Cell-Free Screening, Production and Animal Testing of a STI-Related Chlamydial Major Outer Membrane Protein Supported in Nanolipoproteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. DMPC/Telodendrimer Preparation
2.3. Cell-Free Reaction
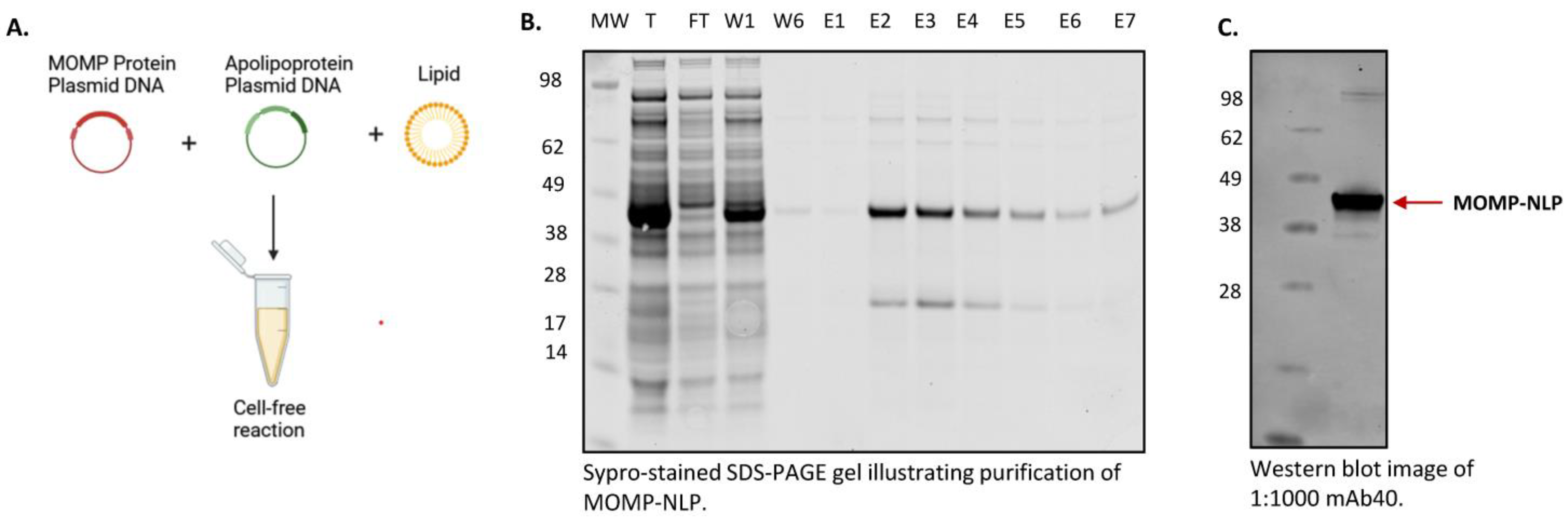
2.4. Affinity Purification of NLP-Related Complexes
2.5. Size Exclusion Chromatography (SEC)
2.6. SDS-PAGE
2.7. Western Blot Analyses
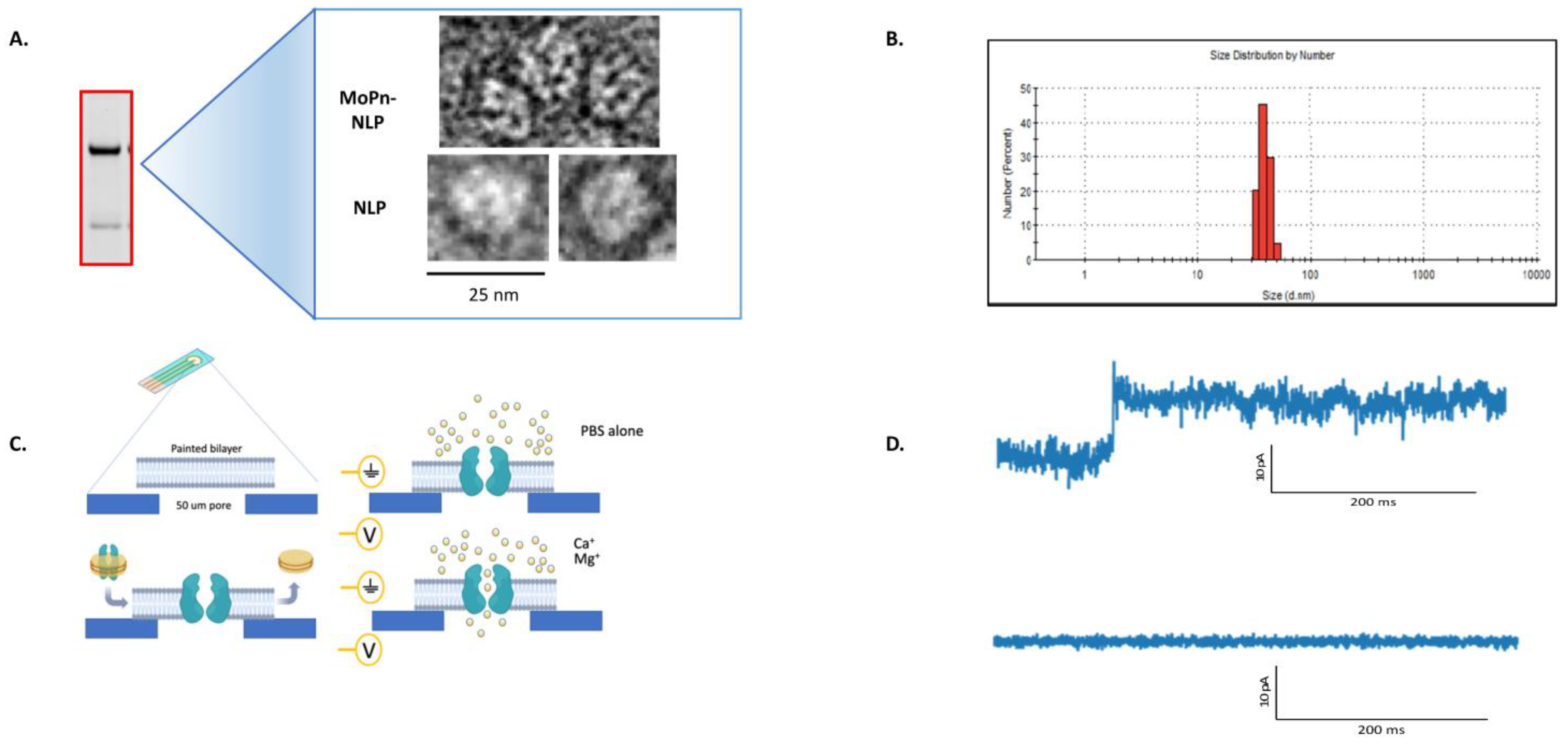
2.8. Dynamic Light Scattering (DLS)
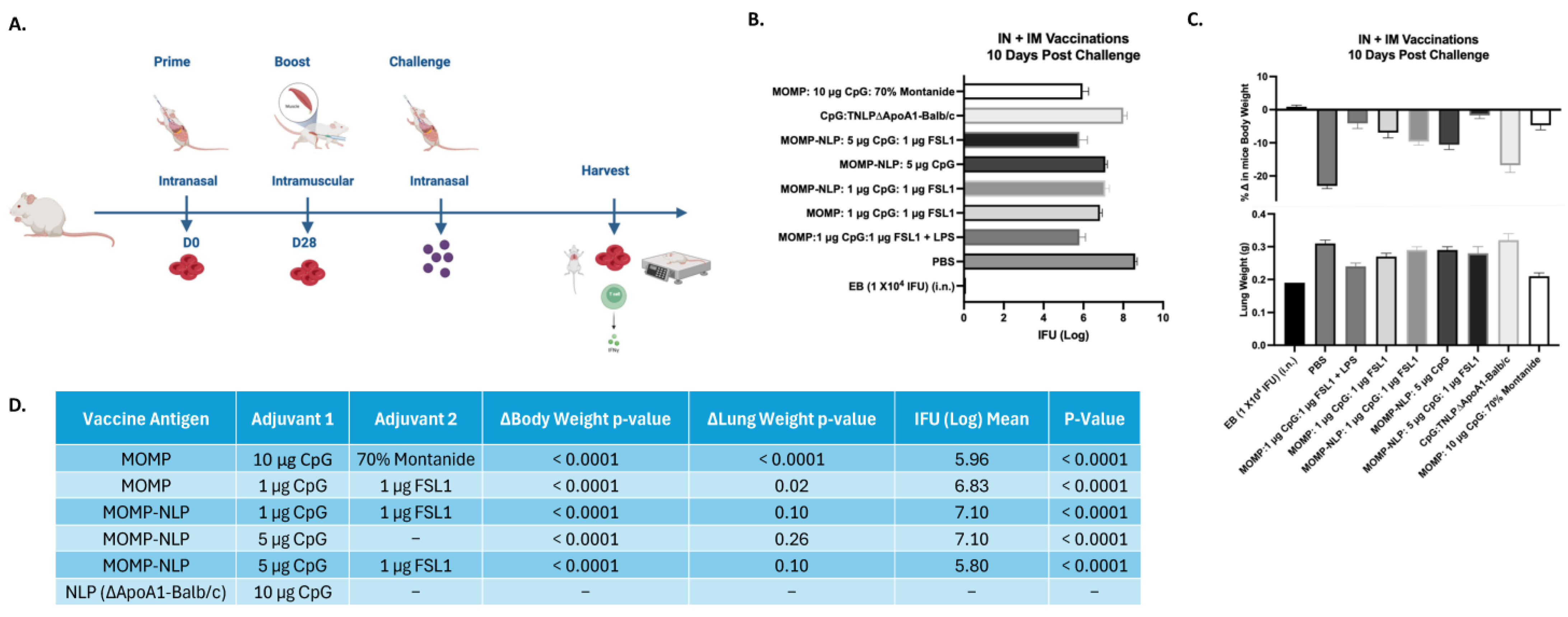
2.9. Animal Vaccinations for Immunological Studies
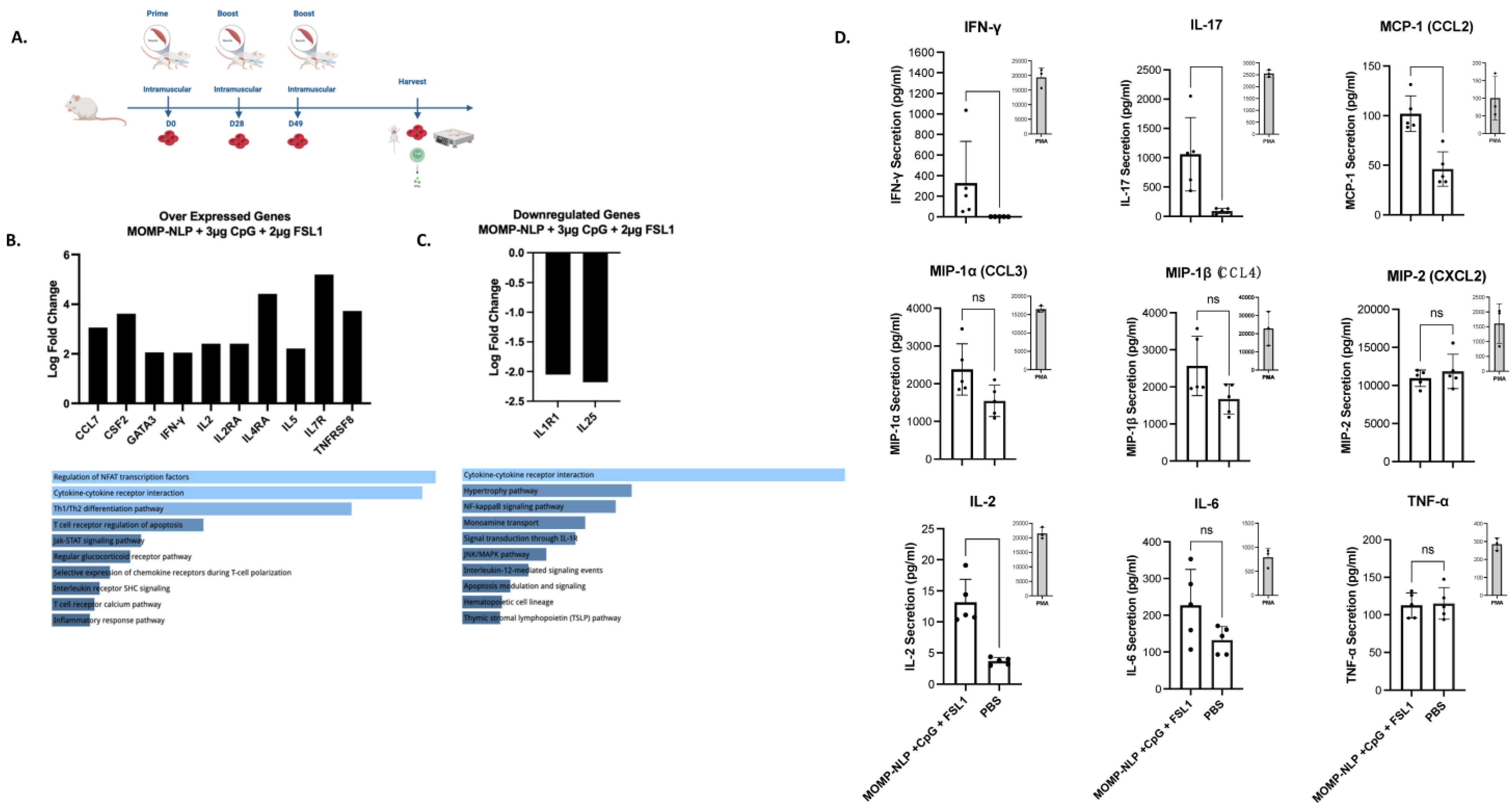
2.10. RNA Extraction and Gene Expression Analysis
2.11. Ex Vivo Splenocyte Restimulation and Multiplex Cytokine/Chemokine Array
2.12. Challenge Studies
2.13. Statistical Analyses
3. Results
3.1. Nanolipoproteins (NLPs) Are a Tool for Studying Membrane Bound Proteins
3.2. Cell-Free Screening Enables Rapid Lipid Testing and Optimization
3.3. Cell-Free Production and Purification of MOMP-NLP Protein Complex
3.4. Characterization of the MOMP-NLP Complex
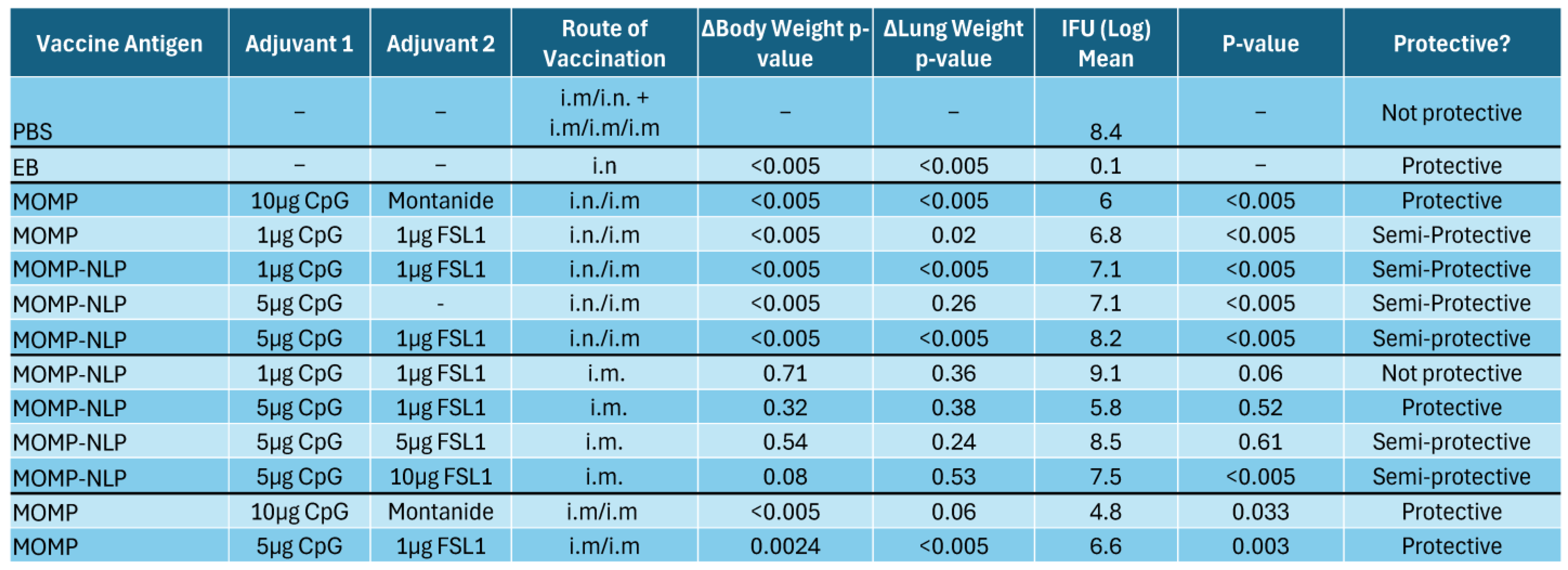
3.5. Effectiveness of NLP Encapsulated MOMP-Based Vaccine Formulations Administered Through Various Routes and Immunization Schedules
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grygiel-Gorniak, B.; Folga, B.A. Chlamydia trachomatis-An Emerging Old Entity? Microorganisms 2023, 11, 1283. [Google Scholar] [CrossRef] [PubMed]
- Footman, A.; Kanney, N.B.; Niccolai, L.M.; Zimet, G.D.; Overton, E.T.; Davies, S.L.; Van Der Pol, B. Perceived Need and Acceptance of a Future Chlamydia Vaccine among Health Care Providers. Sex. Transm. Dis. 2022, 49, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Raccagni, A.R.; Ranzenigo, M.; Bruzzesi, E.; Maci, C.; Castagna, A.; Nozza, S. Neisseria gonorrhoeae Antimicrobial Resistance: The Future of Antibiotic Therapy. J. Clin. Med. 2023, 12, 7767. