Development of Bivalent mRNA Vaccines against SARS-CoV-2 Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Vaccine Design and Production
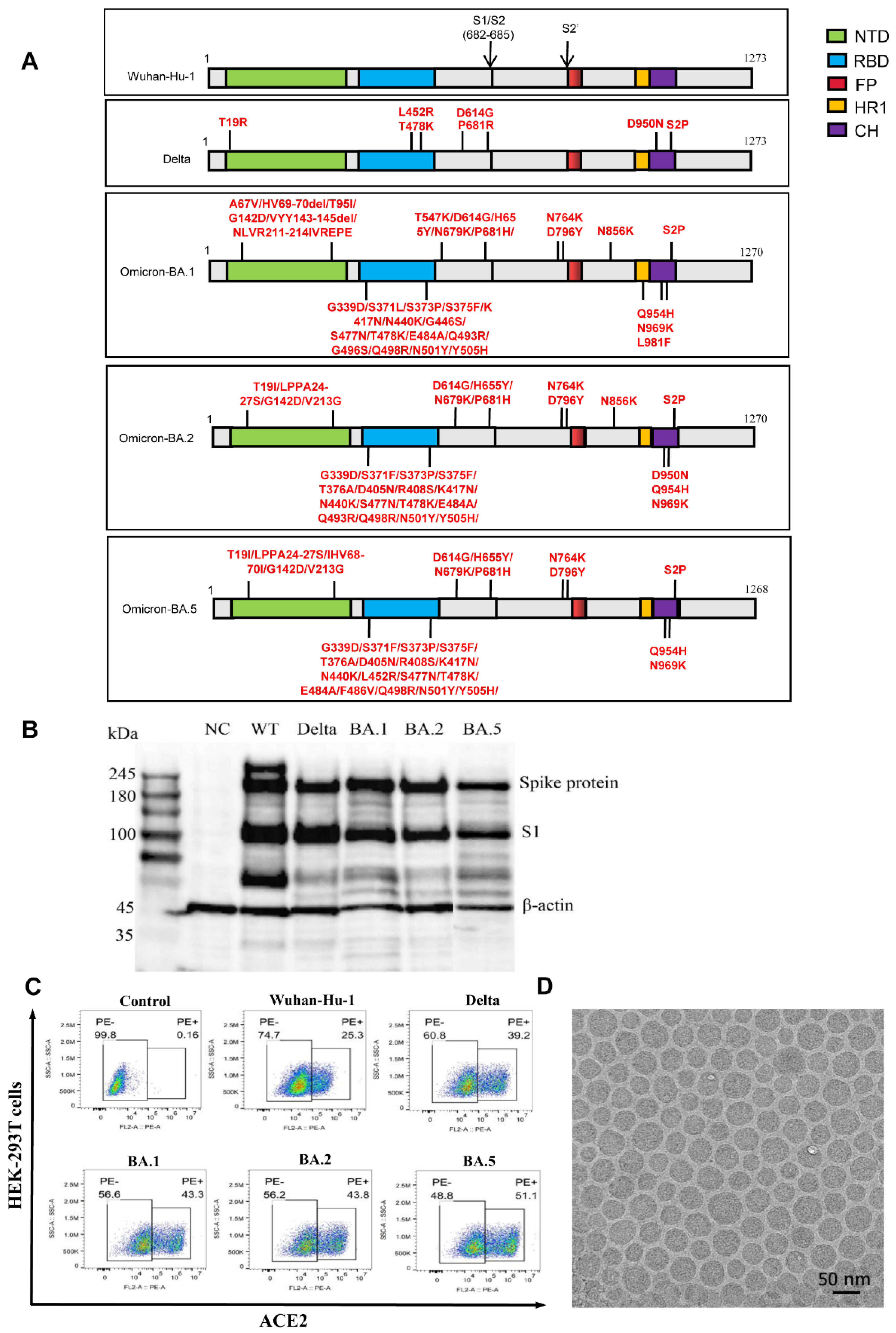
2.2. Verification of S Protein Expression
2.2.1. mRNA Transfection
2.2.2. Vaccine Antigen Detection by Western Blotting
2.2.3. Flow Cytometry Analysis
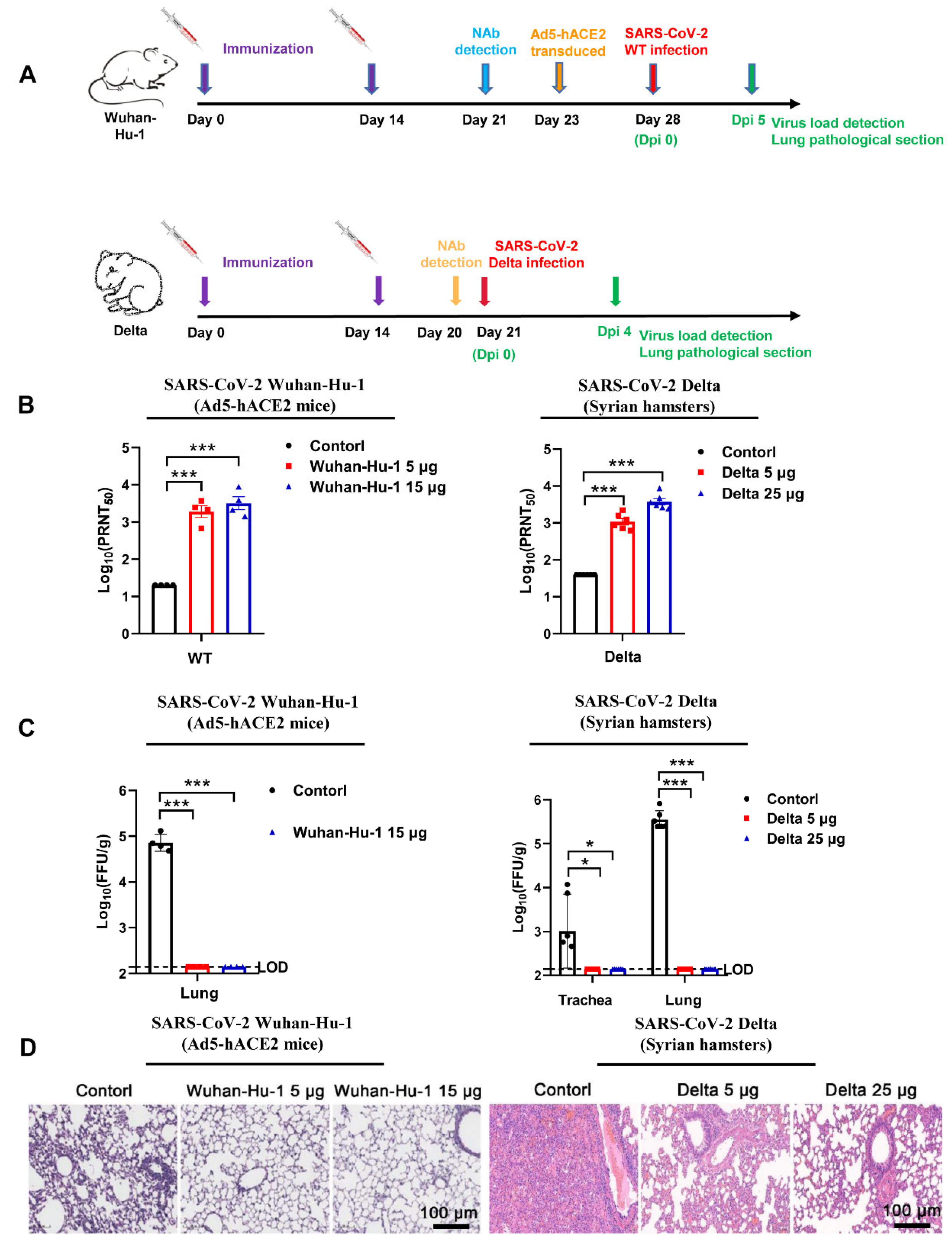
2.3. Immunogenicity Studies
2.3.1. Animal Experiments and Approvals
2.3.2. SARS-CoV-2 Plaque Reduction Neutralization Test (PRNT)
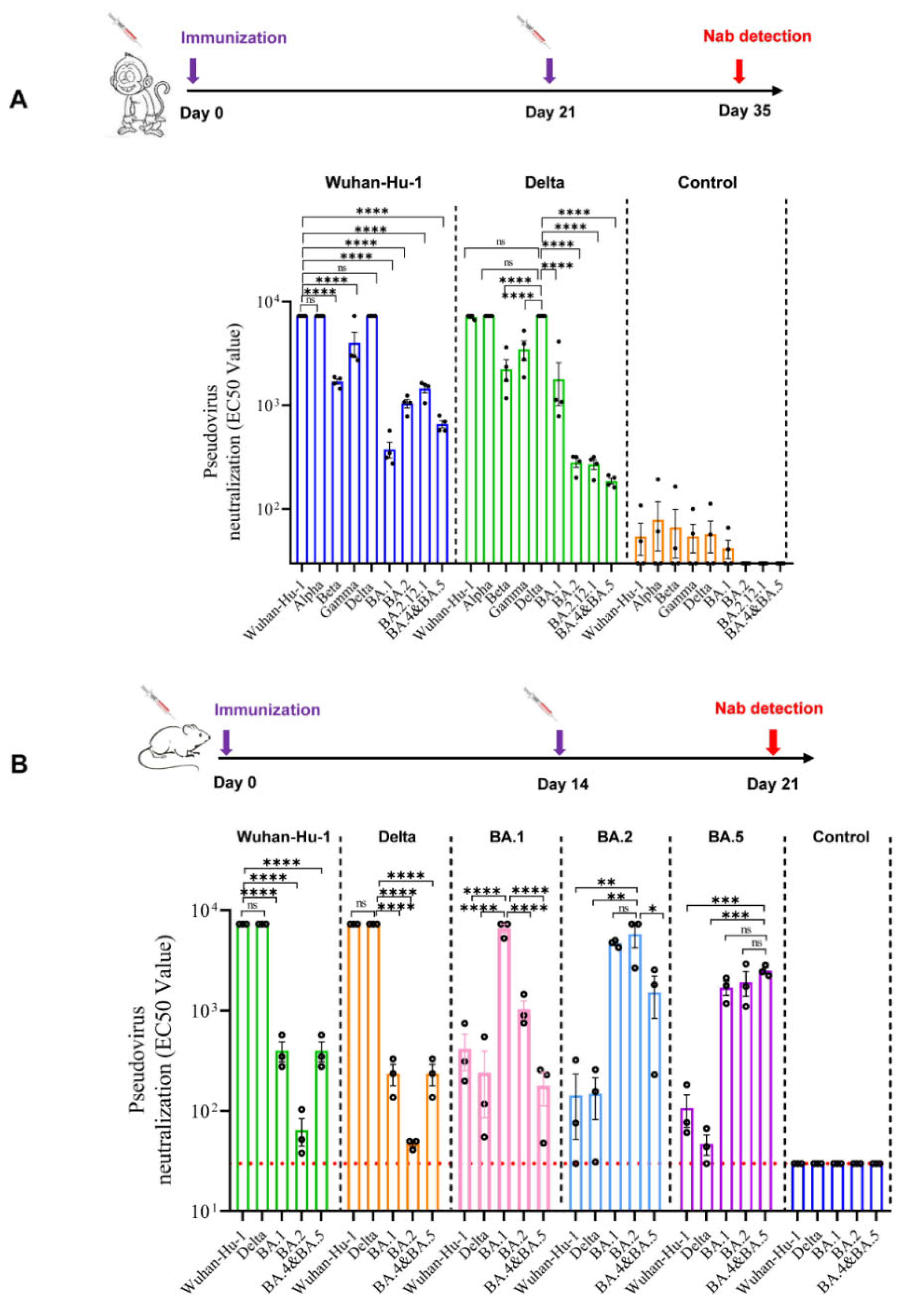
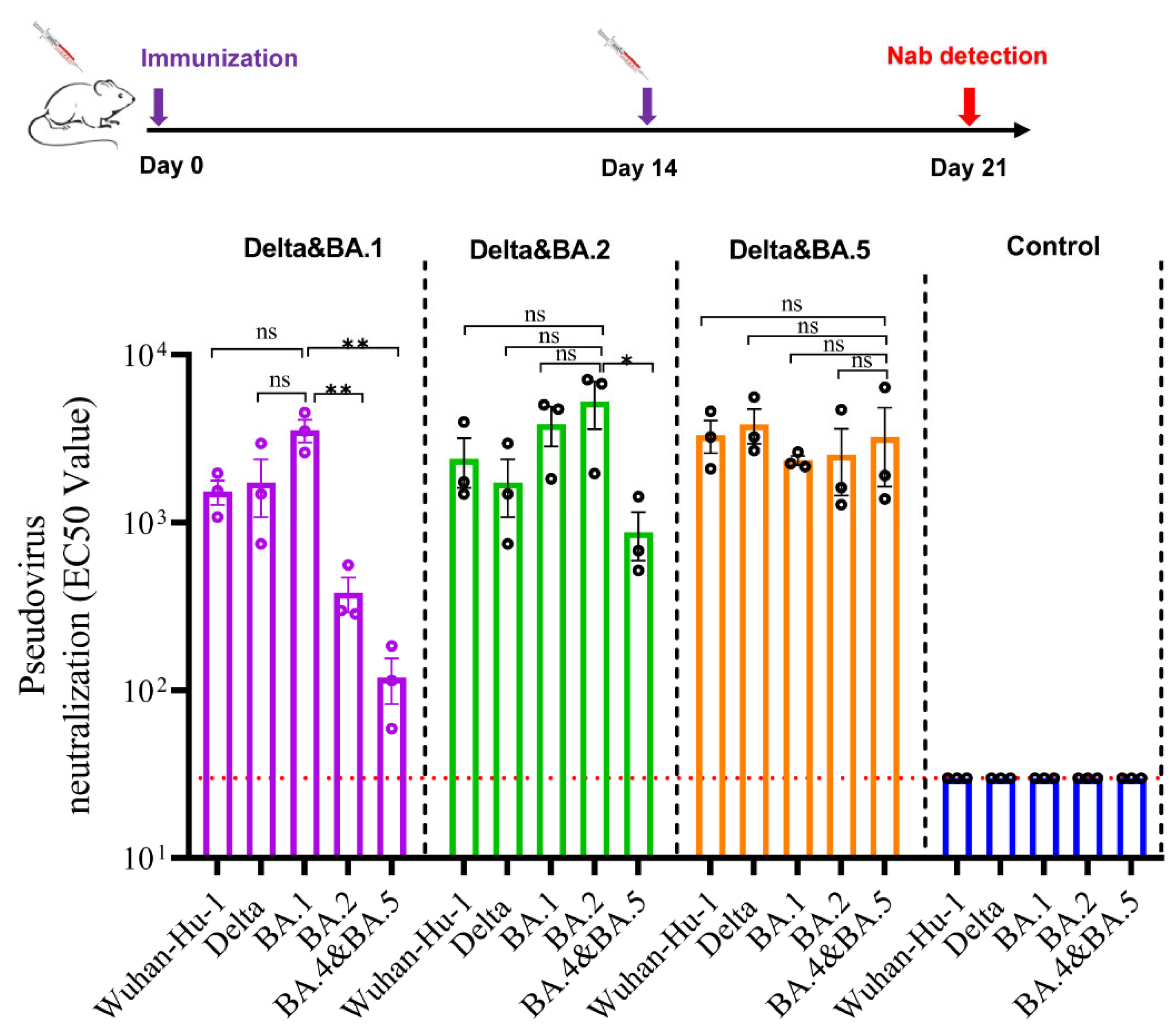
2.3.3. SARS-CoV-2 Pseudovirus Neutralization Assay
2.4. Viral Challenge Study
2.4.1. Viral Infection
2.4.2. Histopathological Examination
2.4.3. SARS-CoV-2 Plaque Assay for Virus Titer Detection
2.5. Statistical Analysis
3. Results
3.1. Design and Expression of the mRNA Vaccines against Different SARS-CoV-2 Variants
3.2. Monovalent Wuhan-Hu-1 and Delta Vaccines Exerted Potent Protection against Corresponding SARS-CoV-2 Strain Challenges
3.3. Profile of Neutralizing Antibodies Produced by Monovalent mRNA Vaccines in Cynomolgus Monkeys
3.4. Profile of Neutralizing Antibodies Produced by Monovalent mRNA Vaccines in Mice
