Insight into the Antioxidant Effect of Fermented and Non-Fermented Spirulina Water and Ethanol Extracts at the Proteome Level Using a Yeast Cell Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Lactic Acid Fermentation of Spirulina
2.2. Spirulina Extract Preparation
2.3. Yeast Culture Preparation and Treating of Cells with Spirulina Extracts
2.4. Preparation of Yeast Cell Lysates
2.5. Protein Digestion to Peptides for LC-MS
2.6. LC-MS Analysis
2.7. MS Raw Data Analysis
2.8. Bioinformatic Analysis of Data
3. Results and Discussion
3.1. Principal Component Analysis (PCA)
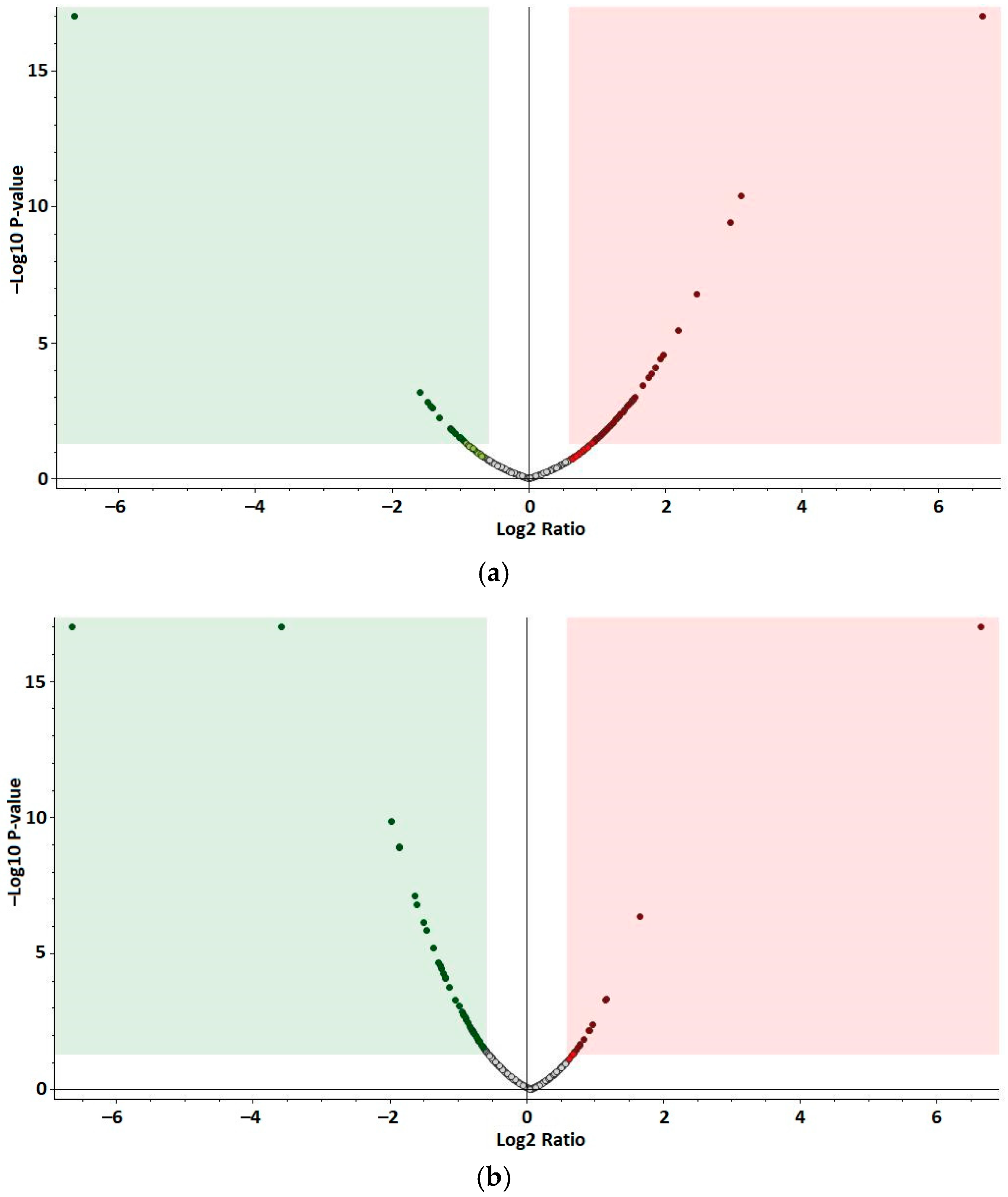
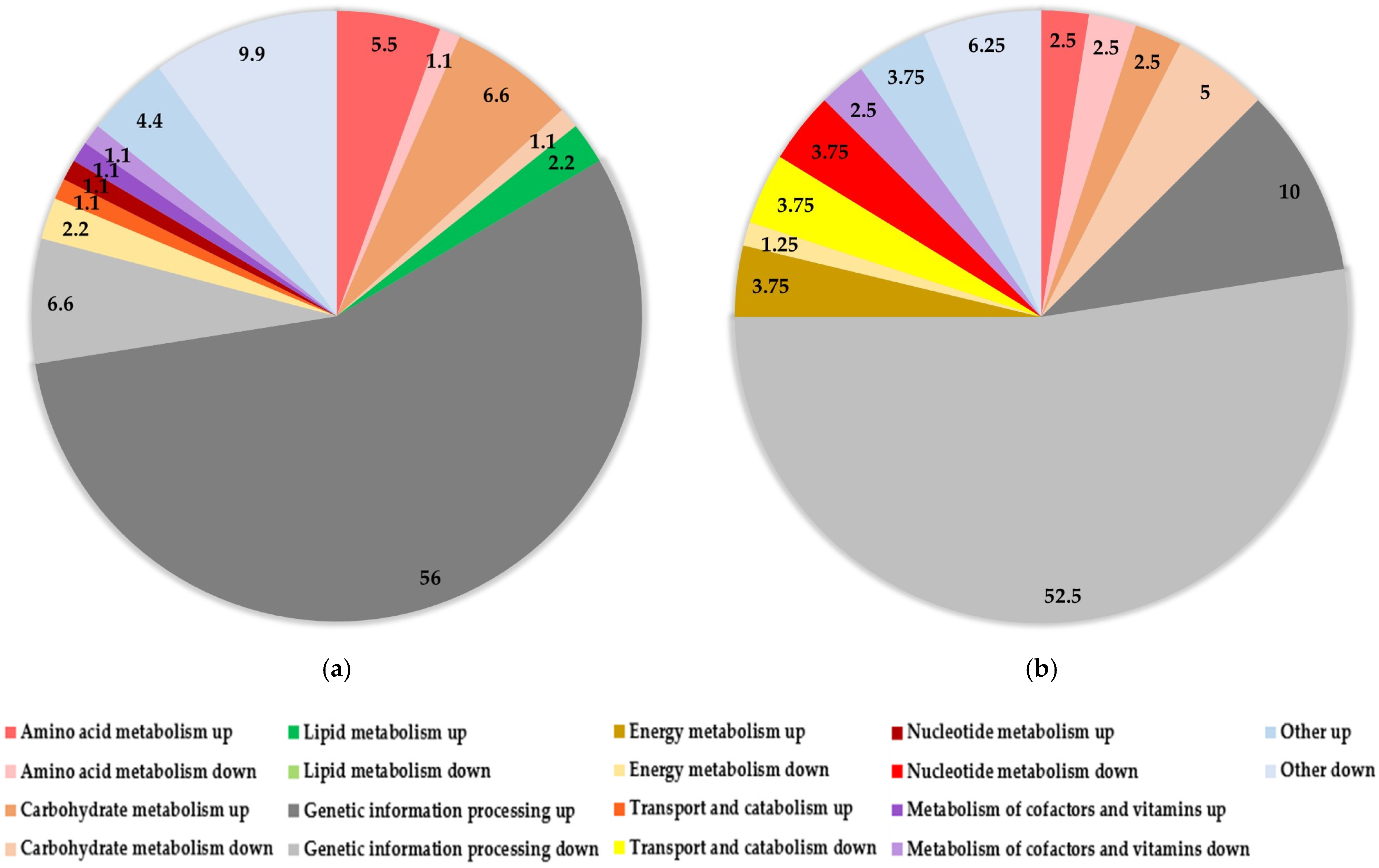
3.2. Quantification and Identification of Differentially Abundant Proteins
3.3. Stress Response Related Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vo, T.-S.; Ngo, D.-H.; Kim, S.-K. Nutritional and pharmaceutical properties of microalgal Spirulina. In Handbook of Marine Microalgae: Biotechnology Advances; Kim, S.-K., Ed.; Elsevier Inc.: London, UK, 2015; pp. 299–308. [Google Scholar]
- Chopra, K.; Bishnoi, M. Antioxidant profile of Spirulina: A blue-green microalga. In Spirulina in Human Nutrition and Health; Gershwin, M.E., Belay, A., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 101–118. [Google Scholar]
- Chu, W.-L.; Lim, Y.-W.; Radhakrishnan, A.K.; Lim, P.-E. Protective effect of aqueous extract from Spirulina platensis against cell death induced by free radicals. BMC Complement. Altern. Med. 2010, 10, 53. [Google Scholar] [CrossRef]
- Liestianty, D.; Rodianawati, I.; Arfah, R.A.; Assa, A.; Patimah; Sundari; Muliadi. Nutritional analysis of Spirulina sp to promote as superfood candidate. IOP Conf. Ser. Mater. Sci. Eng. 2019, 509, 012031. [Google Scholar] [CrossRef]
