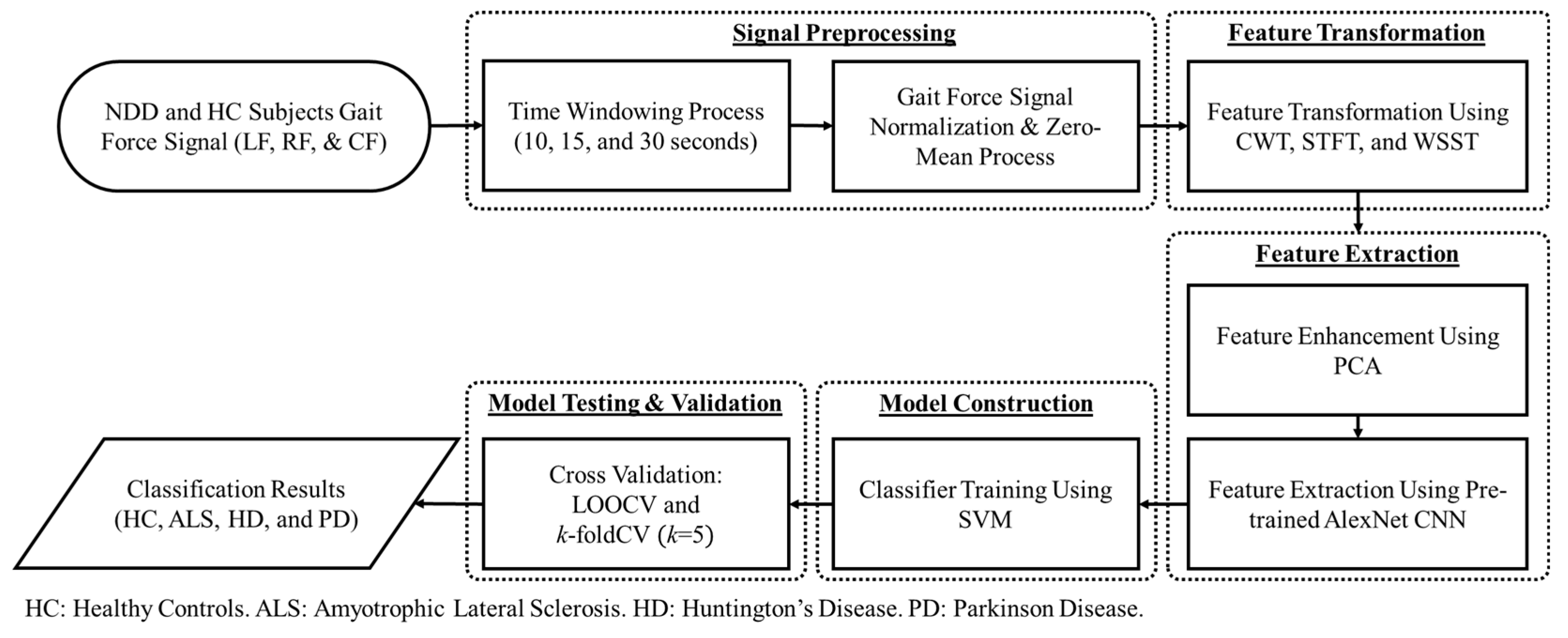
2. Materials and Methods
2.1. Neuro-Degenerative Diseases Gait Dynamics Database
2.2. Signal Preprocessing
2.3. Feature Transformation
2.3.1. Continuous Wavelet Transform (CWT)
2.3.2. Short Time Fourier Transform
2.3.3. Wavelet Synchrosqueezed Transform (WSST)
2.4. Principal Component Analysis (PCA) for Feature Enhancement
2.5. Pre-Trained Convolutional Neural Network (CNN) as Feature Extractor
2.6. Support Vector Machine (SVM) as Classifier
2.7. Cross-Validation
3. Experimental Results
3.1. Classification of the NDD and HC Group
3.2. Classification among the NDD
3.3. Classification of All NDD in One Group with HC Group
3.4. Multi-Class Classification
4. Discussion
4.1. Healthy Control
4.2. Amyotrophic Lateral Sclerosis
4.3. Huntington’s Disease
4.4. Parkinson’s Disease
4.5. Comparison Results with the Existing Literature
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- JPND Research. What is Neurodegenerative Disease? JPND Research. 7 February 2015. Available online: https://bit.ly/2Hkzs9w (accessed on 12 July 2019).
- Lee, A.; Gilbert, R. Epidemiology of Parkinson Disease. Neurol. Clin. 2016, 34, 955–965. [Google Scholar] [CrossRef]
- Parkinson’s Disease Foundation. Statistics on Parkinson’s. Parkinson’s Disease Foundation. 2018. EIN: 13-1866796. Available online: https://bit.ly/2RCeh9H (accessed on 12 July 2019).
