The Cancer Antioxidant Regulation System in Therapeutic Resistance
Abstract
1. Introduction
2. Overview of Therapeutic Resistance in Cancer
3. Antioxidant Defense System and Antioxidant-modulated Therapeutic Resistance
3.1. Antioxidant Transcription Factor Network in Cancer Therapeutic Resistance
3.1.1. KEAP1–NRF2–ARE Axis in Cancer Therapeutic Resistance
- Alterations at the Genetic Level.
- Alterations at the Transcriptional and Translational Levels.
- Alterations at the Posttranslational Modification Level.
- Alterations at the Protein Interaction Level.
3.1.2. Other Antioxidant Transcription Factors in Cancer Therapeutic Resistance
3.2. The GSH Antioxidant System in Cancer Therapeutic Resistance
3.2.1. The GSH Synthesis System in Cancer Therapeutic Resistance
- Enzymes Associated with the De Novo Synthesis of GSH.
- Transporters and Membrane Enzymes.
3.2.2. The GSH Salvage System in Cancer Therapeutic Resistance
3.3. The TRX Antioxidant System in Cancer Therapeutic Resistance
3.4. NADPH Antioxidant System in Cancer Therapeutic Resistance
3.4.1. Pentose Phosphate Pathway
3.4.2. IDH and ME
3.4.3. Other Pathways to Generate NADPH
- One-carbon Metabolism.
- De Novo Synthesis.
- Dehydrogenation of Glutamate.
- NNT.
4. Therapeutic Applications for Targeting the Antioxidant Regulation System
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef]
- Hecht, F.; Zocchi, M.; Alimohammadi, F.; Harris, I.S. Regulation of antioxidants in cancer. Mol. Cell 2024, 84, 23–33. [Google Scholar] [CrossRef]
- Cheung, E.C.; Vousden, K.H. The role of ROS in tumour development and progression. Nat. Rev. Cancer 2022, 22, 280–297. [Google Scholar] [CrossRef]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Zhou, X.; An, B.; Lin, Y.; Ni, Y.; Zhao, X.; Liang, X. Molecular mechanisms of ROS-modulated cancer chemoresistance and therapeutic strategies. Biomed. Pharmacother. 2023, 165, 115036. [Google Scholar] [CrossRef]
- Sies, H.; Belousov, V.V.; Chandel, N.S.; Davies, M.J.; Jones, D.P.; Mann, G.E.; Murphy, M.P.; Yamamoto, M.; Winterbourn, C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022, 23, 499–515. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative stress in cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, J.Q.; Assaraf, Y.G.; Ren, L.; Gupta, P.; Wei, L.; Ashby, C.R., Jr.; Yang, D.H.; Chen, Z.S. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018, 41, 1–25. [Google Scholar] [CrossRef]
- Hennekens, C.H.; Buring, J.E.; Manson, J.E.; Stampfer, M.; Rosner, B.; Cook, N.R.; Belanger, C.; LaMotte, F.; Gaziano, J.M.; Ridker, P.M.; et al. Lack of effect of long-term supplementation with beta carotene on the incidence of malignant neoplasms and cardiovascular disease. New Engl. J. Med. 1996, 334, 1145–1149. [Google Scholar] [CrossRef]
- Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. New Engl. J. Med. 1994, 330, 1029–1035. [Google Scholar] [CrossRef]
- Lippman, S.M.; Klein, E.A.; Goodman, P.J.; Lucia, M.S.; Thompson, I.M.; Ford, L.G.; Parnes, H.L.; Minasian, L.M.; Gaziano, J.M.; Hartline, J.A.; et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: The Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 2009, 301, 39–51. [Google Scholar] [CrossRef]
- Bairati, I.; Meyer, F.; Gélinas, M.; Fortin, A.; Nabid, A.; Brochet, F.; Mercier, J.P.; Têtu, B.; Harel, F.; Mâsse, B.; et al. A randomized trial of antioxidant vitamins to prevent second primary cancers in head and neck cancer patients. J. Natl. Cancer Inst. 2005, 97, 481–488. [Google Scholar] [CrossRef]
- Bairati, I.; Meyer, F.; Gélinas, M.; Fortin, A.; Nabid, A.; Brochet, F.; Mercier, J.P.; Têtu, B.; Harel, F.; Abdous, B.; et al. Randomized trial of antioxidant vitamins to prevent acute adverse effects of radiation therapy in head and neck cancer patients. J. Clin. Oncol. 2005, 23, 5805–5813. [Google Scholar] [CrossRef]
- Meyer, F.; Bairati, I.; Fortin, A.; Gélinas, M.; Nabid, A.; Brochet, F.; Têtu, B. Interaction between antioxidant vitamin supplementation and cigarette smoking during radiation therapy in relation to long-term effects on recurrence and mortality: A randomized trial among head and neck cancer patients. Int. J. Cancer 2008, 122, 1679–1683. [Google Scholar] [CrossRef]
- Ambrosone, C.B.; Zirpoli, G.R.; Hutson, A.D.; McCann, W.E.; McCann, S.E.; Barlow, W.E.; Kelly, K.M.; Cannioto, R.; Sucheston-Campbell, L.E.; Hershman, D.L.; et al. Dietary supplement use during chemotherapy and survival outcomes of patients with breast cancer enrolled in a cooperative group clinical trial (SWOG S0221). J. Clin. Oncol. 2020, 38, 804. [Google Scholar] [CrossRef]
- Bryan, R.T.; Pirrie, S.J.; Abbotts, B.; Maycock, S.; During, V.; Lewis, C.; Grant, M.; Bird, D.; Devall, A.J.; Wallace, D.M.A.; et al. Selenium and Vitamin E for Prevention of Non–Muscle-Invasive Bladder Cancer Recurrence and Progression: A Randomized Clinical Trial. JAMA Netw. Open 2023, 6, e2337494. [Google Scholar] [CrossRef]
- Rock, C.L.; Doyle, C.; Demark-Wahnefried, W.; Meyerhardt, J.; Courneya, K.S.; Schwartz, A.L.; Bandera, E.V.; Hamilton, K.K.; Grant, B.; McCullough, M.; et al. Nutrition and physical activity guidelines for cancer survivors. CA A Cancer J. Clin. 2012, 62, 242–274. [Google Scholar] [CrossRef]
- World Cancer Research Fund and American Institute for Cancer Research: Cancer Survivors: Evidence on Survivors of Breast and Other Cancers. Available online: https://www.wcrf.org/dietandcancer/cancer-survivors (accessed on 21 June 2024).
- Chandel, N.S.; Tuveson, D.A. The promise and perils of antioxidants for cancer patients. New Engl. J. Med. 2014, 371, 177–178. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Marzioni, D.; Mazzucchelli, R. Cellular modulators of the NRF2/KEAP1 signaling pathway in prostate cancer. Front. Biosci. 2023, 28, 143. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xue, L.; Li, H.; Fan, S.; van Hasselt, C.A.; Li, D.; Zeng, X.; Tong, M.C.F.; Chen, G.G. Targeting Nrf2 to treat thyroid cancer. Biomed. Pharmacother. 2024, 173, 116324. [Google Scholar] [CrossRef] [PubMed]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.C.; Baines, A.C.; Gong, Y.; Moore, R., Jr.; Pamuk, G.E.; Saber, H.; Subedee, A.; Thompson, M.D.; Xiao, W.; Pazdur, R.; et al. Trends in the approval of cancer therapies by the FDA in the twenty-first century. Nat. Rev. Drug Discov. 2023, 22, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.S.; Banerji, U. Combine and conquer: Challenges for targeted therapy combinations in early phase trials. Nat. Rev. Clin. Oncol. 2017, 14, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.M.; Tew, K.D. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 2003, 22, 7369–7375. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. New Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kurosu, T.; Kakihana, K.; Mizuchi, D.; Miura, O. The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem. Biophys. Res. Commun. 2004, 319, 1272–1275. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP–dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef]
- Pan, S.; Berk, B.C. Glutathiolation Regulates Tumor Necrosis Factor-α–Induced Caspase-3 Cleavage and Apoptosis: Key Role for Glutaredoxin in the Death Pathway. Circ. Res. 2007, 100, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Kaloni, D.; Diepstraten, S.T.; Strasser, A.; Kelly, G.L. BCL-2 protein family: Attractive targets for cancer therapy. Apoptosis 2023, 28, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Hooten, N.N.; Kompaniez, K.; Barnes, J.; Lohani, A.; Evans, M.K. Poly (ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1). J. Biol. Chem. 2011, 286, 44679–44690. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.R.; Bobola, M.S.; Blank, A.; Schoeler, K.D.; Haroldson, P.D.; Huynh, M.B.; Kolstoe, D.D. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin. Cancer Res. 2002, 8, 3008–3018. [Google Scholar]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef]
- Wu, Y.; Song, Y.; Wang, R.; Wang, T. Molecular mechanisms of tumor resistance to radiotherapy. Mol. Cancer 2023, 22, 96. [Google Scholar] [CrossRef]
- An, L.; Li, M.; Jia, Q. Mechanisms of radiotherapy resistance and radiosensitization strategies for esophageal squamous cell carcinoma. Mol. Cancer 2023, 22, 140. [Google Scholar] [CrossRef]
- Halliwell, B. Understanding mechanisms of antioxidant action in health and disease. Nat. Rev. Mol. Cell Biol. 2024, 25, 13–33. [Google Scholar] [CrossRef]
- Traber, M.G.; Head, B. Vitamin E: How much is enough, too much and why! Free Radic. Biol. Med. 2021, 177, 212–225. [Google Scholar] [CrossRef]
- Blaner, W.S.; Shmarakov, I.O.; Traber, M.G. Vitamin A and vitamin E: Will the real antioxidant please stand up? Annu. Rev. Nutr. 2021, 41, 105–131. [Google Scholar] [CrossRef]
- Padayatty, S.J.; Levine, M. Vitamin C: The known and the unknown and Goldilocks. Oral Dis. 2016, 22, 463–493. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. The antioxidants of human extracellular fluids. Arch. Biochem. Biophys. 1990, 280, 1–8. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Maywald, M.; Rink, L. Zinc in human health and infectious diseases. Biomolecules 2022, 12, 1748. [Google Scholar] [CrossRef]
- Reiter, R.J.; Mayo, J.C.; Tan, D.X.; Sainz, R.M.; Alatorre-Jimenez, M.; Qin, L. Melatonin as an antioxidant: Under promises but over delivers. J. Pineal Res. 2016, 61, 253–278. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- de la Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 2006, 21, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.; Campbell, J.; Brooks, A.; Berger, A.; Lee, W.; Chmielecki, J.; Beer, D.; Cope, L.; Creighton, C.; Danilova, L.; et al. Comprehensive molecular profiling of lung adenocarcinoma: The cancer genome atlas research network. Nature 2014, 511, 543–550. [Google Scholar]
- Ohta, T.; Iijima, K.; Miyamoto, M.; Nakahara, I.; Tanaka, H.; Ohtsuji, M.; Suzuki, T.; Kobayashi, A.; Yokota, J.; Sakiyama, T.; et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008, 68, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Aboulkassim, T.; Tian, X.; Liu, Q.; Qiu, D.; Hancock, M.; Wu, J.H.; Batist, G. A NRF2 inhibitor selectively sensitizes KEAP1 mutant tumor cells to cisplatin and gefitinib by restoring NRF2-inhibitory function of KEAP1 mutants. Cell Rep. 2023, 42, 113104. [Google Scholar] [CrossRef]
- Koppula, P.; Lei, G.; Zhang, Y.; Yan, Y.; Mao, C.; Kondiparthi, L.; Shi, J.; Liu, X.; Horbath, A.; Das, M.; et al. A targetable CoQ-FSP1 axis drives ferroptosis-and radiation-resistance in KEAP1 inactive lung cancers. Nat. Commun. 2022, 13, 2206. [Google Scholar] [CrossRef] [PubMed]