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, C.; Sun, X.; Liu, J.; Zheng, H.; Yang, B.; Tang, W.; Wang, C. Chlamydia infection, PID, and infertility: Further evidence from a case-control study in China. BMC Womens Health 2022, 22, 294. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.C.; Nicola, M.R.C.; Bezerra, N.T.C.; Sardinha, J.C.G.; Morais, J.S.d.S.; Schettini, A.E.P. Genital ulcers caused by sexually transmitted agents. An. Bras. Dermatol. 2022, 97, 551–565. [Google Scholar] [CrossRef]
- Mohseni, M.; Sung, S.; Takov, V. Chlamydia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Rodrigues, R.; Sousa, C.; Vale, N. Chlamydia trachomatis as a Current Health Problem: Challenges and Opportunities. Diagnostics 2022, 12, 1795. [Google Scholar] [CrossRef]
- Mäki-Koivisto, V.; Sinikumpu, S.-P.; Jokelainen, J.; Aho-Laukkanen, E.; Junttila, I.S.; Huilaja, L. Impact of COVID-19 Pandemic on the Incidence of Sexually Transmitted Infections in Northern Finland in 2019 to 2022. Acta Derm. Venereol. 2022, 102, adv00795. [Google Scholar] [CrossRef]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef]
- Witkin, S.S.; Minis, E.; Athanasiou, A.; Leizer, J.; Linhares, I.M. Chlamydia trachomatis: The Persistent Pathogen. Clin. Vaccine Immunol. 2017, 24, e00203-17. [Google Scholar] [CrossRef]
- Bugalhao, J.N.; Mota, L.J. The multiple functions of the numerous Chlamydia trachomatis secreted proteins: The tip of the iceberg. Microb. Cell 2019, 6, 414–449. [Google Scholar] [CrossRef]
- Knapp, K.; Stary, G. Models for sexually transmitted infections. Drug Discov. Today Dis. Models 2020, 32, 1–6. [Google Scholar] [CrossRef]