- Chiò, A.; Logroscino, G.; Traynor, B.; Collins, J.; Simeone, J.; Goldstein, L.; White, L. Global Epidemiology of Amyotrophic Lateral Sclerosis: A Systematic Review of the Published Literature. Neuroepidemiology 2013, 41, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Renton, A.E.; Chio, A.; Traynor, B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014, 17, 17–23. [Google Scholar] [CrossRef]
- Agrawal, M.; Biswas, A. Molecular diagnostics of neurodegenerative disorders. Front. Mol. Biosci. 2015, 2, 54. [Google Scholar] [CrossRef] [Green Version]
- Harvard NeuroDiscovery Center. The Challenge of Neurodegenerative Diseases. Available online: https://bit.ly/2soDGmD (accessed on 12 July 2019).
- Hausdorff, J.M.; Cudkowicz, M.E.; Firtion, R.; Wei, J.Y.; Goldberger, A.L. Gait variability and basal ganglia disorders: Stride-to-stride variations of gait cycle timing in Parkinson’s disease and Huntington’s disease. Mov. Disord. 1998, 13, 428–437. [Google Scholar] [CrossRef]
- Brown, R.H., Jr.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 1602. [Google Scholar] [CrossRef] [Green Version]
- Hausdorff, J.M.; Lertratanakul, A.; Cudkowicz, M.E.; Peterson, A.L.; Kaliton, D.; Goldberger, A.L. Dynamic markers of altered gait rhythm in amyotrophic lateral sclerosis. J. Appl. Physiol. 2000, 88, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Banaie, M.; Sarbaz, Y.; Gharibzadeh, S.; Towhidkhah, F. Huntington’s disease: Modeling the gait disorder and proposing novel treatments. J. Theor. Biol. 2008, 254, 361–367. [Google Scholar] [CrossRef]
- Dayalu, P.; Albin, R.L. Huntington disease: Pathogenesis and treatment. Neurol. Clin. 2015, 33, 101–114. [Google Scholar] [CrossRef]
- Pyo, S.J.; Kim, H.; Kim, I.S.; Park, Y.-M.; Kim, M.-J.; Lee, H.M.; Koh, S.-B. Quantitative gait analysis in patients with huntington’s disease. J. Mov. Disord. 2017, 10, 140–144. [Google Scholar] [CrossRef]
- National Institute of Neurological Disorders and Stroke. Parkinson’s Disease Information Page. 2016. Available online: https://bit.ly/2xTA6rL (accessed on 12 July 2019).
- Hoff, J.; v/d Plas, A.; Wagemans, E.; van Hilten, J. Accelerometric assessment of levodopa-induced dyskinesias in Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2001, 16, 58–61. [Google Scholar] [CrossRef]
- Pistacchi, M. Gait analysis and clinical correlations in early Parkinson’s disease. Funct. Neurol. 2017, 32, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Gao, Q.; Ye, Q. Classification of gait rhythm signals between patients with neuro-degenerative diseases and normal subjects: Experiments with statistical features and different classification models. Biomed. Signal Process. Control. 2015, 18, 254–262. [Google Scholar] [CrossRef]
- Ertuğrul, Ö.F.; Kaya, Y.; Tekin, R.; Almalı, M.N. Detection of Parkinson’s disease by shifted one dimensional local binary patterns from gait. Expert Syst. Appl. 2016, 56, 156–163. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, P.; Luo, X.; Wu, M.; Liao, L.; Yang, S.; Rangayyan, R.M. Measuring signal fluctuations in gait rhythm time series of patients with Parkinson’s disease using entropy parameters. Biomed. Signal Process. Control 2017, 31, 265–271. [Google Scholar] [CrossRef]
- Bilgin, S. The impact of feature extraction for the classification of amyotrophic lateral sclerosis among neurodegenerative diseases and healthy subjects. Biomed. Signal Process. Control. 2017, 31, 288–294. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, C. Classification of neurodegenerative diseases using gait dynamics via deterministic learning. Inf. Sci. 2015, 317, 246–258. [Google Scholar] [CrossRef]
- Zhao, A.; Qi, L.; Dong, J.; Yu, H. Dual channel LSTM based multi-feature extraction in gait for diagnosis of Neurodegenerative diseases. Knowl. Based Syst. 2018, 145, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG arrhythmia classification using STFT-based spectrogram and convolutional neural network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Xie, Q.; Tu, S.; Wang, G.; Lian, Y.; Xu, L. Feature enrichment based convolutional neural network for heartbeat classification from electrocardiogram. IEEE Access 2019, 7, 153751–153760. [Google Scholar] [CrossRef]
- He, W.; Wang, G.; Hu, J.; Li, C.; Guo, B.; Li, F. Simultaneous human health monitoring and time-frequency sparse representation using EEG and ECG signals. IEEE Access 2019, 7, 85985–85994. [Google Scholar] [CrossRef]
- Tadesse, G.A.; Javed, H.; Thanh, N.L.N.; Thai, H.D.H.; Van Tan, L.; Thwaites, L.; Clifton, D.A.; Zhu, T. Multi-modal diagnosis of infectious diseases in the developing world. IEEE J. Biomed. Health Inform. 2020, 24, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Xu, L. Deep bidirectional intelligence: AlphaZero, deep IA-search, deep IA-infer, and TPC causal learning. In Applied Informatics; Springer: Berlin/Heidelberg, Germany, 2018; Volume 5, p. 5. [Google Scholar]
- Salem, M.; Taheri, S.; Yuan, J.-S. ECG Arrhythmia Classification Using Transfer Learning from 2-Dimensional Deep CNN Features. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar]
- Hausdorff, J.M.; Ladin, Z.; Wei, J.Y. Footswitch system for measurement of the temporal parameters of gait. J. Biomech. 1995, 28, 347–351. [Google Scholar] [CrossRef]
- Hausdorff, J.M.; Lertratanakul, A.; Cudkowicz, M.E.; Peterson, A.L.; Kaliton, D.; Goldberger, A.L. Gait Dynamics in Neuro-Degenerative Disease Database. 21 Dec 2000; PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2019, 101, e215–e220. Available online: https://www.physionet.org/content/gaitndd/1.0.0/ (accessed on 12 July 2019).