3.5. Bivalent Vaccines Produced High Level Neutralizing Antibodies against All SARS-CoV-2 Variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci. Total Environ. 2020, 730, 138996. [Google Scholar] [CrossRef] [PubMed]
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J. Clin. Med. 2020, 9, 1225. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm 2021, 2, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Kiso, M.; Iida, S.; Imai, M.; Takashita, E.; Kuroda, M.; Halfmann, P.J.; Loeber, S.; Maemura, T.; Yamayoshi, S.; et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature 2022, 607, 119–127. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, R.; Qiao, S.; Wang, X.; Zhang, W.; Ruan, W.; Dai, L.; Han, P.; Gao, G.F. Omicron SARS-CoV-2 Neutralization from Inactivated and ZF2001 Vaccines. N. Engl. J. Med. 2022, 387, 277–280. [Google Scholar] [CrossRef]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19); StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, G.; Pan, H.; Li, C.; Hu, Y.; Chu, K.; Han, W.; Chen, Z.; Tang, R.; Yin, W.; et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021, 21, 181–192. [Google Scholar] [CrossRef]
- Zhu, F.C.; Guan, X.H.; Li, Y.H.; Huang, J.Y.; Jiang, T.; Hou, L.H.; Li, J.X.; Yang, B.F.; Wang, L.; Wang, W.J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Publisher Correction: Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2021, 590, E26. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Callaway, E. The race for coronavirus vaccines: A graphical guide. Nature 2020, 580, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.C.; Li, Y.H.; Guan, X.H.; Hou, L.H.; Wang, W.J.; Li, J.X.; Wu, S.P.; Wang, B.S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Wack, S.; Patton, T.; Ferris, L.K. COVID-19 vaccine safety and efficacy in patients with immune-mediated inflammatory disease: Review of available evidence. J. Am. Acad. Dermatol. 2021, 85, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Sun, J.; Zhuang, Z.; Zheng, J.; Li, K.; Wong, R.L.; Liu, D.; Huang, J.; He, J.; Zhu, A.; Zhao, J.; et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020, 182, 734–743.e5. [Google Scholar] [CrossRef]
- Wang, Z.J.; Zhang, H.J.; Lu, J.; Xu, K.W.; Peng, C.; Guo, J.; Gao, X.X.; Wan, X.; Wang, W.H.; Shan, C.; et al. Low toxicity and high immunogenicity of an inactivated vaccine candidate against COVID-19 in different animal models. Emerg. Microbes Infect. 2020, 9, 2606–2618. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Sanders, R.W.; Moore, J.P. Virus vaccines: Proteins prefer prolines. Cell Host Microbe 2021, 29, 327–333. [Google Scholar] [CrossRef]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A Bivalent Omicron-Containing Booster Vaccine against Covid-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef]
- Goyal, L.; Zapata, M.; Ajmera, K.; Chaurasia, P.; Pandit, R.; Pandit, T. A Hitchhiker’s Guide to Worldwide COVID-19 Vaccinations: A Detailed Review of Monovalent and Bivalent Vaccine Schedules, COVID-19 Vaccine Side Effects, and Effectiveness Against Omicron and Delta Variants. Cureus 2022, 14, e29837. [Google Scholar] [CrossRef]
- COVID-19 update: Bivalent Pfizer and Moderna COVID-19 vaccines for booster immunization. Med. Lett. Drugs Ther. 2022, 64, 159–160.
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monitor Int. Med. J. Exper. Clin. Res. 2020, 26, e924700. [Google Scholar] [CrossRef]
- Mukherjee, R. Global efforts on vaccines for COVID-19: Since, sooner or later, we all will catch the coronavirus. J. Biosci. 2020, 45, 1–10. [Google Scholar] [CrossRef]
- He, C.; Yang, J.; He, X.; Hong, W.; Lei, H.; Chen, Z.; Shen, G.; Yang, L.; Li, J.; Wang, Z.; et al. A bivalent recombinant vaccine targeting the S1 protein induces neutralizing antibodies against both SARS-CoV-2 variants and wild-type of the virus. MedComm 2021, 2, 430–441. [Google Scholar] [CrossRef]
- Ren, S.Y.; Wang, W.B.; Gao, R.D.; Zhou, A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liu, Q.; Liu, J.; Fang, Z.; Luo, L.; Li, S.; Lei, Y.; Li, Z.; Jin, J.; Xie, R.; et al. Development of Bivalent mRNA Vaccines against SARS-CoV-2 Variants. Vaccines 2022, 10, 1807. https://doi.org/10.3390/vaccines10111807
Li J, Liu Q, Liu J, Fang Z, Luo L, Li S, Lei Y, Li Z, Jin J, Xie R, et al. Development of Bivalent mRNA Vaccines against SARS-CoV-2 Variants. Vaccines. 2022; 10(11):1807. https://doi.org/10.3390/vaccines10111807
Chicago/Turabian StyleLi, Jianglong, Qi Liu, Jun Liu, Zihui Fang, Liping Luo, Shuang Li, Yixin Lei, Zhi Li, Jing Jin, Ronglin Xie, and et al. 2022. "Development of Bivalent mRNA Vaccines against SARS-CoV-2 Variants" Vaccines 10, no. 11: 1807. https://doi.org/10.3390/vaccines10111807
APA StyleLi, J., Liu, Q., Liu, J., Fang, Z., Luo, L., Li, S., Lei, Y., Li, Z., Jin, J., Xie, R., & Peng, Y. (2022). Development of Bivalent mRNA Vaccines against SARS-CoV-2 Variants. Vaccines, 10(11), 1807. https://doi.org/10.3390/vaccines10111807