- Khan, S.A.; Polkinghorne, A.; Waugh, C.; Hanger, J.; Loader, J.; Beagley, K.; Timms, P. Humoral immune responses in koalas (Phascolarctos cinereus) either naturally infected with Chlamydia pecorum or following administration of a recombinant chlamydial major outer membrane protein vaccine. Vaccine 2016, 34, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Kalbina, I.; Wallin, A.; Lindh, I.; Engström, P.; Andersson, S.; Strid, Å. A novel chimeric MOMP antigen expressed in Escherichia coli, Arabidopsis thaliana, and Daucus carota as a potential Chlamydia trachomatis vaccine candidate. Protein Expr. Purif. 2011, 80, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, L.; Angevine, M.; Kim, S.K.; Watkins, D.; DeMars, R. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect. Immun. 2000, 68, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef]
- Jiang, P.; Cai, Y.; Chen, J.; Ye, X.; Mao, S.; Zhu, S.; Xue, X.; Chen, S.; Zhang, L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine 2017, 35, 3096–3103. [Google Scholar] [CrossRef]
- Shelby, M.L.; He, W.; Dang, A.T.; Kuhl, T.L.; Coleman, M.A. Cell-Free Co-Translational Approaches for Producing Mammalian Receptors: Expanding the Cell-Free Expression Toolbox Using Nanolipoproteins. Front. Pharmacol 2019, 10, 744. [Google Scholar] [CrossRef]
- Levine, M.Z.; Gregorio, N.E.; Jewett, M.C.; Watts, K.R.; Oza, J.P. Escherichia coli-Based Cell-Free Protein Synthesis: Protocols for a robust, flexible, and accessible platform technology. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Darwish, M.; Gao, X.; Shatz, W.; Li, H.; Lin, M.; Franke, Y.; Tam, C.; Mortara, K.; Zilberleyb, I.; Hannoush, R.N.; et al. Nanolipoprotein particles for co-delivery of cystine-knot peptides and Fab-based therapeutics. Nanoscale Adv. 2021, 3, 3929–3941. [Google Scholar] [CrossRef]