- Lin, C.-W.; Wen, T.-C.; Setiawan, F. Evaluation of vertical ground reaction forces pattern visualization in neurodegenerative diseases identification using deep learning and recurrence plot image feature extraction. Sensors 2020, 20, 3857. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, J. The continuous wavelet transform: A tool for signal investigation and understanding. Johns Hopkins APL Tech. Dig. 1994, 15, 306. [Google Scholar]
- Allen, J. Short term spectral analysis, synthesis, and modification by discrete Fourier transform. IEEE Trans. Acoust. Speech Signal Process. 1977, 25, 235–238. [Google Scholar] [CrossRef]
- Daubechies, I.; Lu, J.; Wu, H.-T. Synchrosqueezed wavelet transforms: An empirical mode decomposition-like tool. Appl. Comput. Harmon. Anal. 2011, 30, 243–261. [Google Scholar] [CrossRef] [Green Version]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Principal Component Analysis; Nature Publishing Group: Berlin, Germany, 2017; Chapter 1; p. 1. [Google Scholar]
- Jolliffe, I.T. Introduction. In Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Roccetti, M.; Delnevo, G.; Casini, L.; Mirri, S. An alternative approach to dimension reduction for pareto distributed data: A case study. J. Big Data 2021, 8, 1–23. [Google Scholar] [CrossRef]
- Ren, X.-D.; Guo, H.-N.; He, G.-C.; Xu, X.; Di, C.; Li, S.-H. Convolutional neural network based on principal component analysis initialization for image classification. In Proceedings of the 2016 IEEE First International Conference on Data Science in Cyberspace (DSC), Changsha, Chine, 13–16 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 329–334. [Google Scholar]
- Ng, A.; Ngiam, J.; Foo, C.Y.; Mai, Y.; Suen, C. UFLDL Tutorial. 2012. Available online: http://deeplearning.stanford.edu/wiki/index.php/UFLDL_Tutorial (accessed on 19 March 2021).
- Garg, I.; Panda, P.; Roy, K. A low effort approach to structured CNN design using PCA. IEEE Access 2019, 8, 1347–1360. [Google Scholar] [CrossRef]
- Seuret, M.; Alberti, M.; Liwicki, M.; Ingold, R. PCA-initialized deep neural networks applied to document image analysis. In Proceedings of the 2017 14th IAPR International Conference on Document Analysis and Recognition (ICDAR), Kyoto, Japan, 9–15 November 2017; IEEE: Piscataway, NJ, USA, 2017; Volume 1, pp. 877–882. [Google Scholar]
- O’Shea, K.; Nash, R. An introduction to convolutional neural networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.H. Feature extraction and image recognition with convolutional neural networks. J. Phys. Conf. Ser. 2018, 1087, 062032. [Google Scholar] [CrossRef]
- Garcia-Gasulla, D.; Parés, F.; Vilalta, A.; Moreno, J.; Ayguadé, E.; Labarta, J.; Cortés, U.; Suzumura, T. On the behavior of convolutional nets for feature extraction. J. Artif. Intell. Res. 2018, 61, 563–592. [Google Scholar] [CrossRef] [Green Version]
- Rajaraman, S.; Antani, S.K.; Poostchi, M.; Silamut, K.; Hossain, M.A.; Maude, R.J.; Jaeger, S.; Thoma, G.R. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 2018, 6, e4568. [Google Scholar] [CrossRef]
- Hegde, R.B.; Prasad, K.; Hebbar, H.; Singh, B.M.K. Feature extraction using traditional image processing and convolutional neural network methods to classify white blood cells: A study. Australas. Phys. Eng. Sci. Med. 2019, 42, 627–638. [Google Scholar] [CrossRef]