- Bashir, S.; Sharif, K.M.; Butt, M.S.; Shahid, M. Functional properties and amino acid profile of Spirulina platensis protein isolates. Pak. J. Sci. Ind. Res. Ser. B Biol. Sci. 2016, 59, 12–19. [Google Scholar] [CrossRef]
- Gad, A.S.; Khadrawy, Y.A.; El-Nekeety, A.A.; Mohamed, S.R.; Hassan, N.S.; Abdel-Wahhab, M.A. Antioxidant activity and hepatoprotective effects of whey protein and Spirulina in rats. Nutrition 2011, 27, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Czerwonka, A.; Kaławaj, K.; Sławińska-Brych, A.; Lemieszek, M.K.; Bartnik, M.; Wojtanowski, K.K.; Zdzisińska, B.; Rzeski, W. Anticancer effect of the water extract of a commercial Spirulina (Arthrospira platensis) product on the human lung cancer A549 cell line. Biomed. Pharmacother. 2018, 106, 292–302. [Google Scholar] [CrossRef]
- Habib, M.A.B.; Parvin, M.; Huntington, T.C.; Hasan, M.R. A Review on Culture, Production and Use of Spirulina as Food for Humans and Feeds for Domestic Animals and Fish; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008. [Google Scholar]
- Nagaoka, S.; Shimizu, K.; Kaneko, H.; Shibayama, F.; Morikawa, K.; Kanamaru, Y.; Otsuka, A.; Hirahashi, T.; Kato, T. A novel protein C-phycocyanin plays a crucial role in the hypocholesterolemic action of Spirulina platensis concentrate in rats. J. Nutr. 2005, 135, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- de Marco Castro, E.; Shannon, E.; Abu-Ghannam, N. Effect of fermentation on enhancing the nutraceutical properties of Arthrospira platensis (Spirulina). Fermentation 2019, 5, 28. [Google Scholar] [CrossRef]
- Sanjukta, S.; Rai, A.K. Production of bioactive peptides during soybean fermentation and their potential health benefits. Trends Food Sci. Technol. 2016, 50, 1–10. [Google Scholar] [CrossRef]