- Tifrea, D.F.; He, W.; Pal, S.; Evans, A.C.; Gilmore, S.F.; Fischer, N.O.; Rasley, A.; Coleman, M.A.; de la Maza, L.M. Induction of Protection in Mice against a Chlamydia muridarum Respiratory Challenge by a Vaccine Formulated with the Major Outer Membrane Protein in Nanolipoprotein Particles. Vaccines 2021, 9, 755. [Google Scholar] [CrossRef]
- He, W.; Felderman, M.; Evans, A.C.; Geng, J.; Homan, D.; Bourguet, F.; Fischer, N.O.; Li, Y.; Lam, K.S.; Noy, A.; et al. Cell-free production of a functional oligomeric form of a Chlamydia major outer-membrane protein (MOMP) for vaccine development. J. Biol. Chem. 2017, 292, 15121–15132. [Google Scholar] [CrossRef] [PubMed]
- Cappuccio, J.A.; Blanchette, C.D.; Sulchek, T.A.; Arroyo, E.S.; Kralj, J.M.; Hinz, A.K.; Kuhn, E.A.; Chromy, B.A.; Segelke, B.W.; Rothschild, K.J.; et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol. Cell. Proteomics. 2008, 7, 2246–2253. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Dixit, S.; Verma, R.; Duncan, S.A.; Smith, L.; Giambartolomei, G.H.; Singh, S.R.; Dennis, V.A. Encapsulation of Recombinant MOMP in Extended-Releasing PLGA 85:15 Nanoparticles Confer Protective Immunity Against a Chlamydia muridarum Genital Challenge and Re-Challenge. Front. Immunol. 2021, 12, 660932. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Hou, B.; Wang, B.; Lin, X.; Gong, W.; Dong, H.; Zhu, S.; Chen, S.; Xue, X.; Zhao, K.-N.; et al. A multi-epitope vaccine based on Chlamydia trachomatis major outer membrane protein induces specific immunity in mice. Acta Biochim. Biophys. Sin. 2014, 46, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Theodor, I.; Peterson, E.M.; de la Maza, L.M. Immunization with the Chlamydia trachomatis mouse pneumonitis major outer membrane protein can elicit a protective immune response against a genital challenge. Infect. Immun. 2001, 69, 6240–6247. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Gilmore, S.F.; He, W.; Evans, A.C.; Tifrea, D.F.; Pal, S.; Segelke, B.; Peters, S.K.G.; Vannest, B.D.; Fischer, N.O.; Rasley, A.; et al. Cell-free Scaled Production and Adjuvant Addition to a Recombinant Major Outer Membrane Protein from Chlamydia muridarum for Vaccine Development. J. Vis. Exp. 2022, 181, e63028. [Google Scholar] [CrossRef]