- Li, J.; Si, Y.; Xu, T.; Jiang, S. Deep convolutional neural network based ECG classification system using information fusion and one-hot encoding techniques. Math. Probl. Eng. 2018, 2018, 7354081. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, J.; Fan, S.-Z.; Abbod, M.F.; Shieh, J.-S.; Kung, Y.; Lin, L. Spectrum analysis of EEG signals using CNN to model patient’s consciousness level based on anesthesiologists’ experience. IEEE Access 2019, 7, 53731–53742. [Google Scholar] [CrossRef]
- Thanaraj, K.P.; Parvathavarthini, B.; Tanik, U.J.; Rajinikanth, V.; Kadry, S.; Kamalanand, K. Implementation of deep neural networks to classify EEG signals using gramian angular summation field for epilepsy diagnosis. arXiv 2020, arXiv:2003.04534. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H.; Liu, L.; Özsu, M.T. Cross-Validation. Springer Ref. 2011, 532–538. [Google Scholar]
- Wong, T.-T.; Yeh, P.-Y. Reliable Accuracy Estimates from k-Fold Cross Validation. IEEE Trans. Knowl. Data Eng. 2020, 32, 1586–1594. [Google Scholar] [CrossRef]
- Roccetti, M.; Delnevo, G.; Casini, L.; Salomoni, P. A Cautionary Tale for Machine Learning Design: Why we Still Need Human-Assisted Big Data Analysis. Mob. Netw. Appl. 2020, 25, 1075–1083. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Yang, M.; Zheng, H.; Wang, H.; McClean, S. Feature selection and construction for the discrimination of neurodegenerative diseases based on gait analysis. In Proceedings of the 2009 3rd International Conference on Pervasive Computing Technologies for Healthcare, London, UK, 1–3 April 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 1–7. [Google Scholar]
- Nilsson, J.; Thorstensson, A. Adaptability in frequency and amplitude of leg movements during human locomotion at different speeds. Acta Physiol. Scand. 1987, 129, 107–114. [Google Scholar] [CrossRef]
- Pham, T.D. Texture classification and visualization of time series of gait dynamics in patients with neuro-degenerative diseases. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Tang, S.; Fang, F.; Luo, L.; Xu, L.; Bringas-Vega, M.L.; Yao, D.; Kendrick, K.M.; Valdes-Sosa, P.A. Gait rhythm fluctuation analysis for neurodegenerative diseases by empirical mode decomposition. IEEE Trans. Biomed. Eng. 2016, 64, 52–60. [Google Scholar] [CrossRef] [PubMed]




| Layer | Size | Hyperparameter | ||
|---|---|---|---|---|
| Weight | Bias | |||
| Input | Image | - | - | |
| 1 | Convolution | |||
| 2 | ReLU 1 | - | - | |
| 3 | Cross Channel Normalization | - | - | |
| 4 | Max Pooling 2 | - | - | |
| 5 | Grouped Convolution | |||
| 6 | ReLU 1 | - | - | |
| 7 | Cross Channel Normalization | - | - | |
| 8 | Max Pooling 2 | - | - | |
| 9 | Convolution | |||
| 10 | ReLU 1 | - | - | |
| 11 | Grouped Convolution | |||
| 12 | ReLU 1 | - | - | |
| 13 | Grouped Convolution | |||
| 14 | ReLU 1 | - | - | |
| 15 | Max Pooling 2 | - | - | |
| 16 | Fully Connected | |||
| 17 | ReLU 1 | - | - | |
| 18 | Dropout (50%) 2 | - | - | |
| 19 | Fully Connected | |||
| 20 | SVM Classification Model 3 | - | - | - |
| Output | Classification Output | - | - | - |
| Time Window | Total Number of vGRF Spectrogram | Elapsed Time (s) | |
|---|---|---|---|
| LOOCV | k-Fold CV (k = 5) | ||
| 10 s | 1920 | 7933.684 | 33.013 |
| 30 s | 640 | 873.829 | 12.708 |
| 60 s | 320 | 235.986 | 7.473 |