- Limón, R.I.; Peñas, E.; Torino, M.I.; Martínez-Villaluenga, C.; Dueñas, M.; Frias, J. Fermentation enhances the content of bioactive compounds in kidney bean extracts. Food Chem. 2015, 172, 343–352. [Google Scholar] [CrossRef]
- Liu, J.-G.; Hou, C.-W.; Lee, S.-Y.; Chuang, Y.; Lin, C.-C. Antioxidant effects and UVB protective activity of Spirulina (Arthrospira platensis) products fermented with lactic acid bacteria. Process Biochem. 2011, 46, 1405–1410. [Google Scholar] [CrossRef]
- Niccolai, A.; Shannon, E.; Abu-Ghannam, N.; Biondi, N.; Rodolfi, L.; Tredici, M.R. Lactic acid fermentation of Arthrospira platensis (Spirulina) biomass for probiotic-based products. J. Appl. Phycol. 2019, 31, 1077–1083. [Google Scholar] [CrossRef]
- Shalaby, E.A.; Shanab, S.M.M. Comparison of DPPH and ABTS assays for determining antioxidant potential of water and methanol extracts of Spirulina platensis. Indian J. Geo-Mar. Sci. 2013, 42, 556–564. [Google Scholar]
- Santoyo, S.; Herrero, M.; Señorans, F.J.; Cifuentes, A.; Ibáñez, E.; Jaime, L. Functional characterization of pressurized liquid extracts of Spirulina platensis. Eur. Food Res. Technol. 2006, 224, 75–81. [Google Scholar] [CrossRef]
- Shukla, V.; Vashistha, M.; Singh, S.N. Evaluation of antioxidant profile and activity of amalaki (Emblica officinalis), Spirulina and wheat grass. Indian J. Clin. Biochem. 2009, 24, 70–75. [Google Scholar] [CrossRef]
- Ojima, F.; Yamaguchi, Y.; Sakaki, S.; Takenaka, H. Stimulatory Effects of microalgae on the secretion of nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) in 3T3-L1 fibroblasts. Algal Resour. 2019, 12, 61–65. [Google Scholar]