- Binkley, M.S.; Jeon, Y.J.; Nesselbush, M.; Moding, E.J.; Nabet, B.Y.; Almanza, D.; Kunder, C.; Stehr, H.; Yoo, C.H.; Rhee, S.; et al. KEAP1/NFE2L2 mutations predict lung cancer radiation resistance that can be targeted by glutaminase inhibition. Cancer Discov. 2020, 10, 1826–1841. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Arbour, K.C.; Lin, J.J.; Vajdi, A.; Vokes, N.; Hong, L.; Zhang, J.; Tolstorukov, M.Y.; Li, Y.Y.; Spurr, L.F.; et al. Diminished efficacy of programmed death-(ligand) 1 inhibition in STK11-and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J. Thorac. Oncol. 2022, 17, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Zavitsanou, A.M.; Pillai, R.; Hao, Y.; Wu, W.L.; Bartnicki, E.; Karakousi, T.; Rajalingam, S.; Herrera, A.; Karatza, A.; Rashidfarrokhi, A.; et al. KEAP1 mutation in lung adenocarcinoma promotes immune evasion and immunotherapy resistance. Cell Rep. 2023, 42. [Google Scholar] [CrossRef]
- Marzio, A.; Kurz, E.; Sahni, J.M.; Di Feo, G.; Puccini, J.; Jiang, S.; Hirsch, C.A.; Arbini, A.A.; Wu, W.L.; Pass, H.I.; et al. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell 2022, 185, 169–183. [Google Scholar] [CrossRef]
- Shibata, T.; Kokubu, A.; Gotoh, M.; Ojima, H.; Ohta, T.; Yamamoto, M.; Hirohashi, S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008, 135, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Spentzos, D.; Fountzilas, E.; Francoeur, N.; Sanisetty, S.; Grammatikos, A.P.; Hecht, J.L.; Cannistra, S.A. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011, 71, 5081–5089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Singh, A.; Yegnasubramanian, S.; Esopi, D.; Kombairaju, P.; Bodas, M.; Wu, H.; Bova, S.G.; Biswal, S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 2010, 9, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Shibazaki, M.; Kasai, S.; Yasuhira, S.; Watanabe, A.; Inoue, T.; Kageshita, Y.; Tsunoda, K.; Takahashi, K.; Akasaka, T.; et al. A somatic mutation of the KEAP1 gene in malignant melanoma is involved in aberrant NRF2 activation and an increase in intrinsic drug resistance. J. Investig. Dermatol. 2014, 134, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.A.; Arslan, E.; Bartels, M.; Michikawa, C.; Lindemann, A.; Tomczak, K.; Yu, W.; Sandulache, V.; Ma, W.; Shen, L.; et al. Dysregulation and epigenetic reprogramming of NRF2 signaling axis promote acquisition of cisplatin resistance and metastasis in head and neck squamous cell carcinoma. Clin. Cancer Res. 2023, 29, 1344–1359. [Google Scholar] [CrossRef] [PubMed]
- Wamsley, N.T.; Wilkerson, E.M.; Guan, L.; LaPak, K.M.; Schrank, T.P.; Holmes, B.J.; Sprung, R.W.; Gilmore, P.E.; Gerndt, S.P.; Jackson, R.S.; et al. Targeted proteomic quantitation of NRF2 signaling and predictive biomarkers in HNSCC. Mol. Cell. Proteom. 2023, 22, 100647. [Google Scholar] [CrossRef]
- Haque, E.; Śmiech, M.; Łuczyńska, K.; Bouchard, M.F.; Viger, R.; Kono, H.; Pierzchała, M.; Taniguchi, H. NRF2 DLG Domain Mutations Identified in Japanese Liver Cancer Patients Affect the Transcriptional Activity in HCC Cell Lines. Int. J. Mol. Sci. 2021, 22, 5296. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhou, X.; Yu, X.; Chen, X.; Hu, X.; Lu, J.; Zhao, H.; Cao, Q.; Gu, Y.; Yang, Y.; et al. High expression of nuclear NRF2 combined with NFE2L2 alterations predicts poor prognosis in esophageal squamous cell carcinoma patients. Mod. Pathol. 2022, 35, 929–937. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef]
- Liang, C.; Shi, S.; Liu, M.; Qin, Y.; Meng, Q.; Hua, J.; Ji, S.; Zhang, Y.; Yang, J.; Xu, J.; et al. PIN1 maintains redox balance via the c-Myc/NRF2 axis to counteract Kras-induced mitochondrial respiratory injury in pancreatic cancer cells. Cancer Res. 2019, 79, 133–145. [Google Scholar] [CrossRef]
- Liu, P.; Ma, D.; Wang, P.; Pan, C.; Fang, Q.; Wang, J. Nrf2 overexpression increases risk of high tumor mutation burden in acute myeloid leukemia by inhibiting MSH2. Cell Death Dis. 2021, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Muscarella, L.A.; Barbano, R.; D’Angelo, V.; Copetti, M.; Coco, M.; Balsamo, T.; la Torre, A.; Notarangelo, A.; Troiano, M.; Parisi, S.; et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 2011, 6, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Hanada, N.; Takahata, T.; Zhou, Q.; Ye, X.; Sun, R.; Itoh, J.; Ishiguro, A.; Kijima, H.; Mimura, J.; Itoh, K.; et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 2012, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, F.P.; Sparaneo, A.; Centra, F.; Trombetta, D.; Storlazzi, C.T.; Graziano, P.; Maiello, E.; Fazio, V.M.; Muscarella, L.A. Methylation density pattern of KEAP1 gene in lung cancer cell lines detected by quantitative methylation specific PCR and pyrosequencing. Int. J. Mol. Sci. 2019, 20, 2697. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Wu, B.; Yan, J.; Li, X.; Sun, H.; Zhou, D. A possible gene silencing mechanism: Hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem. Biophys. Res. Commun. 2012, 428, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Taheri, Z.; Aghdaei, H.A.; Irani, S.; Modarressi, M.H.; Zahra, N. Evaluation of the epigenetic demethylation of NRF2, a master transcription factor for antioxidant enzymes, in colorectal cancer. Rep. Biochem. Mol. Biol. 2020, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, L.; Tang, N.; Xu, Y.; Ye, X.; Shen, S.; Niu, X.; Lu, S.; Chen, Z. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS Lett. 2014, 588, 3000–3007. [Google Scholar] [CrossRef]
- Zhou, S.; Ye, W.; Zhang, Y.; Yu, D.; Shao, Q.; Liang, J.; Zhang, M. miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am. J. Transl. Res. 2016, 8, 2992. [Google Scholar]
- Yin, Y.; Liu, H.; Xu, J.; Shi, D.; Zhai, L.; Liu, B.; Wang, L.; Liu, G.; Qin, J. miR-144-3p regulates the resistance of lung cancer to cisplatin by targeting Nrf2. Oncol. Rep. 2018, 40, 3479–3488. [Google Scholar] [CrossRef]
- Eades, G.; Yang, M.; Yao, Y.; Zhang, Y.; Zhou, Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011, 286, 40725–40733. [Google Scholar] [CrossRef]
- Akdemir, B.; Nakajima, Y.; Inazawa, J.; Inoue, J. miR-432 induces NRF2 stabilization by directly targeting KEAP1. Mol. Cancer Res. 2017, 15, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.G.; Wang, M.F.; Cao, Y.B.; Li, D.; Li, R.Z.; Fan, X.X.; Khan, I.; Lai, H.L.; Zhang, Y.Z.; Hsiao, W.W.L.; et al. MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3’ UTR and predicts poor survival in non-small cell lung cancer. Cell Death Dis. 2019, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Bi, G.; Liang, J.; Zhao, M.; Zhang, H.; Jin, X.; Lu, T.; Zheng, Y.; Bian, Y.; Chen, Z.; Huang, Y.; et al. miR-6077 promotes cisplatin/pemetrexed resistance in lung adenocarcinoma via CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol. Ther. Nucleic Acids 2022, 28, 366–386. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.J.; Li, F.F.; Xie, Y.X.; Fan, C.Y.; Liu, W.J.; Qiu, J.G.; Jiang, B.H. miR-196a upregulation contributes to gefitinib resistance through inhibiting GLTP expression. Int. J. Mol. Sci. 2022, 23, 1785. [Google Scholar] [CrossRef] [PubMed]
- Ooi, A.; Wong, J.C.; Petillo, D.; Roossien, D.; Perrier-Trudova, V.; Whitten, D.; Min, B.W.H.; Tan, M.H.; Zhang, Z.; Yang, X.J.; et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011, 20, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.; Higgins, M.; Hams, E.; et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.; Hong, M.; Lee, S.; Kumar, M.; Ibrahim, L.; Nutsch, K.; Stanton, C.; Sondermann, P.; Sandoval, B.; Bulos, M.L.; et al. S-lactoyl modification of KEAP1 by a reactive glycolytic metabolite activates NRF2 signaling. Proc. Natl. Acad. Sci. 2023, 120, e2300763120. [Google Scholar] [CrossRef]
- Liu, T.; Lv, Y.F.; Zhao, J.L.; You, Q.D.; Jiang, Z.Y. Regulation of Nrf2 by phosphorylation: Consequences for biological function and therapeutic implications. Free Radic. Biol. Med. 2021, 168, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Sun, Z.; Wang, X.J.; Jiang, T.; Huang, Z.; Fang, D.; Zhang, D.D. Direct interaction between Nrf2 and p21Cip1/WAF1 upregulates the Nrf2-mediated antioxidant response. Mol. Cell 2009, 34, 663–673. [Google Scholar] [CrossRef]
- Jana, S.; Patra, K.; Jana, J.; Mandal, D.P.; Bhattacharjee, S. Nrf-2 transcriptionally activates P21Cip/WAF1 and promotes A549 cell survival against oxidative stress induced by H2O2. Chem. Biol. Interact. 2018, 285, 59–68. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Hast, B.E.; Goldfarb, D.; Mulvaney, K.M.; Hast, M.A.; Siesser, P.F.; Yan, F.; Hayes, D.N.; Major, M.B. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013, 73, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Kumari, S.; Kadian, L.; Anupa, G.; Singh, J.; Kumar, A.; Verma, D.; Pramanik, R.; Kumar, S.; Yadav, R.; et al. Involvement of DPP3 in modulating oncological features and oxidative stress response in esophageal squamous cell carcinoma. Biosci. Rep. 2023, 43, BSR20222472. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhao, D.; Li, H.; Zhang, W.; Lin, Q.; Wang, X.; Zheng, S.; Zhang, L.; Li, L.; Hu, S.; et al. Blocking iASPP/Nrf2/M-CSF axis improves anti-cancer effect of chemotherapy-induced senescence by attenuating M2 polarization. Cell Death Dis. 2022, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Gorrini, C.; Baniasadi, P.S.; Harris, I.S.; Silvester, J.; Inoue, S.; Snow, B.; Joshi, P.A.; Wakeham, A.; Molyneux, S.D.; Martin, B.; et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med. 2013, 210, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Mitsuishi, Y.; Motohashi, H.; Yamamoto, M. The Keap1–Nrf2 system in cancers: Stress response and anabolic metabolism. Front. Oncol. 2012, 2, 200. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Hakomäki, H.; Levonen, A.L. Electrophilic metabolites targeting the KEAP1/NRF2 partnership. Curr. Opin. Chem. Biol. 2024, 78, 102425. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Mei, H.; Yin, Z.; Geng, Y.; Zhang, T.; Wu, G.; Lin, Z. The BET bromodomain inhibitor JQ1 radiosensitizes non-small cell lung cancer cells by upregulating p21. Cancer Lett. 2017, 391, 141–151. [Google Scholar] [CrossRef]
- Tang, Y.; Cui, Y.; Li, Z.; Jiao, Z.; Zhang, Y.; He, Y.; Chen, G.; Zhou, Q.; Wang, W.; Zhou, X.; et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 1–14. [Google Scholar]
- Xiao, G.G.; Wang, M.; Li, N.; Loo, J.A.; Nel, A.E. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J. Biol. Chem. 2003, 278, 50781–50790. [Google Scholar] [CrossRef] [PubMed]
- Maillet, A.; Pervaiz, S. Redox regulation of p53, redox effectors regulated by p53: A subtle balance. Antioxid. Redox Signal. 2012, 16, 1285–1294. [Google Scholar] [CrossRef] [PubMed]
- Krishnaraj, J.; Yamamoto, T.; Ohki, R. p53-dependent cytoprotective mechanisms behind resistance to chemo-radiotherapeutic agents used in cancer treatment. Cancers 2023, 15, 3399. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- NF-κB Target Genes. Available online: https://www.bu.edu/nf-kb/gene-resources/target-genes/ (accessed on 21 June 2024).