| Classification Task | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA (10 s/30 s/60 s) | STFT + PCA (10 s/30 s/60 s) | WSST + PCA (10 s/30 s/60 s) | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| ALS vs. HC | Sen (%) | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 |
| Spec (%) | 99.79/99.38/98.77 | 99.79/99.39/98.82 | 99.79/99.38/98.77 | 99.79/99.39/98.82 | 100/98.77/98.77 | 99.79/98.79/97.65 | |
| Acc (%) | 99.89/99.66/99.31 | 99.89/99.66/99.31 | 99.89/99.66/99.31 | 99.89/99.66/99.31 | 100/99.31/99.31 | 99.89/99.31/98.62 | |
| AUC | 0.9990/0.9969/0.9938 | 1/1/1 | 0.9990/0.9969/0.9938 | 1/1/1 | 1/0.9938/0.9938 | 0.9987/0.9962/0.9846 | |
| HD vs. HC | Sen (%) | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | 99.83/99.49/100 | 99.83/100/100 |
| Spec (%) | 100/100/100 | 100/100/100 | 99.79/100/100 | 99.79/100/100 | 99.79/96.36/89.89 | 100/99.39/91.67 | |
| Acc (%) | 100/100/100 | 100/100/100 | 99.91/100/100 | 99.91/100/100 | 99.81/98.06/95 | 99.91/99.72/95.56 | |
| AUC | 1/1/1 | 1/1/1 | 0.9990/1/1 | 1/1/1 | 0.9981/0.9793/0.9494 | 1/1/1 | |
| PD vs. HC | Sen (%) | 98.38/94.27/94.81 | 99.13/97.47/100 | 93.59/92.36/88.61 | 92.86/91.09/89.70 | 89.68/86.54/79.71 | 94.46/89.96/60.69 |
| Spec (%) | 95.17/98.69/97.44 | 94.53/96.42/96.47 | 91.68/89.76/93.42 | 91.39/91.16/94.62 | 88.06/90.26/76.74 | 92.33/95.07/76.99 | |
| Acc (%) | 96.67/96.45/96.13 | 96.45/96.77/98.06 | 92.58/90.97/90.97 | 91.29/90.97/90.32 | 88.82/88.39/78.06 | 92.47/91.29/61.94 | |
| AUC | 0.9678/0.9648/0.9612 | 0.9992/0.9969/0.9967 | 0.9264/0.9106/0.9101 | 0.9794/0.9659/0.9679 | 0.8887/0.8840/0.7823 | 0.9957/0.9795/0.8763 | |
| Classification Task | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA (10 s/30 s/60 s) | STFT + PCA (10 s/30 s/60 s) | WSST + PCA (10 s/30 s/60 s) | |||||
| PCA | Non-PCA | PCA | Non-PCA | PCA | Non-PCA | ||
| ALS vs. HC | Sen (%) | 100/100/100 | 94.97/96.26/89.02 | 100/100/100 | 87.88/88.49/93.14 | 100/100/100 | 75.69/76.30/86.11 |
| Spec (%) | 99.79/99.39/98.82 | 92.73/95.23/94.21 | 99.79/99.39/98.82 | 93.74/93.65/91.78 | 99.79/98.79/97.65 | 65.49/77.41/62.59 | |
| Acc (%) | 99.89/99.66/99.31 | 93.56/95.52/91.72 | 99.89/99.66/99.31 | 90.69/91.03/91.03 | 99.89/99.31/98.62 | 66.90/69.31/62.07 | |
| AUC | 1/1/1 | 0.9809/0.9871/0.9676 | 1/1/1 | 0.9676/0.9712/0.9596 | 0.9987/0.9962/0.9846 | 0.7753/0.7698/0.6779 | |
| HD vs. HC | Sen (%) | 100/100/100 | 93.28/91.37/93.42 | 100/100/100 | 85.89/89.16/87.45 | 99.83/100/100 | 78.84/76.27/89.42 |
| Spec (%) | 100/100/100 | 81.14/85.05/89.27 | 99.79/100/100 | 81.34/77.68/77.24 | 100/99.39/91.67 | 67.82/61.72/62.52 | |
| Acc (%) | 100/100/100 | 87.04/88.06/91.11 | 99.91/100/100 | 82.31/81.39/77.78 | 99.91/99.72/95.56 | 61.76/67.78/61.11 | |
| AUC | 1/1/1 | 0.9431/0.9366/0.9688 | 1/1/1 | 0.8904/0.9034/0.9031 | 1/1/1 | 0.7186/0.7898/0.7969 | |
| PD vs. HC | Sen (%) | 99.13/97.47/100 | 88.48/89.34/91.98 | 92.86/91.09/89.70 | 83.56/71.14/91.67 | 94.46/89.96/60.69 | 69.61/90.40/78.97 |
| Spec (%) | 94.53/96.42/96.47 | 84.74/89.09/86.32 | 91.39/91.16/94.62 | 80.85/83.56/67.10 | 92.33/95.07/76.99 | 62.60/55.48/54.31 | |
| Acc (%) | 96.45/96.77/98.06 | 86.24/88.06/88.39 | 91.29/90.97/90.32 | 81.08/72.26/67.74 | 92.47/91.29/61.94 | 60.11/53.87/55.48 | |
| AUC | 0.9992/0.9969/0.9967 | 0.9222/0.9481/0.9450 | 0.9794/0.9659/0.9679 | 0.8907/0.8294/0.8954 | 0.9957/0.9795/0.8763 | 0.6908/0.6507/0.7150 | |