- Tifrea, D.F.; Sun, G.; Pal, S.; Zardeneta, G.; Cocco, M.J.; Popot, J.-L.; de la Maza, L.M. Amphipols stabilize the Chlamydia major outer membrane protein and enhance its protective ability as a vaccine. Vaccine 2011, 29, 4623–4631. [Google Scholar] [CrossRef]
- Adachi, J.; Katsura, K.; Seki, E.; Takemoto, C.; Shirouzu, M.; Terada, T.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Cell-Free Protein Synthesis Using S30 Extracts from Escherichia coli RFzero Strains for Efficient Incorporation of Non-Natural Amino Acids into Proteins. Int. J. Mol. Sci. 2019, 20, 492. [Google Scholar] [CrossRef] [PubMed]
- Dopp BJ, L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Cappuccio, J.A.; Hinz, A.K.; Kuhn, E.A.; Fletcher, J.E.; Arroyo, E.S.; Henderson, P.T.; Blanchette, C.D.; Walsworth, V.L.; Corzett, M.H.; Law, R.J.; et al. Cell-free expression for nanolipoprotein particles: Building a high-throughput membrane protein solubility platform. In High Throughput Protein Expression and Purification; Humana Press: Totowa, NJ, USA, 2009; Volume 498, pp. 273–296. [Google Scholar] [CrossRef]
- Sun, G.; Pal, S.; Sarcon, A.K.; Kim, S.; Sugawara, E.; Nikaido, H.; Cocco, M.J.; Peterson, E.M.; de la Maza, L.M. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 2007, 189, 6222–6235. [Google Scholar] [CrossRef] [PubMed]
- Macmillan, L.; Ifere, G.O.; He, Q.; Igietseme, J.U.; Kellar, K.L.; Okenu, D.M.; Eko, F.O. A recombinant multivalent combination vaccine protects against Chlamydia and genital herpes. FEMS Immunol. Med. Microbiol. 2007, 49, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, M.; Li, K.; Dong, X.; Han, J.; Zhang, Q. Cryo-electron tomography of Chlamydia trachomatis gives a clue to the mechanism of outer membrane changes. J. Electron Microsc. 2010, 59, 237–241. [Google Scholar] [CrossRef]