- Yang, L.; Zhou, Y.; Li, Y.; Zhou, J.; Wu, Y.; Cui, Y.; Yang, G.; Hong, Y. Mutations of p53 and KRAS activate NF-κB to promote chemoresistance and tumorigenesis via dysregulation of cell cycle and suppression of apoptosis in lung cancer cells. Cancer Lett. 2015, 357, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, G.; Feng, M.; Zheng, S.; Cao, Z.; Qiu, J.; You, L.; Zheng, L.; Hu, Y.; Zhang, T.; et al. NF-κB in pancreatic cancer: Its key role in chemoresistance. Cancer Lett. 2018, 421, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Godwin, P.; Baird, A.M.; Heavey, S.; Barr, M.; O’byrne, K.; Gately, K. Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front. Oncol. 2013, 3, 120. [Google Scholar] [CrossRef] [PubMed]
- Bejjani, F.; Evanno, E.; Zibara, K.; Piechaczyk, M.; Jariel-Encontre, I. The AP-1 transcriptional complex: Local switch or remote command? Biochim. Biophys. Acta (BBA) Rev. Cancer 2019, 1872, 11–23. [Google Scholar] [CrossRef]
- Soriano, F.X.; Baxter, P.; Murray, L.M.; Sporn, M.B.; Gillingwater, T.H.; Hardingham, G.E. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol. Cells 2009, 27, 279–282. [Google Scholar] [CrossRef]
- Lae Lae Phoo, N.; Sukhamwang, A.; Dejkriengkraikul, P.; Yodkeeree, S. Diclofenac sensitizes signet ring cell gastric carcinoma cells to cisplatin by activating autophagy and inhibition of survival signal pathways. Int. J. Mol. Sci. 2022, 23, 12066. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Sandoval, J.M.; Fattaccioli, A.; Dejeans, N.; Garbe, J.C.; Dieu, M.; Verrax, J.; Renard, P.; Huang, P.; Calderon, P.B. Chromatin remodeling regulates catalase expression during cancer cells adaptation to chronic oxidative stress. Free Radic. Biol. Med. 2016, 99, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.O.; Sánchez-Ramos, C.; Prieto-Arroyo, I.; Urbánek, P.; Steinbrenner, H.; Monsalve, M. Redox regulation of FoxO transcription factors. Redox Biol. 2015, 6, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Harris, I.; Blaser, H.; Moreno, J.; Treloar, A.; Gorrini, C.; Sasaki, M.; Mason, J.; Knobbe, C.; Rufini, A.; Halle, M.; et al. PTPN12 promotes resistance to oxidative stress and supports tumorigenesis by regulating FOXO signaling. Oncogene 2014, 33, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, H.; Matsumoto, M.; Shindo, T.; Saigusa, D.; Kato, H.; Suzuki, K.; Sato, M.; Ishii, Y.; Shimokawa, H.; Igarashi, K. Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J. Biol. Chem. 2020, 295, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, A.L.; Loi, G.; Ivaldo, C.; Passalacqua, M.; Pietra, G.; Mann, G.E.; Nitti, M. HO-1 limits the efficacy of vemurafenib/PLX4032 in BRAFV600E mutated melanoma cells adapted to physiological normoxia or hypoxia. Antioxidants 2022, 11, 1171. [Google Scholar] [CrossRef] [PubMed]
- Paku, M.; Haraguchi, N.; Takeda, M.; Fujino, S.; Ogino, T.; Takahashi, H.; Miyoshi, N.; Uemura, M.; Mizushima, T.; Yamamoto, H.; et al. SIRT3-mediated SOD2 and PGC-1α contribute to chemoresistance in colorectal cancer cells. Ann. Surg. Oncol. 2021, 28, 4720–4732. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Su, W.; Wei, X.; Qu, S.; Zhao, D.; Zhou, J.; Wang, Y.; Guan, Q.; Qin, C.; Xiang, J.; et al. HIF-1α drives resistance to ferroptosis in solid tumors by promoting lactate production and activating SLC1A1. Cell Rep. 2023, 42, 112945. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.W.; Chhoy, P.; Mukhopadhyay, D.; Karner, E.R.; Mercurio, A.M. Targeting prominin2 transcription to overcome ferroptosis resistance in cancer. EMBO Mol. Med. 2021, 13, e13792. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of glutathione in cancer: From mechanisms to therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Liao, K.; Zhou, Y.; Wen, T.; Quan, G.; Pan, X.; Wu, C. Application of glutathione depletion in cancer therapy: Enhanced ROS-based therapy, ferroptosis, and chemotherapy. Biomaterials 2021, 277, 121110. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhou, C.; Ma, Q.; Chen, W.; Atyah, M.; Yin, Y.; Fu, P.; Liu, S.; Hu, B.; Ren, N.; et al. High GCLC level in tumor tissues is associated with poor prognosis of hepatocellular carcinoma after curative resection. J. Cancer 2019, 10, 3333. [Google Scholar] [CrossRef] [PubMed]
- Anderton, B.; Camarda, R.; Balakrishnan, S.; Balakrishnan, A.; Kohnz, R.A.; Lim, L.; Evason, K.J.; Momcilovic, O.; Kruttwig, K.; Huang, Q.; et al. MYC-driven inhibition of the glutamate-cysteine ligase promotes glutathione depletion in liver cancer. EMBO Rep. 2017, 18, 569–585. [Google Scholar] [CrossRef] [PubMed]
- Vaziri-Gohar, A.; Hue, J.J.; Abbas, A.; Graor, H.J.; Hajihassani, O.; Zarei, M.; Titomihelakis, G.; Feczko, J.; Rathore, M.; Chelstowska, S.; et al. Increased glucose availability sensitizes pancreatic cancer to chemotherapy. Nat. Commun. 2023, 14, 3823. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Hua, K.T.; Li, K.C.; Kao, H.F.; Hong, R.L.; Ko, J.Y.; Hsiao, M.; Kuo, M.L.; Tan, C.T. Histone methyltransferase G9a drives chemotherapy resistance by regulating the glutamate–cysteine ligase catalytic subunit in head and neck squamous cell carcinoma. Mol. Cancer Ther. 2017, 16, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Z.X.; Chen, Y.X.; Wu, H.X.; Yin, L.; Zhao, Q.; Luo, H.Y.; Zeng, Z.L.; Qiu, M.Z.; Xu, R.H. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.C.; Chen, C.F.; Ho, C.T.; Liu, J.J.; Liu, T.Z.; Chern, C.L. γ-Glutamylcysteine synthetase (γ-GCS) as a target for overcoming chemo-and radio-resistance of human hepatocellular carcinoma cells. Life Sci. 2018, 198, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.L.; Lin, J.; Ye, Y.; Wu, W.J.; Lin, H.H.; Wei, H.; Huang, M.; Chang, D.W.; Dinney, C.P.; Wu, X. Genetic variations in glutathione pathway genes predict cancer recurrence in patients treated with transurethral resection and bacillus calmette–guerin instillation for non-muscle invasive bladder cancer. Ann. Surg. Oncol. 2015, 22, 4104–4110. [Google Scholar] [CrossRef][Green Version]
- Li, H.; Stokes, W.; Chater, E.; Roy, R.; De Bruin, E.; Hu, Y.; Liu, Z.; Smit, E.F.; Heynen, G.J.; Downward, J.; et al. Decreased glutathione biosynthesis contributes to EGFR T790M-driven erlotinib resistance in non-small cell lung cancer. Cell Discov. 2016, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Ma, Z.; Xu, B.; Li, S.; Yao, Z.; Liang, B.; Wang, J.; Liao, W.; Lin, L.; Wang, C.; et al. Glutamine metabolic microenvironment drives M2 macrophage polarization to mediate trastuzumab resistance in HER2-positive gastric cancer. Cancer Commun. 2023, 43, 909–937. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yin, Y.; Li, Y.; Chen, X.; Chang, Y.; Zhang, H.; Liu, J.; Beasley, J.; McCaw, P.; Zhang, H.; et al. A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2012748118. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M.; et al. The cystine/glutamate antiporter system xc- in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 2013, 18, 522–555. [Google Scholar] [CrossRef]
- Ma, M.Z.; Chen, G.; Wang, P.; Lu, W.H.; Zhu, C.F.; Song, M.; Yang, J.; Wen, S.; Xu, R.H.; Hu, Y.; et al. Xc- inhibitor sulfasalazine sensitizes colorectal cancer to cisplatin by a GSH-dependent mechanism. Cancer Lett. 2015, 368, 88–96. [Google Scholar] [CrossRef]
- Song, Y.; Jang, J.; Shin, T.H.; Bae, S.M.; Kim, J.S.; Kim, K.M.; Myung, S.J.; Choi, E.K.; Seo, H.R. Sulfasalazine attenuates evading anticancer response of CD133-positive hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2017, 36, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Wang, X.; Tian, H.; Wang, Y.; Jin, J.; Shan, Z.; Liu, Y.; Cai, Z.; Tong, X.; et al. Stem cell factor SOX2 confers ferroptosis resistance in lung cancer via upregulation of SLC7A11. Cancer Res. 2021, 81, 5217–5229. [Google Scholar] [CrossRef]
- Conti, L.; Bolli, E.; Di Lorenzo, A.; Franceschi, V.; Macchi, F.; Riccardo, F.; Ruiu, R.; Russo, L.; Quaglino, E.; Donofrio, G.; et al. Immunotargeting of the xCT cystine/glutamate antiporter potentiates the efficacy of HER2-targeted immunotherapies in breast cancer. Cancer Immunol. Res. 2020, 8, 1039–1053. [Google Scholar] [CrossRef]
- Wang, Z.; Ouyang, L.; Liu, N.; Li, T.; Yan, B.; Mao, C.; Xiao, D.; Gan, B.; Liu, S.; Tao, Y. The DUBA-SLC7A11-c-Myc axis is critical for stemness and ferroptosis. Oncogene 2023, 42, 2688–2700. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Tsuchihashi, K.; Ishimoto, T.; Yae, T.; Motohara, T.; Sugihara, E.; Onishi, N.; Masuko, T.; Yoshizawa, K.; Kawashiri, S.; et al. xCT inhibition depletes CD44v-expressing tumor cells that are resistant to EGFR-targeted therapy in head and neck squamous cell carcinoma. Cancer Res. 2013, 73, 1855–1866. [Google Scholar] [CrossRef]
- Miyoshi, S.; Tsugawa, H.; Matsuzaki, J.; Hirata, K.; Mori, H.; Saya, H.; Kanai, T.; Suzuki, H. Inhibiting xCT improves 5-fluorouracil resistance of gastric cancer induced by CD44 variant 9 expression. Anticancer Res. 2018, 38, 6163–6170. [Google Scholar] [CrossRef]
- Zhang, K.R.; Zhang, Y.F.; Lei, H.M.; Tang, Y.B.; Ma, C.S.; Lv, Q.M.; Wang, S.Y.; Lu, L.M.; Shen, Y.; Chen, H.Z.; et al. Targeting AKR1B1 inhibits glutathione de novo synthesis to overcome acquired resistance to EGFR-targeted therapy in lung cancer. Sci. Transl. Med. 2021, 13, eabg6428. [Google Scholar] [CrossRef] [PubMed]
- Fantone, S.; Piani, F.; Olivieri, F.; Rippo, M.R.; Sirico, A.; Di Simone, N.; Marzioni, D.; Tossetta, G. Role of SLC7A11/xCT in Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 587. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, X.; Huang, P. xCT: A critical molecule that links cancer metabolism to redox signaling. Mol. Ther. 2020, 28, 2358–2366. [Google Scholar] [CrossRef]
- Sleire, L.; Skeie, B.; Netland, I.; Førde, H.; Dodoo, E.; Selheim, F.; Leiss, L.; Heggdal, J.; Pedersen, P.; Wang, J.; et al. Drug repurposing: Sulfasalazine sensitizes gliomas to gamma knife radiosurgery by blocking cystine uptake through system Xc-, leading to glutathione depletion. Oncogene 2015, 34, 5951–5959. [Google Scholar] [CrossRef]
- Feng, L.; Zhao, K.; Sun, L.; Yin, X.; Zhang, J.; Liu, C.; Li, B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021, 19, 367. [Google Scholar] [CrossRef]
- Corti, A.; Belcastro, E.; Dominici, S.; Maellaro, E.; Pompella, A. The dark side of gamma-glutamyltransferase (GGT): Pathogenic effects of an ‘antioxidant’enzyme. Free Radic. Biol. Med. 2020, 160, 807–819. [Google Scholar] [CrossRef]
- Daubeuf, S.; Leroy, P.; Paolicchi, A.; Pompella, A.; Wellman, M.; Galteau, M.M.; Visvikis, A. Enhanced resistance of HeLa cells to cisplatin by overexpression of γ-glutamyltransferase. Biochem. Pharmacol. 2002, 64, 207–216. [Google Scholar] [CrossRef]
- Paolicchi, A.; Sotiropuolou, M.; Perego, P.; Daubeuf, S.; Visvikis, A.; Lorenzini, E.; Franzini, M.; Romiti, N.; Chieli, E.; Leone, R.; et al. γ-Glutamyl transpeptidase catalyses the extracellular detoxification of cisplatin in a human cell line derived from the proximal convoluted tubule of the kidney. Eur. J. Cancer 2003, 39, 996–1003. [Google Scholar] [CrossRef]
- Jerremalm, E.; Wallin, I.; Yachnin, J.; Ehrsson, H. Oxaliplatin degradation in the presence of important biological sulphur-containing compounds and plasma ultrafiltrate. Eur. J. Pharm. Sci. 2006, 28, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Pompella, A.; Corti, A.; Visvikis, A. Redox mechanisms in Cisplatin resistance of cancer cells: The twofold role of Gamma-Glutamyltransferase 1 (GGT1). Front. Oncol. 2022, 12, 920316. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; He, S.; He, S.; Wang, Y. Fighting against drug-resistant tumors by the inhibition of γ-glutamyl transferase with supramolecular platinum prodrug nano-assemblies. J. Mater. Chem. B 2021, 9, 4587–4595. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, X.; Gou, S. A cisplatin-based platinum (IV) prodrug containing a glutathione s-transferase inhibitor to reverse cisplatin-resistance in non-small cell lung cancer. J. Inorg. Biochem. 2019, 193, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hayashima, K.; Katoh, H. Expression of gamma-glutamyltransferase 1 in glioblastoma cells confers resistance to cystine deprivation–induced ferroptosis. J. Biol. Chem. 2022, 298. [Google Scholar] [CrossRef]
- Yoo, H.C.; Park, S.J.; Nam, M.; Kang, J.; Kim, K.; Yeo, J.H.; Kim, J.K.; Heo, Y.; Lee, H.S.; Lee, M.Y.; et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 2020, 31, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Law, Y.Y.; Huang, Y.W.; Tran, N.B.; Lin, C.Y.; Lai, C.Y.; Huang, Y.L.; Tsai, C.H.; Ko, C.Y.; Chou, M.C.; et al. Glutamine metabolism controls amphiregulin-facilitated chemoresistance to cisplatin in human chondrosarcoma. Int. J. Biol. Sci. 2023, 19, 5174. [Google Scholar] [CrossRef] [PubMed]
- Najumudeen, A.K.; Ceteci, F.; Fey, S.K.; Hamm, G.; Steven, R.T.; Hall, H.; Nikula, C.J.; Dexter, A.; Murta, T.; Race, A.M.; et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat. Genet. 2021, 53, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhou, R.; Rao, X.; Hong, J.; Li, Q.; Jie, X.; Wang, J.; Xu, Y.; Zhu, K.; Li, Z.; et al. Activated amino acid response pathway generates apatinib resistance by reprograming glutamine metabolism in non-small-cell lung cancer. Cell Death Dis. 2022, 13, 636. [Google Scholar] [CrossRef]