| Classification Task | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA (10 s/30 s/60 s) | STFT + PCA (10 s/30 s/60 s) | WSST + PCA (10 s/30 s/60 s) | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| ALS vs. HD | Sen (%) | 97.95/96.83/100 | 97.99/98.46/100 | 96.93/93.43/92.19 | 99.01/96.18/95.79 | 96.70/87.60/90.32 | 97.88/82.21/89 |
| Spec (%) | 98.67/96.08/93.46 | 98.67/96.61/95.32 | 98.16/98.96/94.06 | 97.10/97.54/93.06 | 98.49/91.54/91.26 | 96.30/95.38/88.49 | |
| Acc (%) | 98.38/96.36/95.76 | 98.38/97.27/96.97 | 97.68/96.67/93.33 | 97.78/96.97/93.33 | 97.78/90/90.91 | 96.87/86.67/85.45 | |
| AUC | 0.9831/0.9645/0.9673 | 0.9953/0.9958/0.9800 | 0.9755/0.9620/0.9312 | 0.9876/0.9851/0.9792 | 0.9760/0.8957/0.9079 | 0.9934/0.9687/0.9419 | |
| PD vs. ALS | Sen (%) | 99.78/99.34/98.68 | 99.78/99.35/98.75 | 99.78/99.34/98.68 | 99.78/99.35/98.75 | 99.78/98.04/94.94 | 99.78/98.75/97.50 |
| Spec (%) | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | 100/100/100 | |
| Acc (%) | 99.88/99.64/99.29 | 99.88/99.64/99.29 | 99.88/99.64/99.29 | 99.88/99.64/99.29 | 99.88/99.64/97.14 | 99.88/99.29/98.57 | |
| AUC | 0.9989/0.9967/0.9934 | 1/1/0.9923 | 0.9989/0.9967/0.9934 | 1/1/0.9923/0.9923 | 0.9989/0.9902/0.9747 | 0.9987/0.9962/0.9923 | |
| HD vs. PD | Sen (%) | 98.82/96.14/94.17 | 98.52/95.28/96.23 | 94.93/93.30/96.08 | 94.61/94.78/94.85 | 94.21/89.16/86.60 | 89.79/93.43/96.80 |
| Spec (%) | 96.72/99.30/95.83 | 97.82/99.31/97.42 | 95.44/96.45/97.26 | 97.01/95.75/96.40 | 93.05/87.07/79.49 | 99.51/82.95/84.09 | |
| Acc (%) | 97.90/97.43/94.86 | 98.19/96.86/96.57 | 95.14/94.57/96.57 | 95.52/94.57/94.86 | 93.71/88.29/83.43 | 93.14/86/89.14 | |
| AUC | 0.9777/0.9772/0.9500 | 0.9982/0.9953/0.9940 | 0.9519/0.9488/0.9667 | 0.9912/0.9926/0.9953 | 0.9363/0.8812/0.8304 | 0.9979/0.9540/0.9767 | |
| NDD vs. HC | Sen (%) | 99.58/98.96/97.94 | 100/99.38/97.58 | 96.77/95.91/96.69 | 98.06/96.60/97.15 | 93.85/98.49/93.64 | 95.30/93.91/100 |
| Spec (%) | 95.76/96.88/97.40 | 95.29/96.47/97.39 | 93.52/92.72/92.31 | 92.09/93.73/94.29 | 91.51/86.44/77.38 | 99.29/88.54/72.83 | |
| Acc (%) | 98.59/98.44/97.81 | 98.75/98.59/97.50 | 95.99/95.16//95.63 | 96.41/95.63/95.94 | 93.33/95.16/89.38 | 96.09/90.78/89.38 | |
| AUC | 0.9767/0.9792/0.9767 | 0.9991/0.9959/0.9689 | 0.9515/0.9431/0.9450 | 0.9844/0.9847/0.9742 | 0.9268/0.9246/0.8551 | 0.9978/0.9938/0.9943 | |
| Classification Task | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA (10 s/30 s/60 s) | STFT + PCA (10 s/30 s/60 s) | WSST + PCA (10 s/30 s/60 s) | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| ALS vs. HD | Sen (%) | 97.99/98.46/100 | 81.75/88.74/92.62 | 99.01/96.18/95.79 | 86.74/83.68/82.84 | 97.88/82.21/89 | 48.35/55.47/55.68 |
| Spec (%) | 98.67/96.61/95.32 | 95.82/93.63/96.09 | 97.10/97.54/93.06 | 87.75/91.06/92.05 | 96.30/95.38/88.49 | 92.85/85.99/83.86 | |
| Acc (%) | 98.38/97.27/96.97 | 88.89/90.61/94.55 | 97.78/96.97/93.33 | 86.97/86.67/83.64 | 96.87/86.67/85.45 | 56.56/52.12/61.82 | |
| AUC | 0.9953/0.9958/0.9800 | 0.9584/0.9551/0.9788 | 0.9876/0.9851/0.9792 | 0.9410/0.9510/0.9519 | 0.9934/0.9687/0.9419 | 0.6849/0.6187/0.7612 | |
| PD vs. ALS | Sen (%) | 99.78/99.35/98.75 | 91.99/80.17/86.08 | 99.78/99.35/98.75 | 86.12/87.07/85.79 | 99.78/98.75/97.50 | 74.55/63.51/84.04 |
| Spec (%) | 100/100/100 | 88.84/85.75/79.51 | 100/100/100 | 83.91/74.38/76.07 | 100/100/100 | 63.60/86.06/69.66 | |
| Acc (%) | 99.88/99.64/99.29 | 90.36/81.43/80 | 99.88/99.64/99.29 | 84.17/77.50/77.14 | 99.88/99.29/98.57 | 64.40/64.64/57.14 | |