- Mangia, A.; Serra, N.; Cocomazzi, G.; Giambra, V.; Antinucci, S.; Maiorana, A.; Giuliani, F.; Montomoli, E.; Cantaloni, P.; Manenti, A.; et al. Cellular and Humoral Immune Responses and Breakthrough Infections After Two Doses of BNT162b Vaccine in Healthcare Workers (HW) 180 Days After the Second Vaccine Dose. Front. Public Health 2022, 10, 847384. [Google Scholar] [CrossRef]
- Pal, S.; Cruz-Fisher, M.I.; Cheng, C.; Carmichael, J.R.; Tifrea, D.F.; Tatarenkova, O.; de la Maza, L.M. Vaccination with the recombinant major outer membrane protein elicits long-term protection in mice against vaginal shedding and infertility following a Chlamydia muridarum genital challenge. npj Vaccines 2020, 5, 90. [Google Scholar] [CrossRef]
- Abisoye-Ogunniyan, A.; Carrano, I.M.; Weilhammer, D.R.; Gilmore, S.F.; Fischer, N.O.; Pal, S.; de la Maza, L.M.; Coleman, M.A.; Rasley, A. A Survey of Preclinical Studies Evaluating Nanoparticle-Based Vaccines Against Non-Viral Sexually Transmitted Infections. Front. Pharmacol. 2021, 12, 768461. [Google Scholar] [CrossRef]
- Awate, S.; Babiuk, L.A.; Mutwiri, G. Mechanisms of action of adjuvants. Front. Immunol. 2013, 4, 114. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, K. Overview of vaccine adjuvants. Med. Drug Discov. 2021, 11, 100103. [Google Scholar] [CrossRef]
- Zuo, Z.; Zou, Y.; Li, Q.; Guo, Y.; Zhang, T.; Wu, J.; He, C.; Eko, F.O. Intranasal immunization with inactivated chlamydial elementary bodies formulated in VCG-chitosan nanoparticles induces robust immunity against intranasal Chlamydia psittaci challenge. Sci. Rep. 2021, 11, 10389. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Fajardo-Cavazos, P.; Setlow, P. Levels of mRNAs which code for small, acid-soluble spore proteins and their LacZ gene fusions in sporulating cells of Bacillus subtilis. Nucleic Acids Res. 1988, 16, 6567–6583. [Google Scholar] [CrossRef] [PubMed]