- Wu, L.-C.; Ho, J.-A.A.; Shieh, M.-C.; Lu, I.-W. Antioxidant and antiproliferative activities of Spirulina and Chlorella water extracts. J. Agric. Food Chem. 2005, 53, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.-J.; Seo, Y.-J.; Choi, J.; Lee, H.Y.; Kang, D.-H.; Kim, K.-J.; Lee, B.-Y. Spirulina maxima extract prevents neurotoxicity via promoting activation of BDNF/CREB signaling pathways in neuronal cells and mice. Molecules 2017, 22, 1363. [Google Scholar] [CrossRef]
- Putri, A.K.; Dimarti, S.C.; Yuniati, R.; Susilaningsih, N. Cytotoxicity and antiproliferation of phycocyanin from Spirulina platensis extract on WiDr colon cancer cell line. Biosaintifika J. Biol. Biol. Educ. 2020, 12, 42–49. [Google Scholar] [CrossRef]
- Cepoi, L.; Rudi, L.; Miscu, V.; Cojocari, A.; Chiriac, T.; Sadovnic, D. Antioxidative activity of ethanol extracts from Spirulina platensis and Nostoc linckia measured by various methods. Analele Univ. din Oradea Fasc. Biol. 2009, Tom XVI, 43–48. [Google Scholar]
- Gabr, G.A.; El-Sayed, S.M.; Hikal, M.S. Antioxidant activities of phycocyanin: A bioactive compound from Spirulina platensis. J. Pharm. Res. Int. 2020, 32, 73–85. [Google Scholar] [CrossRef]
- Guthrie, G.; Fink, G.R. Guide to Yeast Genetics and Molecular Biology, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1991; Volume 194, pp. 1–863. [Google Scholar]
- Ma, D. Applications of yeast in drug discovery. Prog. Drug Res. 2001, 57, 117–162. [Google Scholar] [CrossRef] [PubMed]
- Menacho-Márquez, M.; Murguía, J.R. Yeast on drugs: Saccharomyces cerevisiae as a tool for anticancer drug research. Clin. Transl. Oncol. 2007, 9, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Jamnik, P.; Mahnič, N.; Mrak, A.; Pogačnik, L.; Jeršek, B.; Niccolai, A.; Masten, J.; Ogrinc, N.; Dušak, L.; Ferjančič, B.; et al. Fermented Biomass of Arthrospira platensis as a Potential Food Ingredient. Unpublished; manuscript in preparation.