| AUC | 1/1/0.9923 | 0.9513/0.8835/0.8195 | 1/1/0.9923/0.9923 | 0.9315/0.8892/0.8938 | 0.9987/0.9962/0.9923 | 0.7588/0.6604/0.6641 | |
| HD vs. PD | Sen (%) | 98.52/95.28/96.23 | 84.34/79.74/86.86 | 94.61/94.78/94.85 | 72.76/76.98/75.95 | 89.79/93.43/96.80 | 72.56/63.81/79.10 |
| Spec (%) | 97.82/99.31/97.42 | 77.75/78.58/75.16 | 97.01/95.75/96.40 | 71.33/70.22/66.98 | 99.51/82.95/84.09 | 54.34/59.58/63.71 | |
| Acc (%) | 98.19/96.86/96.57 | 80.29/77.14/78.86 | 95.52/94.57/94.86 | 70.48/72.57/69.71 | 93.14/86/89.14 | 56.48/59.71/61.71 | |
| AUC | 0.9982/0.9953/0.9940 | 0.8869/0.8692/0.8457 | 0.9912/0.9926/0.9953 | 0.7580/0.7922/0.7817 | 0.9979/0.9540/0.9767 | 0.6515/0.5985/0.6963 | |
| NDD vs. HC | Sen (%) | 100/99.38/97.58 | 90.57/94.41/88.94 | 98.06/96.60/97.15 | 89.69/89.21/90.01 | 95.30/93.91/100 | 83.12/89.43/83.38 |
| Spec (%) | 95.29/96.47/97.39 | 77.63/74.75/83.47 | 92.09/93.73/94.29 | 65.08/58.69/66.96 | 99.29/88.54/72.83 | 46.99/38.03/70.67 | |
| Acc (%) | 98.75/98.59/97.50 | 87.19/87.97/84.38 | 96.41/95.63/95.94 | 82.71/78.28/74.38 | 96.09/90.78/89.38 | 65.99/48.91/69.06 | |
| AUC | 0.9991/0.9959/0.9689 | 0.8938/0.9308/0.8634 | 0.9844/0.9847/0.9742 | 0.8478/0.8280/0.8909 | 0.9978/0.9938/0.9943 | 0.6424/0.7777/0.7500 | |
| Classification Class | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA | STFT + PCA | WSST + PCA | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| HC | Sen (%) | 98.75 | 97.71 | 92.50 | 94.79 | 90 | 87.71 |
| Spec (%) | 99.03 | 98.89 | 97.92 | 97.64 | 96.18 | 97.71 | |
| Acc (%) | 98.96 | 98.59 | 96.56 | 96.93 | 94.64 | 95.21 | |
| AUC | 0.9889 | 0.9830 | 0.9521 | 0.9622 | 0.9309 | 0.9271 | |
| ALS | Sen (%) | 98.21 | 97.95 | 97.44 | 97.69 | 97.18 | 95.90 |
| Spec (%) | 99.35 | 99.80 | 99.35 | 98.76 | 98.95 | 99.02 | |
| Acc (%) | 99.11 | 99.43 | 98.96 | 98.54 | 98.59 | 98.39 | |
| AUC | 0.9878 | 0.9888 | 0.9839 | 0.9823 | 0.9807 | 0.9746 | |
| HD | Sen (%) | 97 | 97 | 95 | 93.83 | 89.83 | 89.33 |
| Spec (%) | 98.86 | 99.17 | 96.52 | 96.74 | 96.89 | 98.11 | |
| Acc (%) | 98.28 | 98.49 | 96.04 | 95.83 | 94.69 | 95.36 | |
| AUC | 0.9793 | 0.9808 | 0.9576 | 0.9529 | 0.9336 | 0.9372 | |
| PD | Sen (%) | 95.33 | 96.22 | 85.33 | 86.22 | 86.22 | 83.78 |
| Spec (%) | 98.64 | 97.21 | 96.19 | 96.73 | 91.90 | 92.52 | |
| Acc (%) | 97.86 | 96.98 | 93.65 | 94.27 | 90.57 | 90.47 | |
| AUC | 0.9699 | 0.9808 | 0.9076 | 0.9148 | 0.8906 | 0.8815 | |
| Classification Class | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA | STFT + PCA | WSST + PCA | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| HC | Sen (%) | 96.25 | 97.50 | 89.38 | 92.50 | 86.25 | 91.88 |
| Spec (%) | 99.17 | 98.54 | 97.92 | 97.71 | 96.88 | 96.46 | |
| Acc (%) | 98.44 | 98.28 | 95.78 | 96.41 | 94.22 | 95.31 | |
| AUC | 0.9771 | 0.9802 | 0.9365 | 0.9510 | 0.9156 | 0.9417 | |
| ALS | Sen (%) | 96.92 | 95.38 | 96.92 | 96.92 | 93.08 | 98.46 |
| Spec (%) | 98.63 | 98.82 | 98.63 | 98.24 | 95.88 | 91.76 | |
| Acc (%) | 98.28 | 98.13 | 98.28 | 97.97 | 95.31 | 93.13 | |
| AUC | 0.9778 | 0.9710 | 0.9778 | 0.9758 | 0.9448 | 0.9511 | |
| HD | Sen (%) | 93.50 | 93.50 | 94 | 94.50 | 76.50 | 84.50 |
| Spec (%) | 97.27 | 97.73 | 97.50 | 95 | 95.68 | 95 | |
| Acc (%) | 96.09 | 96.41 | 96.41 | 94.84 | 89.69 | 91.72 | |
| AUC | 0.9539 | 0.9561 | 0.9575 | 0.9475 | 0.8609 | 0.8975 | |