- Wen, Q.; Huang, M.; Xie, J.; Liu, R.; Miao, Q.; Huang, J.; Zhang, J.; Qi, M.; Wu, C.; Qi, Q.; et al. lncRNA SYTL5-OT4 promotes vessel co-option by inhibiting the autophagic degradation of ASCT2. Drug Resist. Updat. 2023, 69, 100975. [Google Scholar] [CrossRef]
- Nachef, M.; Ali, A.K.; Almutairi, S.M.; Lee, S.H. Targeting SLC1A5 and SLC3A2/SLC7A5 as a potential strategy to strengthen anti-tumor immunity in the tumor microenvironment. Front. Immunol. 2021, 12, 624324. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Li, L.; Peng, H.; Liu, L.; Heymann, D.; Robert, C.; Vallette, F.; Shen, S. Drug-tolerant persister cells in cancer: The cutting edges and future directions. Nat. Rev. Clin. Oncol. 2023, 20, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tan, Y.; Huang, C.; Wei, X. Redox signaling in drug-tolerant persister cells as an emerging therapeutic target. EBioMedicine 2023, 89, 104483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qin, S.; Chen, Y.; Zhou, L.; Yang, M.; Tang, Y.; Zuo, J.; Zhang, J.; Mizokami, A.; Nice, E.C.; et al. Inhibition of NPC1L1 disrupts adaptive responses of drug-tolerant persister cells to chemotherapy. EMBO Mol. Med. 2022, 14, e14903. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Viswanathan, V.S.; Ryan, M.J.; Bole, D.; Eaton, J.K.; Matov, A.; Galeas, J.; Dhruv, H.D.; Berens, M.E.; Schreiber, S.L.; et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017, 551, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.S.; Ryan, M.J.; Dhruv, H.D.; Gill, S.; Eichhoff, O.M.; Seashore-Ludlow, B.; Kaffenberger, S.D.; Eaton, J.K.; Shimada, K.; Aguirre, A.J.; et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017, 547, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Liu, X.; Zhang, Y.; Lei, G.; Yan, Y.; Lee, H.; Koppula, P.; Wu, S.; Zhuang, L.; Fang, B.; et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021, 593, 586–590. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, L.; Shang, W.; Yang, Z.; Li, T.; Liu, F.; Shao, W.; Lv, L.; Chai, L.; Qu, L.; et al. Wnt/beta-catenin signaling confers ferroptosis resistance by targeting GPX4 in gastric cancer. Cell Death Differ. 2022, 29, 2190–2202. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, X.; Sun, W.; Xu, F.; Kou, H.; Hu, W.; Zhang, Y.; Jiang, Q.; Tang, J.; Xu, Y. RelB-activated GPX4 inhibits ferroptosis and confers tamoxifen resistance in breast cancer. Redox Biol. 2023, 68, 102952. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, H.L.; Li, J.; Ye, Z.P.; Du, T.; Li, L.C.; Guo, Y.Q.; Yang, D.; Li, Z.L.; Cao, J.H.; et al. Tubastatin A potently inhibits GPX4 activity to potentiate cancer radiotherapy through boosting ferroptosis. Redox Biol. 2023, 62, 102677. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiao, Y.; Ding, J.H.; Jin, X.; Ma, D.; Li, D.Q.; Shi, J.X.; Huang, W.; Wang, Y.P.; Jiang, Y.Z.; et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023, 35, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Hassannia, B.; Vandenabeele, P.; Berghe, T.V. Targeting ferroptosis to iron out cancer. Cancer Cell 2019, 35, 830–849. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Carlisle, A.E.; Peppers, A.; Park, S.J.; Doshi, M.B.; Spears, M.E.; Kim, D. xCT-driven expression of GPX4 determines sensitivity of breast cancer cells to ferroptosis inducers. Antioxidants 2021, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.A.; Kalinina, E.; Nuzhina, J.; Volodina, Y.; Shtil, A.; Tatarskiy, V. Potentiation of cisplatin cytotoxicity in resistant ovarian cancer SKOV3/cisplatin cells by quercetin pre-treatment. Int. J. Mol. Sci. 2023, 24, 10960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, Y.; He, Y.; Xiao, Y.; Wang, Q.; Zhao, Y.; Zhang, T.; Wu, C.; Xie, Y.; Zhou, J.; et al. GPX1-associated prognostic signature predicts poor survival in patients with acute myeloid leukemia and involves in immunosuppression. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2022, 1868, 166268. [Google Scholar] [CrossRef]
- Lei, F.J.; Chiang, J.Y.; Chang, H.J.; Chen, D.C.; Wang, H.L.; Yang, H.A.; Wei, K.Y.; Huang, Y.C.; Wang, C.C.; Wei, S.T.; et al. Cellular and exosomal GPx1 are essential for controlling hydrogen peroxide balance and alleviating oxidative stress in hypoxic glioblastoma. Redox Biol. 2023, 65, 102831. [Google Scholar] [CrossRef]
- Meng, Q.; Shi, S.; Liang, C.; Liang, D.; Hua, J.; Zhang, B.; Xu, J.; Yu, X. Abrogation of glutathione peroxidase-1 drives EMT and chemoresistance in pancreatic cancer by activating ROS-mediated Akt/GSK3β/Snail signaling. Oncogene 2018, 37, 5843–5857. [Google Scholar] [CrossRef]
- Feng, Q.; Li, X.; Sun, W.; Sun, M.; Li, Z.; Sheng, H.; Xie, F.; Zhang, S.; Shan, C. Targeting G6PD reverses paclitaxel resistance in ovarian cancer by suppressing GSTP1. Biochem. Pharmacol. 2020, 178, 114092. [Google Scholar] [CrossRef]
- Simic, P.; Pljesa, I.; Nejkovic, L.; Jerotic, D.; Coric, V.; Stulic, J.; Kokosar, N.; Popov, D.; Savic-Radojevic, A.; Pazin, V.; et al. Glutathione transferase P1: Potential therapeutic target in ovarian cancer. Medicina 2022, 58, 1660. [Google Scholar] [CrossRef] [PubMed]
- Laborde, E. Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ. 2010, 17, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Chen, N.; Feng, Z.; Luo, L.; Yin, Z. GSTP1 negatively regulates Stat3 activation in epidermal growth factor signaling. Oncol. Lett. 2013, 5, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Tew, K.D. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994, 54, 4313–4320. [Google Scholar] [CrossRef] [PubMed]
- Stefan, S.M.; Wiese, M. Small-molecule inhibitors of multidrug resistance-associated protein 1 and related processes: A historic approach and recent advances. Med. Res. Rev. 2019, 39, 176–264. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.L.; Wu, X.H.; Chen, C.; Wang, K.; Huang, L.Y.; Xia, J.; Liu, Y.; Shan, X.F.; Tang, N. SLC27A5 promotes sorafenib-induced ferroptosis in hepatocellular carcinoma by downregulating glutathione reductase. Cell Death Dis. 2023, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.D.; Ferrarone, J.; Liu, A.; Brandstädter, C.; Munuganti, R.; Farnsworth, D.A.; Lu, D.; Luu, J.; Sihota, T.; Jansen, S.; et al. Characterization of a small molecule inhibitor of disulfide reductases that induces oxidative stress and lethality in lung cancer cells. Cell Rep. 2022, 38, 110343. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, X.; Wang, L.; Zhang, R.; Pu, X.; Wu, S.; Li, L.; Tong, P.; Wang, J.; Meng, Q.H.; et al. Inhibition of thioredoxin/thioredoxin reductase induces synthetic lethality in lung cancers with compromised glutathione homeostasis. Cancer Res. 2019, 79, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, M.; Podolski-Renić, A.; Krasavin, M.; Pešić, M. The role of the thioredoxin detoxification system in cancer progression and resistance. Front. Mol. Biosci. 2022, 9, 883297. [Google Scholar] [CrossRef]
- Patwari, P.; Higgins, L.J.; Chutkow, W.A.; Yoshioka, J.; Lee, R.T. The interaction of thioredoxin with Txnip: Evidence for formation of a mixed disulfide by disulfide exchange. J. Biol. Chem. 2006, 281, 21884–21891. [Google Scholar] [CrossRef]
- Lee, T.H.; Jin, J.O.; Yu, K.J.; Kim, H.S.; Lee, P.C.W. Inhibition of peroxiredoxin 2 suppresses Wnt/β-catenin signaling in gastric cancer. Biochem. Biophys. Res. Commun. 2019, 512, 250–255. [Google Scholar] [CrossRef]
- Hao, J.; Song, Z.; Su, J.; Li, L.; Zou, L.; Zou, K. The PRX-1/TLR4 axis promotes hypoxia-induced radiotherapy resistance in non-small cell lung cancer by targeting the NF-κB/p65 pathway. Cell. Signal. 2023, 110, 110806. [Google Scholar] [CrossRef]
- Peng, L.; Jiang, J.; Chen, H.N.; Zhou, L.; Huang, Z.; Qin, S.; Jin, P.; Luo, M.; Li, B.; Shi, J.; et al. Redox-sensitive cyclophilin A elicits chemoresistance through realigning cellular oxidative status in colorectal cancer. Cell Rep. 2021, 37, 110069. [Google Scholar] [CrossRef]
- Cerda, M.B.; Lloyd, R.; Batalla, M.; Giannoni, F.; Casal, M.; Policastro, L. Silencing peroxiredoxin-2 sensitizes human colorectal cancer cells to ionizing radiation and oxaliplatin. Cancer Lett. 2017, 388, 312–319. [Google Scholar] [CrossRef]
- Song, I.S.; Jeong, Y.J.; Jung, Y.; Park, Y.H.; Shim, S.; Kim, S.J.; Eom, D.W.; Hong, S.M.; Lee, P.C.; Kim, S.U.; et al. The sulfiredoxin-peroxiredoxin redox system regulates the stemness and survival of colon cancer stem cells. Redox Biol. 2021, 48, 102190. [Google Scholar] [CrossRef]
- Klopotowska, M.; Bajor, M.; Graczyk-Jarzynka, A.; Kraft, A.; Pilch, Z.; Zhylko, A.; Firczuk, M.; Baranowska, I.; Lazniewski, M.; Plewczynski, D.; et al. PRDX-1 supports the survival and antitumor activity of primary and CAR-modified NK cells under oxidative stress. Cancer Immunol. Res. 2022, 10, 228–244. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, S.; Li, Y.; Liu, Z.; Mi, L.; Cai, Y.; Wang, X.; Chen, L.; Ran, H.; Xiao, D.; et al. Suppression of mitochondrial ROS by prohibitin drives glioblastoma progression and therapeutic resistance. Nat. Commun. 2021, 12, 3720. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, Y.; Liu, Y.; Zhou, J.; Wang, X.; Zheng, H.; Xu, Z.; Zhang, J.; Zhou, Y.; Qian, L.; et al. ent-Kaurane diterpenoids induce apoptosis and ferroptosis through targeting redox resetting to overcome cisplatin resistance. Redox Biol. 2021, 43, 101977. [Google Scholar] [CrossRef]
- Peng, L.; Xiong, Y.; Wang, R.; Xiang, L.; Zhou, H.; Fu, Z. The critical role of peroxiredoxin-2 in colon cancer stem cells. Aging 2021, 13, 11170. [Google Scholar] [CrossRef]
- Ding, N.; Jiang, H.; Thapa, P.; Hao, Y.; Alshahrani, A.; Allison, D.; Izumi, T.; Rangnekar, V.M.; Liu, X.; Wei, Q. Peroxiredoxin IV plays a critical role in cancer cell growth and radioresistance through the activation of the Akt/GSK3 signaling pathways. J. Biol. Chem. 2022, 298. [Google Scholar] [CrossRef]
- Wang, R.; Mi, Y.; Ni, J.; Wang, Y.; Ding, L.; Ran, X.; Sun, Q.; Tan, S.Y.; Koeffler, H.P.; Feng, N.; et al. Identification of PRDX5 as A Target for The Treatment of Castration-Resistant Prostate Cancer. Adv. Sci. 2024, 11, 2304939. [Google Scholar] [CrossRef] [PubMed]
- Salovska, B.; Kondelova, A.; Pimkova, K.; Liblova, Z.; Pribyl, M.; Fabrik, I.; Bartek, J.; Vajrychova, M.; Hodny, Z. Peroxiredoxin 6 protects irradiated cells from oxidative stress and shapes their senescence-associated cytokine landscape. Redox Biol. 2022, 49, 102212. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, P.; Hu, W.; Chen, D. New insights into the roles of peroxiredoxins in cancer. Biomed. Pharmacother. 2023, 164, 114896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Zhao, Z.; Cai, W.; Fang, J. Thioredoxin signaling pathways in cancer. Antioxid. Redox Signal. 2023, 38, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Guo, Q.; Luo, Z.; Wang, Y.; Weng, J.; Chen, Y.; Liang, W.; Li, Y.; Zhang, Y.; Chen, K.; et al. TXN inhibitor impedes radioresistance of colorectal cancer cells with decreased ALDH1L2 expression via TXN/NF-κB signaling pathway. Br. J. Cancer 2022, 127, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Fang, D.; Ji, Z.; Zhong, Z.; Zhou, B.; Ye, L.; Jiang, L.; Sun, X. Inhibition of thioredoxin-1 enhances the toxicity of glycolysis inhibitor 2-deoxyglucose by downregulating SLC1A5 expression in colorectal cancer cells. Cell. Oncol. 2023, 47, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Yoo, S.Y.; Bartholomeusz, G.; Graham, R.A.; Majidi, M.; Yan, S.; Meng, J.; Ji, L.; Coombes, K.; Minna, J.D.; et al. KEAP1-dependent synthetic lethality induced by AKT and TXNRD1 inhibitors in lung cancer. Cancer Res. 2013, 73, 5532–5543. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Ding, R.; Qu, X.; Li, Y.; Shen, T.; Wang, L.; Li, R.; Zhang, J.; Ru, Y.; Bu, X.; et al. BCR-ABL triggers a glucose-dependent survival program during leukemogenesis through the suppression of TXNIP. Cell Death Dis. 2023, 14, 287. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, X.; Yuan, Y.; Jiang, J.; Zhang, P.; Zhang, B. Identification of a novel mechanism for reversal of doxorubicin-induced chemotherapy resistance by TXNIP in triple-negative breast cancer via promoting reactive oxygen-mediated DNA damage. Cell Death Dis. 2022, 13, 338. [Google Scholar] [CrossRef]
- Zhu, T.; Hu, Z.; Wang, Z.; Ding, H.; Li, R.; Wang, J.; Wang, G. microRNA-301b-3p from mesenchymal stem cells-derived extracellular vesicles inhibits TXNIP to promote multidrug resistance of gastric cancer cells. Cell Biol. Toxicol. 2023, 39, 1923–1937. [Google Scholar] [CrossRef]
- Li, W.; Xin, X.; Li, X.; Geng, J.; Sun, Y. Exosomes secreted by M2 macrophages promote cancer stemness of hepatocellular carcinoma via the miR-27a-3p/TXNIP pathways. Int. Immunopharmacol. 2021, 101, 107585. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.Q.; Lin, J.F.; Tian, T.; Xie, D.; Xu, R.H. NADPH homeostasis in cancer: Functions, mechanisms and therapeutic implications. Signal Transduct. Target. Ther. 2020, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Chen, Z.; Wu, K.; Huang, S.M.; Wu, W.L.; LeBoeuf, S.E.; Pillai, R.G.; Rabinowitz, J.D.; Papagiannakopoulos, T. Activation of the NRF2 antioxidant program sensitizes tumors to G6PD inhibition. Sci. Adv. 2021, 7, eabk1023. [Google Scholar] [CrossRef] [PubMed]