- Longo, V.D.; Gralla, E.B.; Valentine, J.S. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 12275–12280. [Google Scholar] [CrossRef]
- Moradas-Ferreira, P.; Costa, V.; Piper, P.; Mager, W. The molecular defences against reactive oxygen species in yeast. Mol. Microbiol. 1996, 19, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Gralla, E.B.; Kosman, D.J. Molecular genetics of superoxide dismutases in yeasts and related fungi. Adv. Genet. 1992, 30, 251–319. [Google Scholar] [CrossRef] [PubMed]
- Cigut, T.; Polak, T.; Gašperlin, L.; Raspor, P.; Jamnik, P. Antioxidative activity of propolis extract in yeast cells. J. Agric. Food Chem. 2011, 59, 11449–11455. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Agustini, T.W.; Suzery, M.; Sutrisnanto, D.; Ma’ruf, W.F.; Hadiyanto, H. Comparative study of bioactive substances extracted from fresh and dried Spirulina sp. Procedia Environ. Sci. 2015, 23, 282–289. [Google Scholar] [CrossRef]
- Lee, E.H.; Park, J.-E.; Choi, Y.-J.; Huh, K.-B.; Kim, W.-Y. A Randomized study to establish the effects of spirulina in type 2 diabetes mellitus patients. Nutr. Res. Pract. 2008, 2, 295–300. [Google Scholar] [CrossRef]
- Ravi, M.; De, S.L.; Azharuddin, S.; Paul, S.F.D. The beneficial effects of Spirulina focusing on its immunomodulatory and antioxidant properties. Nutr. Diet. Suppl. 2010, 2, 73–83. [Google Scholar] [CrossRef]
- Hamid, A.A.; Shah, Z.M.; Muse, R.; Mohamed, S. Characterisation of antioxidative activities of various extracts of Centella asiatica (L) Urban. Food Chem. 2002, 77, 465–469. [Google Scholar] [CrossRef]
- Mello, B.C.B.S.; Hubinger, M.D. Antioxidant activity and polyphenol contents in Brazilian green propolis extracts prepared with the use of ethanol and water as solvents in different pH values. Int. J. Food Sci. Technol. 2012, 47, 2510–2518. [Google Scholar] [CrossRef]
- Herrero, M.; Martín-Álvarez, P.J.; Señoráns, F.J.; Cifuentes, A.; Ibáñez, E. Optimization of accelerated solvent extraction of antioxidants from Spirulina platensis microalga. Food Chem. 2005, 93, 417–423. [Google Scholar] [CrossRef]
- Đorđević, T.M.; Šiler-Marinković, S.S.; Dimitrijević-Branković, S.I. Effect of fermentation on antioxidant properties of some cereals and pseudo cereals. Food Chem. 2010, 119, 957–963. [Google Scholar] [CrossRef]
- Katina, K.; Laitila, A.; Juvonen, R.; Liukkonen, K.-H.; Kariluoto, S.; Piironen, V.; Landberg, R.; Åman, P.; Poutanen, K. Bran fermentation as a means to enhance technological properties and bioactivity of rye. Food Microbiol. 2007, 24, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Othman, N.B.; Roblain, D.; Chammen, N.; Thonart, P.; Hamdi, M. Antioxidant phenolic compounds loss during the fermentation of Chétoui olives. Food Chem. 2009, 116, 662–669. [Google Scholar] [CrossRef]
- Rai, A.K.; Jeyaram, K. Health benefits of functional proteins in fermented foods. In Health Benefits of Fermented Foods and Beverages; Tamang, J.P., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 455–474. [Google Scholar]
- Bhat, V.B.; Madyastha, K.M. Scavenging of peroxynitrite by phycocyanin and phycocyanobilin from Spirulina platensis: Protection against oxidative damage to DNA. Biochem. Biophys. Res. Commun. 2001, 285, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Petelinc, T.; Polak, T.; Jamnik, P. Insight into the molecular mechanisms of propolis activity using a subcellular proteomic approach. J. Agric. Food Chem. 2013, 61, 11502–11510. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.M.; Bravim, F.; Soares, J.; Nati, T.; Broach, J.R.; Fernandes, A.A.R.; Fernandes, P.M.B. High hydrostatic pressure upregulate central carbon metabolism genes in a distillery yeast strain. BMC Proc. 2014, 8, P207. [Google Scholar] [CrossRef]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta BBA Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Halprin, K.M.; Ohkawara, A. The measurement of glutathione in human epidermis using glutathione reductase. J. Investig. Dermatol. 1967, 48, 149–152. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, J.; Guo, R.; Li, J.; Yu, L.; Xu, D. Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 2010, 10, 199–208. [Google Scholar] [CrossRef]