| PD | Sen (%) | 92.67 | 94 | 88 | 84.67 | 83.33 | 76.67 |
| Spec (%) | 98.16 | 94.69 | 93.88 | 96.53 | 90.20 | 93.06 | |
| Acc (%) | 96.88 | 94.53 | 92.50 | 93.75 | 88.59 | 89.22 | |
| AUC | 0.9541 | 0.9435 | 0.9094 | 0.9060 | 0.8677 | 0.8486 | |
| Classification Class | Evaluation Parameter | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| CWT + PCA | STFT + PCA | WSST + PCA | |||||
| LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | LOOCV | k-Fold CV (k = 5) | ||
| HC | Sen (%) | 95 | 98.75 | 92.50 | 97.50 | 78.75 | 76.25 |
| Spec (%) | 99.17 | 97.92 | 97.50 | 98.04 | 92.92 | 88.75 | |
| Acc (%) | 98.13 | 98.13 | 96.25 | 94.38 | 89.38 | 85.63 | |
| AUC | 0.9708 | 0.9833 | 0.9500 | 0.9542 | 0.8583 | 0.8250 | |
| ALS | Sen (%) | 92.31 | 93.85 | 90.77 | 86.15 | 78.46 | 75.38 |
| Spec (%) | 98.04 | 96.86 | 97.25 | 98.82 | 98.82 | 90.20 | |
| Acc (%) | 96.88 | 96.25 | 95.94 | 96.25 | 94.69 | 87.19 | |
| AUC | 0.9517 | 0.9535 | 0.9401 | 0.9249 | 0.8864 | 0.8279 | |
| HD | Sen (%) | 98 | 95 | 95 | 94 | 75 | 92 |
| Spec (%) | 93.64 | 95.45 | 94.09 | 95 | 91.36 | 87.27 | |
| Acc (%) | 95 | 95.31 | 94.38 | 94.69 | 86.25 | 88.75 | |
| AUC | 0.9582 | 0.9523 | 0.9455 | 0.9450 | 0.8318 | 0.8964 | |
| PD | Sen (%) | 89.33 | 89.33 | 85.33 | 82.67 | 68 | 57.33 |
| Spec (%) | 97.55 | 97.96 | 93.88 | 95.51 | 83.27 | 88.98 | |
| Acc (%) | 95.63 | 95.94 | 91.88 | 92.50 | 79.69 | 81.56 | |
| AUC | 0.9344 | 0.9365 | 0.8961 | 0.8909 | 0.7563 | 0.7316 | |
| Studies | Evaluation Parameter | Classification Task | |||
|---|---|---|---|---|---|
| ALS vs. HC | HD vs. HC | PD vs. HC | NDD vs. HC | ||
| Zeng et al. [22] | Sen (%) | 92.31 | 85 | 87.50 | - |
| Spec (%) | 87.50 | 81.25 | 86.67 | - | |
| Acc (%) | 89.66 | 87.10 | 87.10 | - | |
| Zhao et al. [23] | Acc (%) | 97.43 | 94.96 | 97.33 | 96.42 |
| Ren et al. [64] | AUC | 0.8980 | 0.8810 | 0.9010 | - |
| Pham, T.D. [63] | Sen (%) | 100 | 100 | 100 | - |
| Spec (%) | 100 | 100 | 100 | - | |
| Acc (%) | 100 | 100 | 100 | - | |
| AUC | 1 | 1 | 1 | - | |
| The Proposed Method | Sen (%) | 100 | 100 | 99.08 | 98.96 |
| Spec (%) | 100 | 100 | 95.97 | 96.88 | |
| Acc (%) | 100 | 100 | 97.42 | 98.44 | |
| AUC | 1 | 1 | 0.9752 | 0.9792 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Setiawan, F.; Lin, C.-W. Identification of Neurodegenerative Diseases Based on Vertical Ground Reaction Force Classification Using Time–Frequency Spectrogram and Deep Learning Neural Network Features. Brain Sci. 2021, 11, 902. https://doi.org/10.3390/brainsci11070902
Setiawan F, Lin C-W. Identification of Neurodegenerative Diseases Based on Vertical Ground Reaction Force Classification Using Time–Frequency Spectrogram and Deep Learning Neural Network Features. Brain Sciences. 2021; 11(7):902. https://doi.org/10.3390/brainsci11070902
Chicago/Turabian StyleSetiawan, Febryan, and Che-Wei Lin. 2021. "Identification of Neurodegenerative Diseases Based on Vertical Ground Reaction Force Classification Using Time–Frequency Spectrogram and Deep Learning Neural Network Features" Brain Sciences 11, no. 7: 902. https://doi.org/10.3390/brainsci11070902
APA StyleSetiawan, F., & Lin, C.-W. (2021). Identification of Neurodegenerative Diseases Based on Vertical Ground Reaction Force Classification Using Time–Frequency Spectrogram and Deep Learning Neural Network Features. Brain Sciences, 11(7), 902. https://doi.org/10.3390/brainsci11070902