- Aurora, A.B.; Khivansara, V.; Leach, A.; Gill, J.G.; Martin-Sandoval, M.; Yang, C.; Kasitinon, S.Y.; Bezwada, D.; Tasdogan, A.; Gu, W.; et al. Loss of glucose 6-phosphate dehydrogenase function increases oxidative stress and glutaminolysis in metastasizing melanoma cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2120617119. [Google Scholar] [CrossRef] [PubMed]
- Min, H.Y.; Lee, H.J.; Suh, Y.A.; Pei, H.; Kwon, H.; Jang, H.J.; Yun, H.J.; Moon, H.G.; Lee, H.Y. Targeting epidermal growth factor receptor in paclitaxel-resistant human breast and lung cancer cells with upregulated glucose-6-phosphate dehydrogenase. Br. J. Cancer 2022, 127, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Silic-Benussi, M.; Sharova, E.; Ciccarese, F.; Cavallari, I.; Raimondi, V.; Urso, L.; Corradin, A.; Kotler, H.; Scattolin, G.; Buldini, B.; et al. mTOR inhibition downregulates glucose-6-phosphate dehydrogenase and induces ROS-dependent death in T-cell acute lymphoblastic leukemia cells. Redox Biol. 2022, 51, 102268. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ke, M.; Qi, M.; Han, Z.; Cao, Y.; Deng, Z.; Qian, J.; Yang, Y.; Gu, C. G6PD promotes cell proliferation and dexamethasone resistance in multiple myeloma via increasing anti-oxidant production and activating Wnt/β-catenin pathway. Exp. Hematol. Oncol. 2022, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, S.; Shi, X.; Lv, J.; Wu, X.; Zhang, Y.; Wang, H.; He, J.; Zhu, Y.; Ju, Y.; et al. Metabolic classification suggests the GLUT1/ALDOB/G6PD axis as a therapeutic target in chemotherapy-resistant pancreatic cancer. Cell Rep. Med. 2023, 4, 101162. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Xu, M.; Ge, Y.; Huang, G.; Chen, D.; Ye, X.; Xiao, Y.; Zhu, H.; Yin, R.; Shen, H.; et al. Inhibiting G6PD by quercetin promotes degradation of EGFR T790M mutation. Cell Rep. 2023, 42, 113417. [Google Scholar] [CrossRef]
- Mele, L.; la Noce, M.; Paino, F.; Regad, T.; Wagner, S.; Liccardo, D.; Papaccio, G.; Lombardi, A.; Caraglia, M.; Tirino, V.; et al. Glucose-6-phosphate dehydrogenase blockade potentiates tyrosine kinase inhibitor effect on breast cancer cells through autophagy perturbation. J. Exp. Clin. Cancer Res. 2019, 38, 160. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, P.; Zhou, X.; Qiang, Y.; Fan, J.; Lin, Y.; Chen, Y.; Guo, J.; Wang, F.; Xue, H.; et al. Deficiency of the X-inactivation escaping gene KDM5C in clear cell renal cell carcinoma promotes tumorigenicity by reprogramming glycogen metabolism and inhibiting ferroptosis. Theranostics 2021, 11, 8674. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; He, Q.; Li, J.; Yang, Z.; Ahmad, M.; Lin, Y.; Wu, D.; Zheng, L.; Li, J.; Wang, B.; et al. A GSTP1-mediated lactic acid signaling promotes tumorigenesis through the PPP oxidative branch. Cell Death Dis. 2023, 14, 463. [Google Scholar] [CrossRef] [PubMed]
- Sarfraz, I.; Rasul, A.; Hussain, G.; Shah, M.A.; Zahoor, A.F.; Asrar, M.; Selamoglu, Z.; Ji, X.Y.; Adem, Ş.; Sarker, S.D. 6-Phosphogluconate dehydrogenase fuels multiple aspects of cancer cells: From cancer initiation to metastasis and chemoresistance. BioFactors 2020, 46, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, Z.; Yu, B.; Ning, Z.; Lu, Z.; Li, L.; Long, F.; Hu, Q.; Zhong, C.; Zhang, Y.; et al. ATP13A2 activates the pentose phosphate pathway to promote colorectal cancer growth though TFEB-PGD axis. Clin. Transl. Med. 2023, 13, e1272. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, W.; Tao, B.; Wang, X.; Yang, Z.; Zhang, Y.; Wang, C.; Liu, R.; Gao, H.; Liang, J.; et al. Tyrosine phosphorylation activates 6-phosphogluconate dehydrogenase and promotes tumor growth and radiation resistance. Nat. Commun. 2019, 10, 991. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Sun, X.; Zhang, X.; Chen, D. 6PGD upregulation is associated with chemo-and immuno-resistance of renal cell carcinoma via AMPK signaling-dependent NADPH-mediated metabolic reprograming. Am. J. Med. Sci. 2020, 360, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Li, H.M.; Wang, X.Y.; Xia, R.; Li, X.; Ma, Y.J.; Wang, M.; Zhang, H.S. TIGAR drives colorectal cancer ferroptosis resistance through ROS/AMPK/SCD1 pathway. Free Radic. Biol. Med. 2022, 182, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Bailleul, J.; Ruan, Y.; Abdulrahman, L.; Scott, A.J.; Yazal, T.; Sung, D.; Park, K.; Hoang, H.; Nathaniel, J.; Chu, F.I.; et al. M2 isoform of pyruvate kinase rewires glucose metabolism during radiation therapy to promote an antioxidant response and glioblastoma radioresistance. Neuro-Oncology 2023, 25, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Yang, P.; Li, Z. Pyruvate kinase M2: A multifarious enzyme in non-canonical localization to promote cancer progression. Biochim. Biophys. Acta (BBA) Rev. Cancer 2019, 1871, 331–341. [Google Scholar] [CrossRef]
- Leca, J.; Fortin, J.; Mak, T.W. Illuminating the cross-talk between tumor metabolism and immunity in IDH-mutated cancers. Curr. Opin. Biotechnol. 2021, 68, 181–185. [Google Scholar] [CrossRef]
- Pirozzi, C.J.; Yan, H. The implications of IDH mutations for cancer development and therapy. Nat. Rev. Clin. Oncol. 2021, 18, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Calvert, A.E.; Chalastanis, A.; Wu, Y.; Hurley, L.A.; Kouri, F.M.; Bi, Y.; Kachman, M.; May, J.L.; Bartom, E.; Hua, Y.; et al. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep. 2017, 19, 1858–1873. [Google Scholar] [CrossRef] [PubMed]
- Barnabas, G.D.; Lee, J.S.; Shami, T.; Harel, M.; Beck, L.; Selitrennik, M.; Jerby-Arnon, L.; Erez, N.; Ruppin, E.; Geiger, T. Serine biosynthesis is a metabolic vulnerability in IDH2-driven breast cancer progression. Cancer Res. 2021, 81, 1443–1456. [Google Scholar] [CrossRef] [PubMed]
- Wahl, D.R.; Dresser, J.; Wilder-Romans, K.; Parsels, J.D.; Zhao, S.G.; Davis, M.; Zhao, L.; Kachman, M.; Wernisch, S.; Burant, C.F.; et al. Glioblastoma therapy can be augmented by targeting IDH1-mediated NADPH biosynthesis. Cancer Res. 2017, 77, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Vaziri-Gohar, A.; Cassel, J.; Mohammed, F.S.; Zarei, M.; Hue, J.J.; Hajihassani, O.; Graor, H.J.; Srikanth, Y.V.; Karim, S.A.; Abbas, A.; et al. Limited nutrient availability in the tumor microenvironment renders pancreatic tumors sensitive to allosteric IDH1 inhibitors. Nat. Cancer 2022, 3, 852–865. [Google Scholar] [CrossRef]
- Varn, F.S.; Johnson, K.C.; Martinek, J.; Huse, J.T.; Nasrallah, M.P.; Wesseling, P.; Cooper, L.A.; Malta, T.M.; Wade, T.E.; Sabedot, T.S.; et al. Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell 2022, 185, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Murnan, K.M.; Horbinski, C.; Stegh, A.H. Redox Homeostasis and Beyond: The Role of Wild-Type Isocitrate Dehydrogenases for the Pathogenesis of Glioblastoma. Antioxid. Redox Signal. 2023, 39, 923–941. [Google Scholar] [CrossRef] [PubMed]
- Alzial, G.; Renoult, O.; Paris, F.; Gratas, C.; Clavreul, A.; Pecqueur, C. Wild-type isocitrate dehydrogenase under the spotlight in glioblastoma. Oncogene 2022, 41, 613–621. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef]
- Murai, S.; Ando, A.; Ebara, S.; Hirayama, M.; Satomi, Y.; Hara, T. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis 2017, 6, e329. [Google Scholar] [CrossRef]
- Ying, M.; You, D.; Zhu, X.; Cai, L.; Zeng, S.; Hu, X. Lactate and glutamine support NADPH generation in cancer cells under glucose deprived conditions. Redox Biol. 2021, 46, 102065. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.; Baddour, J.; Muller, F.; Wu, C.C.; Wang, H.; Liao, W.T.; Lan, Z.; Chen, A.; Gutschner, T.; Kang, Y.; et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 2017, 542, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.X.; Ju, H.Q.; Liu, Z.X.; Chen, D.L.; Wang, Y.; Zhao, Q.; Wu, Q.N.; Zeng, Z.L.; Qiu, H.B.; Hu, P.S.; et al. ME1 regulates NADPH homeostasis to promote gastric cancer growth and metastasis. Cancer Res. 2018, 78, 1972–1985. [Google Scholar] [CrossRef] [PubMed]
- Ran, M.; Zhou, Y.; Guo, Y.; Huang, D.; Zhang, S.L.; Tam, K.Y. Cytosolic malic enzyme and glucose-6-phosphate dehydrogenase modulate redox balance in NSCLC with acquired drug resistance. FEBS J. 2023, 290, 4792–4809. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Zhang, C.; Xiao, M.; Li, X.; Chen, W.; Jiang, Y.; Yuan, Y.; Zhang, Y.; Zou, Y.; Deng, L.; et al. Redox metabolism maintains the leukemogenic capacity and drug resistance of AML cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2210796120. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ye, J.; Kamphorst, J.J.; Shlomi, T.; Thompson, C.B.; Rabinowitz, J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014, 510, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.; Henriksson, M.; Helleday, T.; Green, A.C. Targeting MTHFD2 to exploit cancer-specific metabolism and the DNA damage response. Cancer Res. 2024, 84, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Moran, D.M.; Trusk, P.B.; Pry, K.; Paz, K.; Sidransky, D.; Bacus, S.S. KRAS mutation status is associated with enhanced dependency on folate metabolism pathways in non–small cell lung cancer cells. Mol. Cancer Ther. 2014, 13, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, P.M.; Vazquez, A.; Kerrigan, J.E.; Bertino, J.R. Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol. Cancer Res. 2015, 13, 1361–1366. [Google Scholar] [CrossRef]
- Krupenko, S.A.; Krupenko, N.I. ALDH1L1 and ALDH1L2 folate regulatory enzymes in cancer. In Alcohol and Cancer: PAdvances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; pp. 127–143. [Google Scholar]
- Tsang, Y.H.; Dogruluk, T.; Tedeschi, P.M.; Wardwell-Ozgo, J.; Lu, H.; Espitia, M.; Nair, N.; Minelli, R.; Chong, Z.; Chen, F.; et al. Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat. Commun. 2016, 7, 10500. [Google Scholar] [CrossRef]
- Ilter, D.; Drapela, S.; Schild, T.; Ward, N.P.; Adhikari, E.; Low, V.; Asara, J.; Oskarsson, T.; Lau, E.K.; DeNicola, G.M.; et al. NADK-mediated de novo NADP (H) synthesis is a metabolic adaptation essential for breast cancer metastasis. Redox Biol. 2023, 61, 102627. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, G.; Ben-Sahra, I.; Lockwood, S.E.; Timson, R.C.; Byles, V.; Henning, G.T.; Gao, P.; Selfors, L.M.; Asara, J.M.; Manning, B.D. Direct stimulation of NADP+ synthesis through Akt-mediated phosphorylation of NAD kinase. Science 2019, 363, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Schild, T.; McReynolds, M.R.; Shea, C.; Low, V.; Schaffer, B.E.; Asara, J.M.; Piskounova, E.; Dephoure, N.; Rabinowitz, J.D.; Gomes, A.P.; et al. NADK is activated by oncogenic signaling to sustain pancreatic ductal adenocarcinoma. Cell Rep. 2021, 35, 109238. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, D.; Wu, Y.; Zhou, H.; Diao, W.; Liu, G.; Li, Q. A feedback loop of PPP and PI3K/AKT signal pathway drives regorafenib-resistance in HCC. Cancer Metab. 2023, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, F.; Zhou, Y.; Hou, X.; Liu, S.; Li, X.; Zhang, Y.; Jing, X.; Yang, L. LncRNA XLOC_006390 promotes pancreatic carcinogenesis and glutamate metabolism by stabilizing c-Myc. Cancer Lett. 2020, 469, 419–428. [Google Scholar] [CrossRef]
- Jin, L.; Li, D.; Alesi, G.N.; Fan, J.; Kang, H.B.; Lu, Z.; Boggon, T.J.; Jin, P.; Yi, H.; Wright, E.R.; et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell 2015, 27, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, M.; Li, H.; Rao, X.; Ao, L.; Wang, H.; Yao, L.; Wang, X.; Hong, X.; Wang, J.; et al. Therapeutic targeting of glutamate dehydrogenase 1 that links metabolic reprogramming and Snail-mediated epithelial–mesenchymal transition in drug-resistant lung cancer. Pharmacol. Res. 2022, 185, 106490. [Google Scholar] [CrossRef]
- Li, S.; Zhuang, Z.; Wu, T.; Lin, J.C.; Liu, Z.X.; Zhou, L.F.; Dai, T.; Lu, L.; Ju, H.Q. Nicotinamide nucleotide transhydrogenase-mediated redox homeostasis promotes tumor growth and metastasis in gastric cancer. Redox Biol. 2018, 18, 246–255. [Google Scholar] [CrossRef]
- Ho, H.Y.; Lin, Y.T.; Lin, G.; Wu, P.R.; Cheng, M.L. Nicotinamide nucleotide transhydrogenase (NNT) deficiency dysregulates mitochondrial retrograde signaling and impedes proliferation. Redox Biol. 2017, 12, 916–928. [Google Scholar] [CrossRef]
- Ward, N.P.; Kang, Y.P.; Falzone, A.; Boyle, T.A.; DeNicola, G.M. Nicotinamide nucleotide transhydrogenase regulates mitochondrial metabolism in NSCLC through maintenance of Fe-S protein function. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, Y.Y.; Pan, Y.Q.; Zheng, X.J.; Liao, K.; Mo, H.Y.; Sheng, H.; Wu, Q.N.; Liu, Z.X.; Zeng, Z.L.; et al. IL-1β-associated NNT acetylation orchestrates iron-sulfur cluster maintenance and cancer immunotherapy resistance. Mol. Cell 2023, 83, 1887–1902. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Jiang, S.; Ma, X.; Jiang, Z.; Pan, Y.; Li, X.; Zhang, L.; Zhou, H.; Chen, S.; Xing, X.; et al. CRISPR-based DNA methylation editing of NNT rescues the cisplatin resistance of lung cancer cells by reducing autophagy. Arch. Toxicol. 2023, 97, 441–456. [Google Scholar] [CrossRef]
- Available online: https://clinicaltrials.gov/ (accessed on 21 June 2024).