- Fairley, S.J.; Singh, S.R.; Yilma, A.N.; Waffo, A.B.; Subbarayan, P.; Dixit, S.; Taha, M.A.; Cambridge, C.D.; Dennis, V.A. Chlamydia trachomatis recombinant MOMP encapsulated in PLGA nanoparticles triggers primarily T helper 1 cellular and antibody immune responses in mice: A desirable candidate nanovaccine. Int. J. Nanomed. 2013, 8, 2085–2099. [Google Scholar] [CrossRef] [PubMed]
- Macian, F. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef]
- Lee, J.U.; Kim, L.K.; Choi, J.M. Revisiting the Concept of Targeting NFAT to Control T Cell Immunity and Autoimmune Diseases. Front. Immunol. 2018, 9, 2747. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Mukherjee, R. Cytokine-mediated communication: A quantitative appraisal of immune complexity. Nat. Rev. Immunol. 2019, 19, 205–217. [Google Scholar] [CrossRef]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509–517. [Google Scholar] [CrossRef]
- Lee, G.R. Molecular Mechanisms of T Helper Cell Differentiation and Functional Specialization. Immune Netw. 2023, 23, e4. [Google Scholar] [CrossRef]
- Butcher, M.J.; Zhu, J. Recent advances in understanding the Th1/Th2 effector choice. Fac. Rev. 2021, 10, 30. [Google Scholar] [CrossRef]
- Valizadeh, A.; Khosravi, A.; Zadeh, L.J.; Parizad, E.G. Role of IL-25 in Immunity. J. Clin. Diagn. Res. 2015, 9, OE01–OE04. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, F.; Tao, H.; Nawaz, W.; Chen, D.; Wu, Z. The potential roles of interleukin-25 in infectious diseases. Front. Immunol. 2022, 13, 986118. [Google Scholar] [CrossRef] [PubMed]