- Radovic, S.; Rapisarda, V.A.; Tosato, V.; Bruschi, C.V. Functional and comparative characterization of Saccharomyces cerevisiae RVB1 and RVB2 genes with bacterial Ruv homologues. FEMS Yeast Res. 2007, 7, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. Yap1-dependent oxidative stress response provides a link to riboflavin production in Ashbya gossypii. Fungal Genet. Biol. 2012, 49, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Izawa, S.; Inoue, Y.; Kimura, A. Oxidative stress response in yeast: Effect of glutathione on adaptation to hydrogen peroxide stress in Saccharomyces ceerevisiae. FEBS Lett. 1995, 368, 73–76. [Google Scholar] [CrossRef]
- Ashoori, M.; Saedisomeolia, A. Riboflavin (vitamin B2) and oxidative stress: A review. Br. J. Nutr. 2014, 111, 1985–1991. [Google Scholar] [CrossRef]
- Choi, W.Y.; Kang, D.H.; Heo, S.-J.; Lee, H.Y. Enhancement of the neuroprotective effect of fermented Spirulina maxima associated with antioxidant activities by ultrasonic extraction. Appl. Sci. 2018, 8, 2469. [Google Scholar] [CrossRef]


| Gene Symbol | UniProt Accession No. | Protein Name | Stress Response Related Activity |
|---|---|---|---|
| GSH2 | Q08220 | Glutathione synthetase (Gsh2) | Glutathione biosynthesis |
| GND2 | P53319 | 6-phosphogluconate dehydrogenase, decarboxylating 2 (Gnd2) | Glutathione metabolism |
| PHO3 | P24031 | Constitutive acid phosphatase (Pho3) | Thiamine, riboflavin metabolism |
| RIB5 | P38145 | Riboflavin synthase (Rib5) | Riboflavin biosynthesis |
| YBL036C | P38197 | Pyridoxal phosphate homeostasis protein (Ybl036c) | Homeostatic regulation of the active form of vitamin B6 |
| SDH8 | P38345 | Succinate dehydrogenase assembly factor 4, Mitochondrial (Sdh8) | Response to reactive oxygen species |
| RPL13B | P40212 | 60S ribosomal protein L13-B (Rpl13b) | Autophagy |
| RPL14A | P36105 | 60S ribosomal protein L14-A (Rpl14a) | Autophagy |
| CUE5 | Q08412 | Ubiquitin-binding protein CUE5 (Cue5) | Autophagy |
| YPT1 | P01123 | GTP-binding protein YPT1 (Ypt1) | Autophagy |
| SHP1 | P34223 | UBX domain-containing protein 1 (Shp1) | Autophagosome assembly, proteasome-mediated ubiquitin-dependent protein catabolic process, piecemeal microautophagy of the nucleus |
| TUB2 | P02557 | Tubulin beta chain (Tub2) | Phagosome function |
| TUB1 | P09733 | Tubulin alpha-1 chain (Tub1) | Phagosome function |
| RVB1 | Q03940 | RuvB-like protein 1 (Rvb1) | DNA damage, DNA repair |
| CUB1 | Q08977 | Cu(2+) suppressing and bleomycin sensitive protein 1 (Cub1) | DNA repair and/or proteasome function |
| NAS2 | P40555 | Probable 26S proteasome regulatory subunit p27 (Nas2) | Proteasome regulatory complex assembly |
| NAS6 | P50086 | Probable 26S proteasome regulatory subunit p28 (Nas6) | Proteasome regulatory complex assembly |
| GUA1 | P38625 | GMP synthase (glutamine-hydrolyzing) (Gua1) | Purine biosynthesis |
| ADE13 | Q05911 | Adenylosuccinate lyase (Ade13) | Purine biosynthesis, AMP, IMP biosynthesis |
| URA6 | P15700 | Uridylate kinase (Ura6) | Pyrimidine biosynthesis |
| NDI1 | P32340 | Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial (Ndi1) | Positive regulation of the apoptotic process |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masten Rutar, J.; Cillero-Pastor, B.; Mohren, R.; Poklar Ulrih, N.; Ogrinc, N.; Jamnik, P. Insight into the Antioxidant Effect of Fermented and Non-Fermented Spirulina Water and Ethanol Extracts at the Proteome Level Using a Yeast Cell Model. Antioxidants 2021, 10, 1366. https://doi.org/10.3390/antiox10091366
Masten Rutar J, Cillero-Pastor B, Mohren R, Poklar Ulrih N, Ogrinc N, Jamnik P. Insight into the Antioxidant Effect of Fermented and Non-Fermented Spirulina Water and Ethanol Extracts at the Proteome Level Using a Yeast Cell Model. Antioxidants. 2021; 10(9):1366. https://doi.org/10.3390/antiox10091366
Chicago/Turabian StyleMasten Rutar, Jasmina, Berta Cillero-Pastor, Ronny Mohren, Nataša Poklar Ulrih, Nives Ogrinc, and Polona Jamnik. 2021. "Insight into the Antioxidant Effect of Fermented and Non-Fermented Spirulina Water and Ethanol Extracts at the Proteome Level Using a Yeast Cell Model" Antioxidants 10, no. 9: 1366. https://doi.org/10.3390/antiox10091366