- Paiboonrungruang, C.; Xiong, Z.; Lamson, D.; Li, Y.; Bowman, B.; Chembo, J.; Huang, C.; Li, J.; Livingston, E.W.; Frank, J.E.; et al. Small molecule screen identifies pyrimethamine as an inhibitor of NRF2-driven esophageal hyperplasia. Redox Biol. 2023, 67, 102901. [Google Scholar] [CrossRef] [PubMed]
- Montero, A.; Diaz-Montero, C.; Deutsch, Y.; Hurley, J.; Koniaris, L.; Rumboldt, T.; Yasir, S.; Jorda, M.; Garret-Mayer, E.; Avisar, E.; et al. Phase 2 study of neoadjuvant treatment with NOV-002 in combination with doxorubicin and cyclophosphamide followed by docetaxel in patients with HER-2 negative clinical stage II–IIIc breast cancer. Breast Cancer Res. Treat. 2012, 132, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Krasner, C.; Seiden, M.; Penson, R.; Roche, M.; Kendall, D.; Young, J.; Matulonis, U.; Pereira, L.; Berlin, S. NOV-002 plus carboplatin in platinum-resistant ovarian cancer. J. Clin. Oncol. 2008, 26, 5593. [Google Scholar] [CrossRef]
- Hammel, P.; Fabienne, P.; Mineur, L.; Metges, J.P.; Andre, T.; De La Fouchardiere, C.; Louvet, C.; El Hajbi, F.; Faroux, R.; Guimbaud, R.; et al. Erythrocyte-encapsulated asparaginase (eryaspase) combined with chemotherapy in second-line treatment of advanced pancreatic cancer: An open-label, randomized Phase IIb trial. Eur. J. Cancer 2020, 124, 91–101. [Google Scholar] [CrossRef]
- Bachet, J.B.; Blons, H.; Hammel, P.; Hariry, I.E.; Portales, F.; Mineur, L.; Metges, J.P.; Mulot, C.; Bourreau, C.; Cain, J.; et al. Circulating tumor DNA is prognostic and potentially predictive of eryaspase efficacy in second-line in patients with advanced pancreatic adenocarcinoma. Clin. Cancer Res. 2020, 26, 5208–5216. [Google Scholar] [CrossRef] [PubMed]
- Villablanca, J.G.; Volchenboum, S.L.; Cho, H.; Kang, M.H.; Cohn, S.L.; Anderson, C.P.; Marachelian, A.; Groshen, S.; Tsao-Wei, D.; Matthay, K.K.; et al. A phase I new approaches to neuroblastoma therapy study of buthionine sulfoximine and melphalan with autologous stem cells for recurrent/refractory high-risk neuroblastoma. Pediatr. Blood Cancer 2016, 63, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Motzer, R.; Emamekhoo, H.; Matrana, M.; Percent, I.; Hsieh, J.J.; Hussain, A.; Vaishampayan, U.; Liu, S.; McCune, S.; et al. Telaglenastat plus everolimus in advanced renal cell carcinoma: A randomized, double-blinded, placebo-controlled, phase II ENTRATA trial. Clin. Cancer Res. 2022, 28, 3248–3255. [Google Scholar] [CrossRef]
- Tannir, N.M.; Agarwal, N.; Porta, C.; Lawrence, N.J.; Motzer, R.; McGregor, B.; Lee, R.J.; Jain, R.K.; Davis, N.; Appleman, L.J.; et al. Efficacy and safety of telaglenastat plus cabozantinib vs placebo plus cabozantinib in patients with advanced renal cell carcinoma: The CANTATA randomized clinical trial. JAMA Oncol. 2022, 8, 1411–1418. [Google Scholar] [CrossRef]
- Ramanathan, R.K.; Abbruzzese, J.; Dragovich, T.; Kirkpatrick, L.; Guillen, J.M.; Baker, A.F.; Pestano, L.A.; Green, S.; Von Hoff, D.D. A randomized phase II study of PX-12, an inhibitor of thioredoxin in patients with advanced cancer of the pancreas following progression after a gemcitabine-containing combination. Cancer Chemother. Pharmacol. 2011, 67, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, R.K.; Kirkpatrick, D.L.; Belani, C.P.; Friedland, D.; Green, S.B.; Chow, H.S.; Cordova, C.A.; Stratton, S.P.; Sharlow, E.R.; Baker, A.; et al. A Phase I pharmacokinetic and pharmacodynamic study of PX-12, a novel inhibitor of thioredoxin-1, in patients with advanced solid tumors. Clin. Cancer Res. 2007, 13, 2109–2114. [Google Scholar] [CrossRef] [PubMed]
- Halatsch, M.E.; Kast, R.E.; Karpel-Massler, G.; Mayer, B.; Zolk, O.; Schmitz, B.; Scheuerle, A.; Maier, L.; Bullinger, L.; Mayer-Steinacker, R.; et al. A phase Ib/IIa trial of 9 repurposed drugs combined with temozolomide for the treatment of recurrent glioblastoma: CUSP9v3. Neuro-Oncol. Adv. 2021, 3, vdab075. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.; Oronsky, B.; Scicinski, J.; Scribner, C.L.; Knox, S.J.; Ning, S.; Peehl, D.M.; Korn, R.; Stirn, M.; Carter, C.A.; et al. Safety and activity of RRx-001 in patients with advanced cancer: A first-in-human, open-label, dose-escalation phase 1 study. Lancet Oncol. 2015, 16, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.; Oronsky, B.; Caroen, S.; Quinn, M.; Williams, J.; Cabrales, P.; Abrouk, N. Phase 1 pilot study of RRx-001+ nivolumab in patients with advanced metastatic cancer (PRIMETIME). Front. Immunol. 2023, 14, 1104753. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Parmar, H.A.; Schipper, M.; Devasia, T.; Aryal, M.P.; Kesari, S.; O’Day, S.; Morikawa, A.; Spratt, D.E.; Junck, L.; et al. BRAINSTORM: A multi-institutional phase 1/2 Study of RRx-001 in combination with whole brain radiation therapy for patients with brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 2020, 107, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Fine, H.; Reid, T.; Caroen, S.; Oronsky, B.; Abrouk, N.; Butowski, N. A multicenter, phase 1, dose escalation clinical trial (G-FORCE-1) of XRT, RRx-001 and temozolomide followed by temozolomide+/-RRx-001 in newly diagnosed glioblastoma. Front. Oncol. 2023, 13, 1176448. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.R.; Abrouk, N.; Caroen, S.; Oronsky, B.; Stirn, M.; Larson, C.; Beale, K.; Knox, S.J.; Fisher, G. ROCKET: Phase II randomized, active-controlled, multicenter trial to assess the safety and efficacy of RRx-001+ irinotecan vs. single-agent regorafenib in Third/Fourth line colorectal cancer. Clin. Color. Cancer 2023, 22, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Morgensztern, D.; Rose, M.; Waqar, S.N.; Morris, J.; Ma, P.C.; Reid, T.; Brzezniak, C.E.; Zeman, K.G.; Padmanabhan, A.; Hirth, J.; et al. RRx-001 followed by platinum plus etoposide in patients with previously treated small-cell lung cancer. Br. J. Cancer 2019, 121, 211–217. [Google Scholar] [CrossRef]