- Slepenkin, A.; Pal, S.; Rasley, A.; Coleman, M.A.; de la Maza, L.M. Safety and efficacy of C. muridarum vaccines adjuvanted with CpG-1826 and four concentrations of Montanide-ISA-720-VG. npj Vaccines 2024, 9, 104. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohagheghi, M.; Abisoye-Ogunniyan, A.; Evans, A.C.; Peterson, A.E.; Bude, G.A.; Hoang-Phou, S.; Vannest, B.D.; Hall, D.; Rasley, A.; Weilhammer, D.R.; et al. Cell-Free Screening, Production and Animal Testing of a STI-Related Chlamydial Major Outer Membrane Protein Supported in Nanolipoproteins. Vaccines 2024, 12, 1246. https://doi.org/10.3390/vaccines12111246
Mohagheghi M, Abisoye-Ogunniyan A, Evans AC, Peterson AE, Bude GA, Hoang-Phou S, Vannest BD, Hall D, Rasley A, Weilhammer DR, et al. Cell-Free Screening, Production and Animal Testing of a STI-Related Chlamydial Major Outer Membrane Protein Supported in Nanolipoproteins. Vaccines. 2024; 12(11):1246. https://doi.org/10.3390/vaccines12111246
Chicago/Turabian StyleMohagheghi, Mariam, Abisola Abisoye-Ogunniyan, Angela C. Evans, Alexander E. Peterson, Gregory A. Bude, Steven Hoang-Phou, Byron Dillon Vannest, Dominique Hall, Amy Rasley, Dina R. Weilhammer, and et al. 2024. "Cell-Free Screening, Production and Animal Testing of a STI-Related Chlamydial Major Outer Membrane Protein Supported in Nanolipoproteins" Vaccines 12, no. 11: 1246. https://doi.org/10.3390/vaccines12111246
APA StyleMohagheghi, M., Abisoye-Ogunniyan, A., Evans, A. C., Peterson, A. E., Bude, G. A., Hoang-Phou, S., Vannest, B. D., Hall, D., Rasley, A., Weilhammer, D. R., Fischer, N. O., He, W., Robinson, B. V., Pal, S., Slepenkin, A., de la Maza, L., & Coleman, M. A. (2024). Cell-Free Screening, Production and Animal Testing of a STI-Related Chlamydial Major Outer Membrane Protein Supported in Nanolipoproteins. Vaccines, 12(11), 1246. https://doi.org/10.3390/vaccines12111246