- Oronsky, B.; Reid, T.R.; Larson, C.; Caroen, S.; Quinn, M.; Burbano, E.; Varner, G.; Thilagar, B.; Brown, B.; Coyle, A.; et al. REPLATINUM Phase III randomized study: RRx-001+ platinum doublet versus platinum doublet in third-line small cell lung cancer. Future Oncol. 2019, 15, 3427–3433. [Google Scholar] [CrossRef]
- Ji, J.; Ma, S.; Zhu, Y.; Zhao, J.; Tong, Y.; You, Q.; Jiang, Z. ARE-PROTACs Enable Co-degradation of an Nrf2–MafG Heterodimer. J. Med. Chem. 2023, 66, 6070–6081. [Google Scholar] [CrossRef] [PubMed]
- Douer, D.; Gökbuget, N.; Stock, W.; Boissel, N. Optimizing use of L-asparaginase–based treatment of adults with acute lymphoblastic leukemia. Blood Rev. 2022, 53, 100908. [Google Scholar] [CrossRef]
- dos Reis Oliveira, C.; Pereira, J.C.; Barros Ibiapina, A.; Roseno Martins, I.R.; de Castro e Sousa, J.M.; Ferreira, P.M.P.; Carneiro da Silva, F.C. Buthionine sulfoximine and chemoresistance in cancer treatments: A systematic review with meta-analysis of preclinical studies. J. Toxicol. Environ. Health Part B 2023, 26, 417–441. [Google Scholar] [CrossRef] [PubMed]
- Conche, C.; Finkelmeier, F.; Pešić, M.; Nicolas, A.M.; Böttger, T.W.; Kennel, K.B.; Denk, D.; Ceteci, F.; Mohs, K.; Engel, E.; et al. Combining ferroptosis induction with MDSC blockade renders primary tumours and metastases in liver sensitive to immune checkpoint blockade. Gut 2023, 72, 1774–1782. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Scicinski, J.; Reid, T.; Oronsky, A.; Carter, C.; Oronsky, N.; Cabrales, P. RRx-001, a novel clinical-stage chemosensitizer, radiosensitizer, and immunosensitizer, inhibits glucose 6-phosphate dehydrogenase in human tumor cells. Discov. Med. 2016, 21, 251–265. [Google Scholar]
- Jayabalan, N.; Oronsky, B.; Cabrales, P.; Reid, T.; Caroen, S.; Johnson, A.M.; Birch, N.A.; O’Sullivan, J.D.; Gordon, R. A review of RRx-001: A late-stage multi-indication inhibitor of NLRP3 activation and chronic inflammation. Drugs 2023, 83, 389–402. [Google Scholar] [CrossRef]
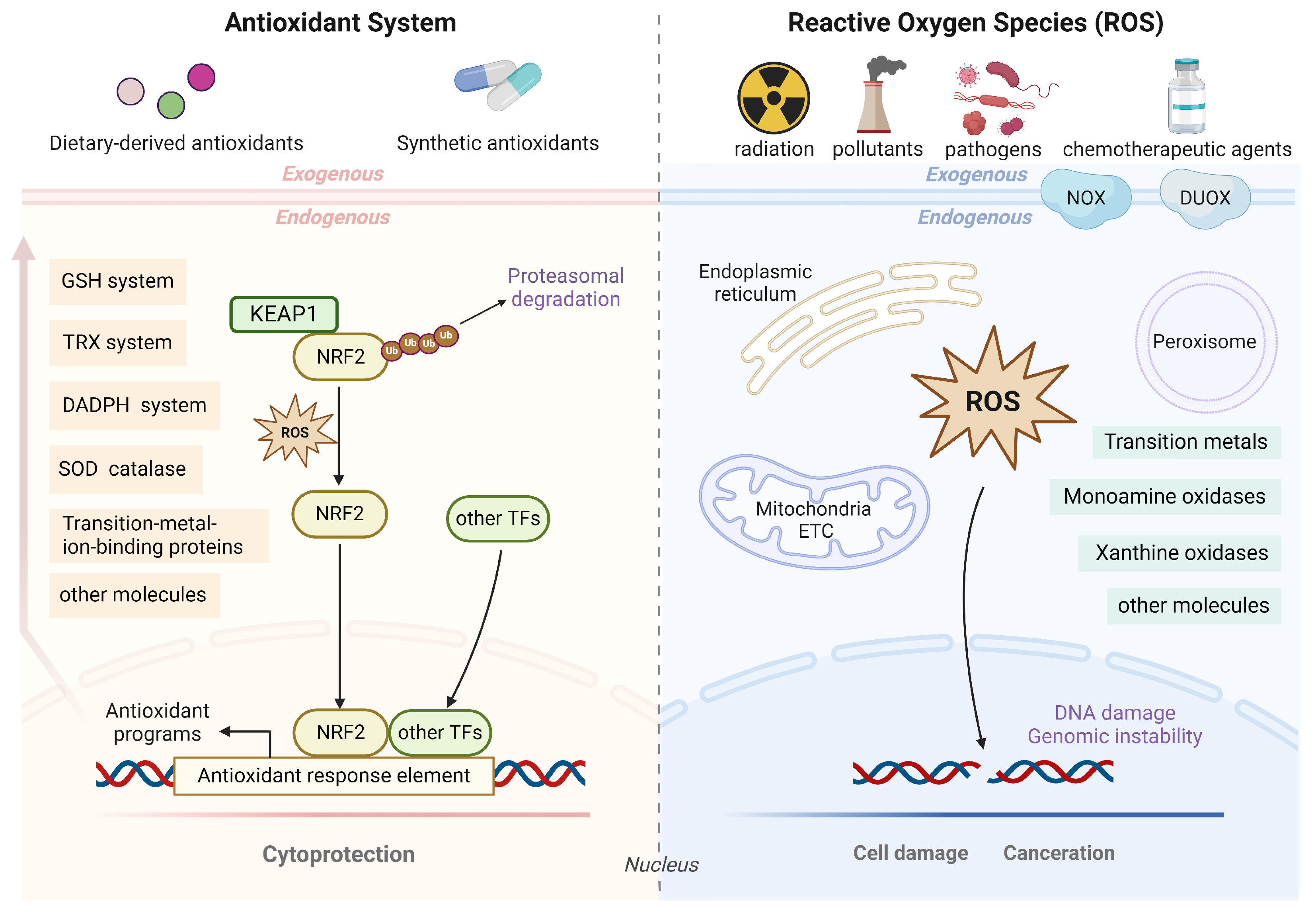
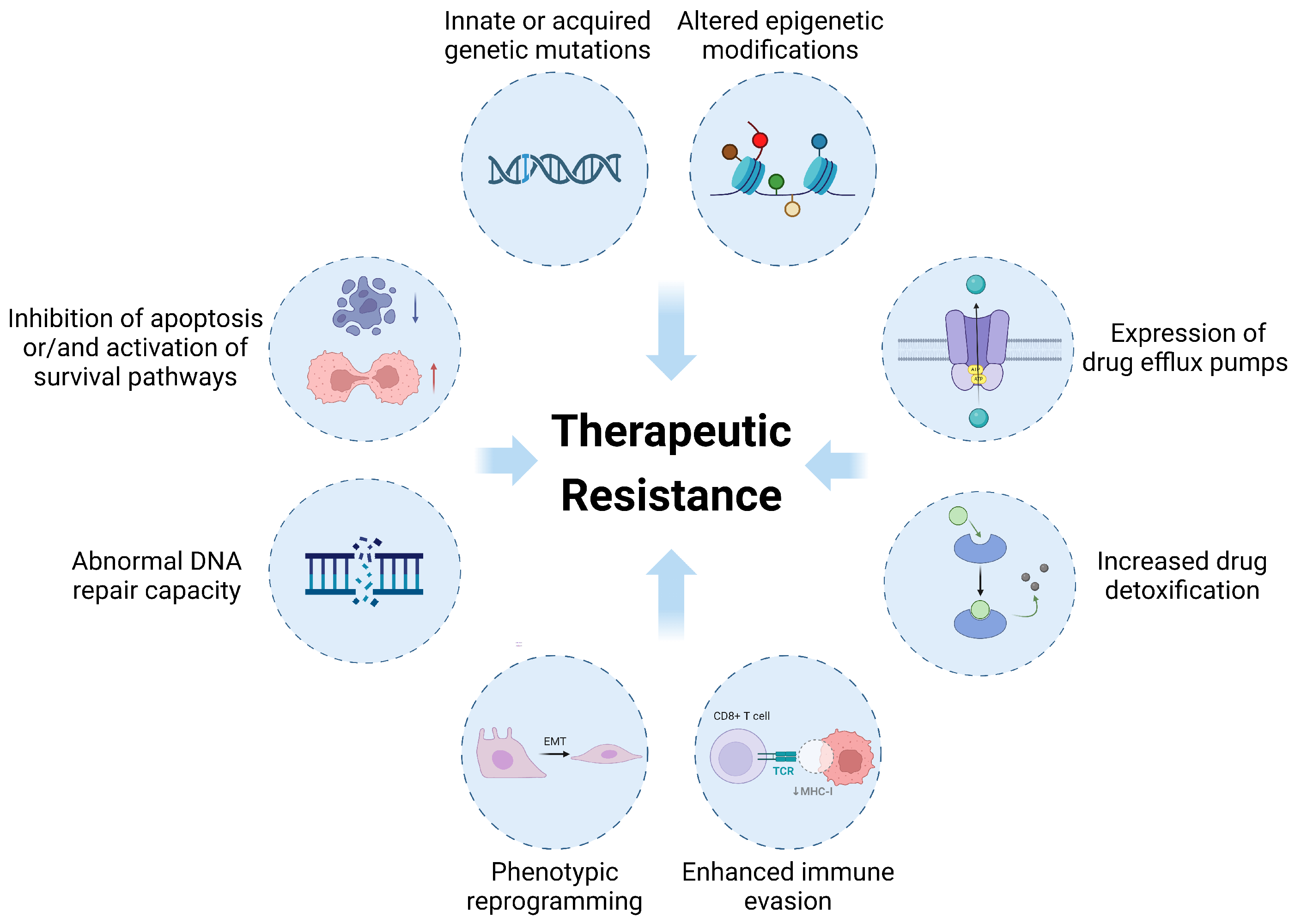
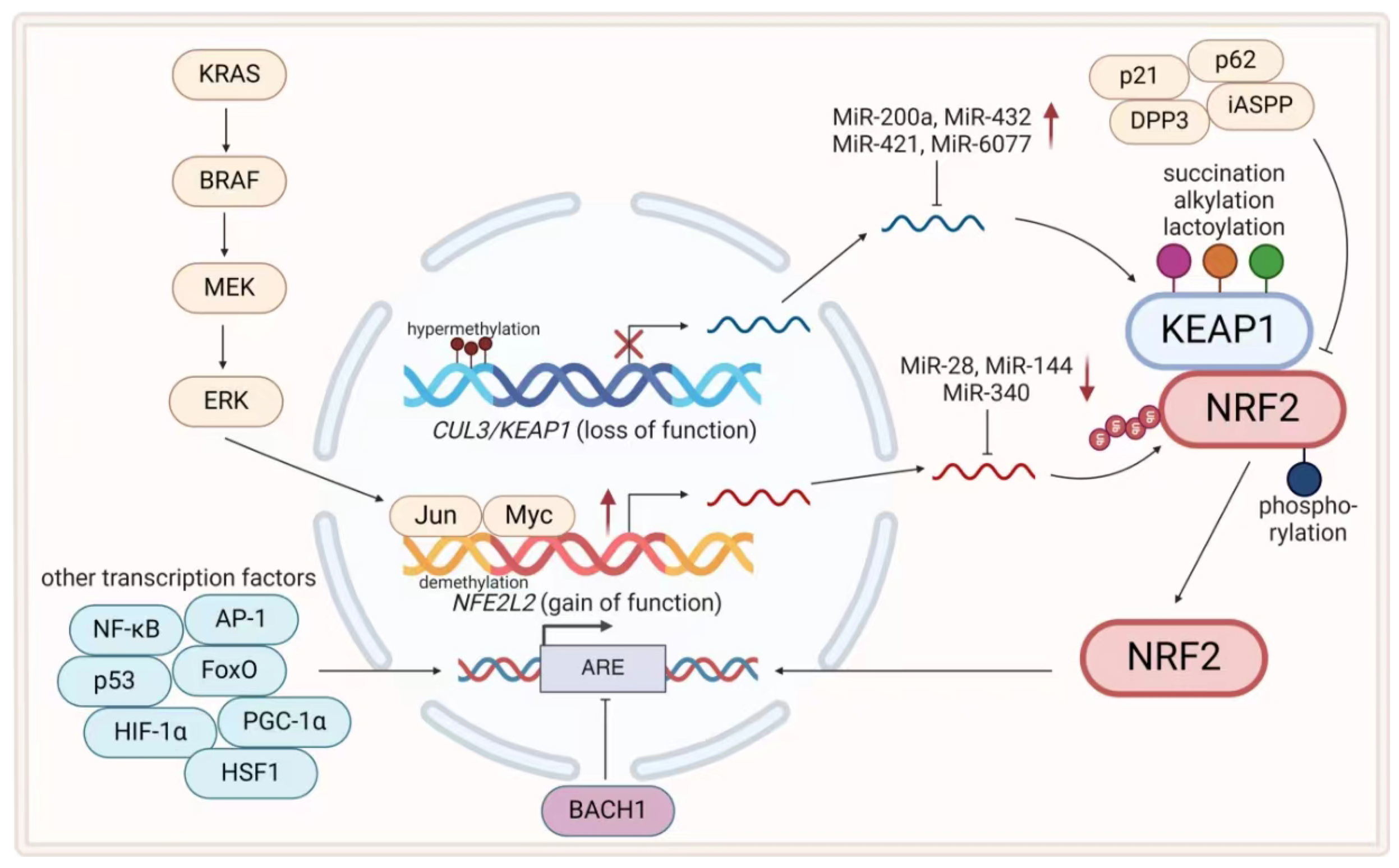
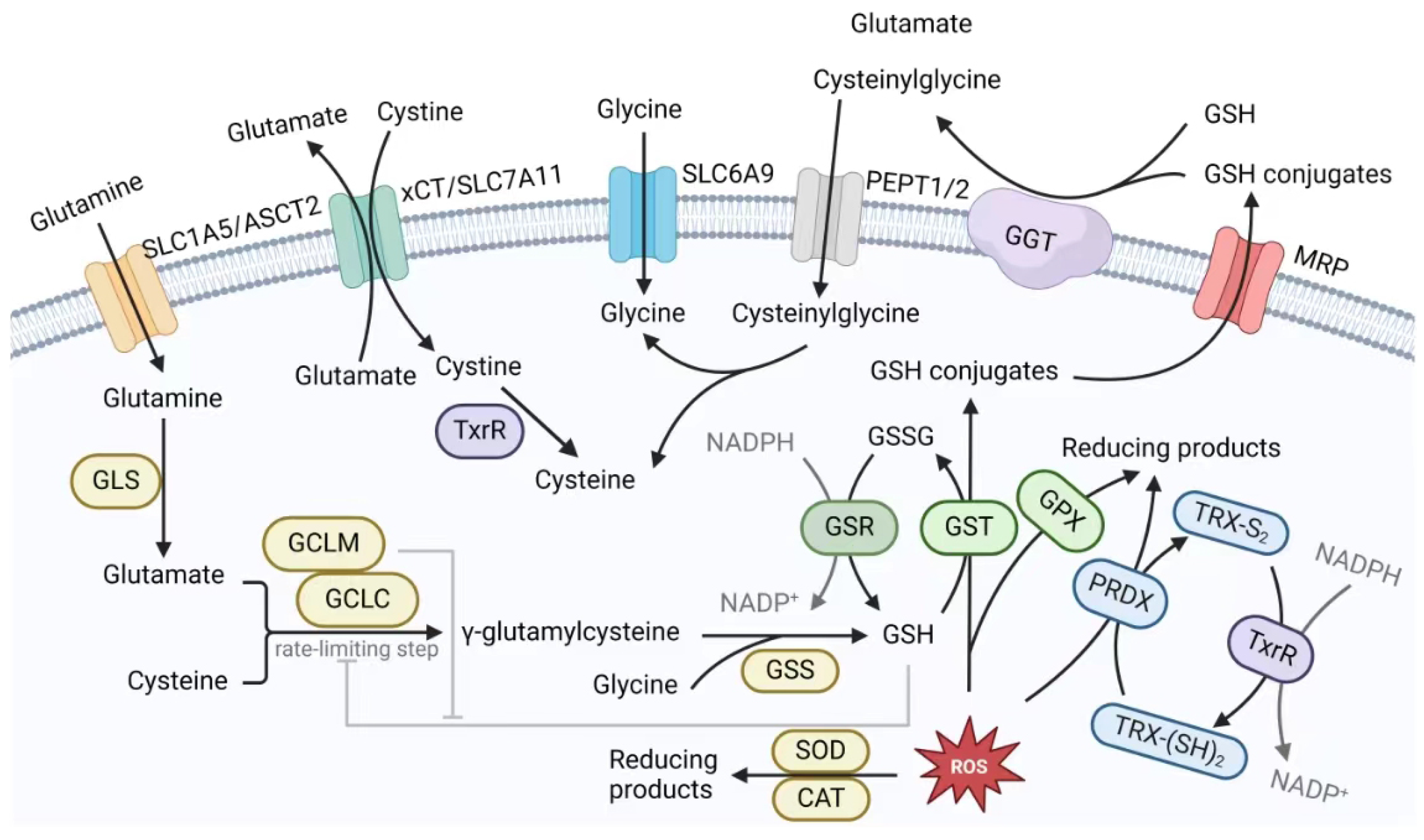
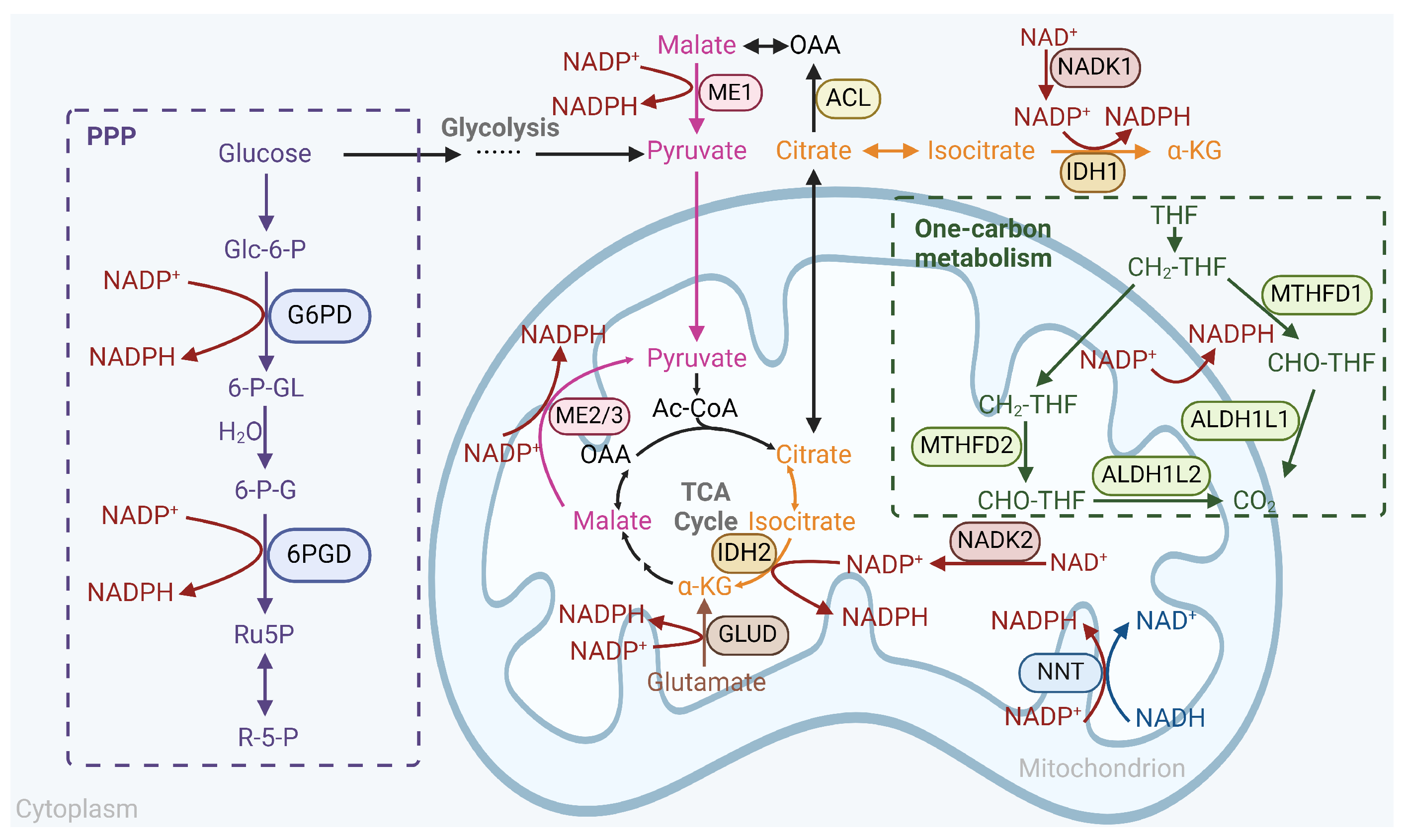
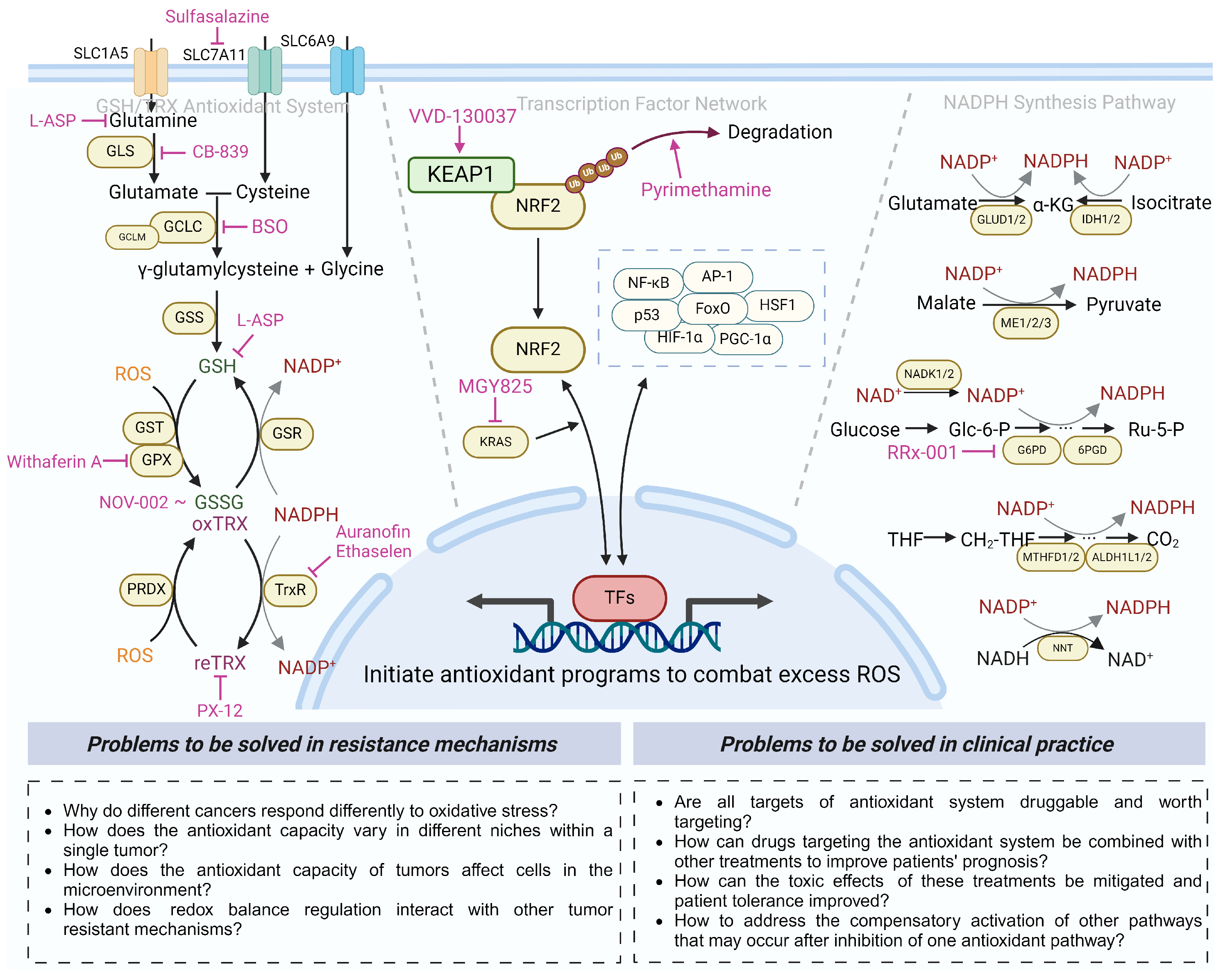
| Cancer Types | Therapies | Specific Resistance Mechanisms | Refs. |
|---|---|---|---|
| Genetic alterations | |||
| Lung cancer | Cisplatin | Gain-of-function mutations in NFE2L2 or loss-of-function mutations in KEAP1 | [52,53,54,55,56] |
| Radiotherapy | Depletion of KEAP1 and impairment of ferroptosis | [57,58] | |
| PD-(L)1 inhibition | KEAP1 mutation, damaged immune cell infiltration and CD8+ T-cell immunity | [59,60,61] | |
| Gefitinib | KEAP1 mutation and elevated NRF2 levels | [56] | |
| Gallbladder cancer | 5-Fluorouracil | KEAP1 mutation and activation of NRF2 pathways | [62] |
| Ovarian cancer | Platinum-based drugs | [63] | |
| Prostate cancer | Paclitaxel, cisplatin, etopside, and irradiation | [64] | |
| Melanoma | Cisplatin, dacarbazine | [65] | |
| HNSCC | Cisplatin | Enhanced antioxidant capacity due to gain-of-function mutations in NFE2L2 or loss-of-function mutations in KEAP1 | [66,67] |
| HCC | NA | NFE2L2 mutations and increased transcriptional activity of NRF2 | [68] |
| ESCC | NA | [69] | |
| Transcriptional alterations | |||
| Pancreatic cancer | NA | Activation of oncogenic proteins such as KRAS/ERK/NRF2 pathway | [70,71] |
| AML | Cytarabine | Overexpression of NRF2 inhibited MSH2, which induced gene instability-dependent resistance | [72] |
| Glioma | Radiotherapy, temozolomide | Hypermethylation of CpG islands in promoters of KEAP1 catalyzed by DNMTs, which suppressed transcription of KEAP1 mRNA | [73] |
| Colorectal cancer | NA | [74] | |
| Lung cancer | NA | [75,76] | |
| Colorectal cancer | NA | Demethylation of NFE2L2 promoter | [77] |
| Lung cancer | NA | EZH2 deficiency caused demethylation of histone H3K27, which promoted transcription of NRF2 | [78] |
| Translational alterations | |||
| HCC | 5-Fluorouracil | MiR-144(-3p), a microRNA that degraded NRF2 mRNA effectively, was lower in cancer cells compared with normal cells | [79] |
| Lung cancer | Cisplatin | [80] | |
| Breast cancer | NA | MiR-200a degraded KEAP1 mRNA and then induced NRF2-dependent gene expression | [81] |
| ESCC | Cisplatin, 5-fluorouracil | MiR-432 directly targeted KEAP1 transcripts and then stabilized the NRF2 proteins | [82] |
| Lung cancer | Paclitaxel, cisplatin, pemetrexed | MiR-6077 inhibited CDKN1A-CDK1-mediated cell cycle arrest and targeted KEAP1, facilitating NRF2-SLC7A11/NQO1 antioxidant axis | [83,84] |
| Gefitinib | NRF2-miR-196a axis was upregulated to suppress GLTP | [85] | |
| Posttranslational Modification | |||
| Renal cell carcinoma | NA | KEAP1 succination due to accumulation of fumarate in fumarate hydratase-deficient cells | [86] |
| NA | NA | KEAP1 alkylation by the excessive TCA cycle metabolite itaconate | [87] |
| NA | NA | KEAP1 lactoylation due to accumulation of glycolysis metabolite glyceraldehyde 3-phosphate | [88] |
| NA | NA | KEAP1 phosphorylation by multiple kinases | [89] |
| Protein–protein interaction | |||
| Lung cancer | Oxidative stress induced by H2O2 | p21 stabilized NRF2 by competing with KEAP1 and binding NRF2; NRF2 also promotes CDKN1A gene transcription | [90,91] |
| HCC | Sorafenib, erastin | p62 bound to KEAP1 and confined it to autophagosomes, which stabilized NRF2 | [92,93] |
| ESCC | Cisplatin, paclitaxel | DPP3 bound to KEAP1 and freed NRF2 | [94,95] |
| Colon cancer | Doxorubicin | iASPP bound to KEAP1 and interrupted KEAP1/NRF2 interaction | [96] |
| Breast cancer | NA | BRAC1 interacted with NRF2 and promoted its stability and activation | [97] |
| Drug Name | Mechanism of Action | Cancer Types | Current Status | Trial ID/Refs. |
|---|---|---|---|---|
| Drugs targeting KEAP1/NRF2 system | ||||
| Pyrimethamine | Inhibitor of NRF2 by promoting NRF2 ubiquitination and degradation | Locally advanced (stage III-IV) HNSCC (single agent) | Phase I, recruiting | NCT05678348 [267] |
| MGY825 | Inhibitor of KRAS | Advanced NSCLC harboring NFE2L2/KEAP1/CUL3 mutations (single agent) | Phase I, recruiting | NCT05275868 |
| VVD-130037 (BAY 3605349) | Activator of KEAP1 | Advanced solid tumors without KEAP1 mutations (single agent) | Phase I, recruiting | NCT05954312 |
| Drugs targeting GSH/TRX system | ||||
| NOV-002 | Oxidized glutathione mimic that alters intracellular GSH/GSSG ratio | HER2-negative IIB-IIIC breast cancer (combination with doxorubicin and cyclophosphamide)/ovarian cancer | Phase II, completed | NCT00499122 [268] NCT00345540 [269] |
| L-asparaginase | Various mechanisms, includ- ing hydrolyzing glutamine and GSH depletion | Locally advanced or metastatic pancreatic cancer (combination with modified FOLFIRINOX chemotherapy) | Phase I, active | NCT04292743 |
| Advanced or metastatic pancreatic cancer (combination with gemcitabine or FOLFOX) | Phase IIb, completed | NCT02195180 [270,271] | ||
| Advanced or metastatic pancreatic cancer (combination with gemcitabine or irinotecan-based chemotherapy) | Phase III, completed | NCT03665441 | ||
| Buthionine sulfoximine (BSO) | Irreversible inhibitor of GCLC that inhibits de novo GSH synthesis | High-risk neuroblastoma (combination with melphalan) | Phase I, completed | NCT00005835 [272] |
| Telaglenastat hydrochloride (CB-839 HCl) | Inhibitor of glutaminase, in- terfering with glutamate-de- pendent cellular metabolism and redox status | Advanced tumors include TNBC, NSCLC, RCC, mesothelioma, CRC, hematological tumors (sin- gle agent or combination with other therapies) | Phase I, recruit- ing, active or com- pleted | NCT02071862 |
| NCT02071888 | ||||
| NCT03965845 | ||||
| NCT03263429 | ||||
| NCT03831932 | ||||
| NCT02071927 | ||||
| NCT03047993 | ||||
| Advanced or metastatic solid tumors with specific mutations (single agent) | Phase II, active | NCT03872427 | ||
| Melanoma, ccRCC, NSCLC (combination with nivolumab) | Phase I/II, terminated | NCT02771626 | ||
| Advanced or metastatic RCC (combination with everolimus or cabozantinib) | Phase II, completed | NCT03163667 [273] | ||
| NCT03428217 [274] | ||||
| Sulfasalazine | Approved as anti-inflam- matory agent, competitive in- hibitor of Cys/Glu trans- porter xCT, inducing lipid peroxidation and ferroptosis | AML (combination with standard-care induction therapy) | Phase I/II, recruiting | NCT05580861 |
| Recurrent glioblastoma (combination with -knife radiosurgery)/glioma (single agent) | Phase I, completed | NCT04205357 NCT01577966 | ||
| Metastatic colorectal cancer (single agent) | Phase I, recruiting | NCT06134388 | ||
| Withaferin A | Inhibitor of GPX4 | High-grade relapsed or metastatic osteosarcoma (single agent) | Phase I/II, unknown status | NCT00689195 |
| PX-12 | Inhibitor of thioredoxin-1 (TRX-1) | Advanced pancreatic cancer (stage IV) (single agent) | Phase II, terminated | NCT00417287 [275] |
| Drugs targeting GSH/TRX system | ||||
| PX-12 | Inhibitor of thioredoxin-1 (TRX-1) | Advanced or metastatic solid tumors (single agent) | Phase I, completed | NCT00736372 [276] |
| Auranofin | Approved as antirheum- atic drug, with activity tar- geting TrxR | Recurrent ovarian cancer (combination with sirolimus)/advanced or recurrent SCLC or NSCLC (combination with sirolimus) | Phase I/II, active | NCT03456700 NCT01737502 |
| Relapsed or refractory CLL (single agent)/recurrent glioblastoma (9 repurposed drugs combined with temozolomide) | Phase I/II, completed | NCT01419691 NCT02770378 [277] | ||
| Ethaselen | Selective inhibitor of TrxR | High-TrxR-expressing advanced NSCLC (single agent) | Phase I, completed | NCT02166242 |
| Drugs targeting NADPH generation system | ||||
| RRx-001 | Novel epigenetic modula- tor that produces NO and ROS under hypoxic condi- tions and inhibits G6PD and NLRP3 activity | Brain metastases (combination with whole-brain radiotherapy)/high-grade glioma (combination with radiotherapy and temozolomide)/advanced cancers | Phase I/II, completed | NCT01359982 [278] NCT02518958 [279] NCT02215512 [280] NCT02871843 [281] |
| Metastatic CRC (combination with irinotecan)/platinum-resistant SCLC (combination with platinum and etoposide) | Phase II, completed | NCT02096354 [282] NCT02489903 [283] | ||
| Third-line or beyond SCLC (combination with platinum regimens) | Phase III, active | NCT03699956 [284] NCT05566041 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, X.; Mu, C.; Zheng, R.; Zhang, Z.; Zhang, Q.; Liang, T. The Cancer Antioxidant Regulation System in Therapeutic Resistance. Antioxidants 2024, 13, 778. https://doi.org/10.3390/antiox13070778
Gu X, Mu C, Zheng R, Zhang Z, Zhang Q, Liang T. The Cancer Antioxidant Regulation System in Therapeutic Resistance. Antioxidants. 2024; 13(7):778. https://doi.org/10.3390/antiox13070778
Chicago/Turabian StyleGu, Xuanhao, Chunyang Mu, Rujia Zheng, Zhe Zhang, Qi Zhang, and Tingbo Liang. 2024. "The Cancer Antioxidant Regulation System in Therapeutic Resistance" Antioxidants 13, no. 7: 778. https://doi.org/10.3390/antiox13070778
APA StyleGu, X., Mu, C., Zheng, R., Zhang, Z., Zhang, Q., & Liang, T. (2024). The Cancer Antioxidant Regulation System in Therapeutic Resistance. Antioxidants, 13(7), 778. https://doi.org/10.3390/antiox13070778









