Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Is a Promising Signature to Predict Prognosis and Therapies for Hepatocellular Carcinoma (HCC)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Public Data Collection Acquisition
2.2. Consensus Clustering Analysis
2.3. Comparison of the Different Molecular Classifications of HCC
2.4. MAM-Score and TME-Score Calculations
2.5. Prognostic Model Validation
2.6. Single-Cell Data Analysis
2.7. Single-Cell Gene Set Enrichment Analysis
2.8. Mapping Specific Regulon Networks by SCENIC
2.9. CellChat Analysis
2.10. Regulon Analysis
2.11. Calculation of Microenvironment Cell Abundance and Immune Infiltration in the Low- and High-MAM-Score Groups
2.12. Prediction of the Response to Immunotherapy in HCC
2.13. Prediction of the Response to Chemotherapy in HCC
2.14. Statistical Analysis
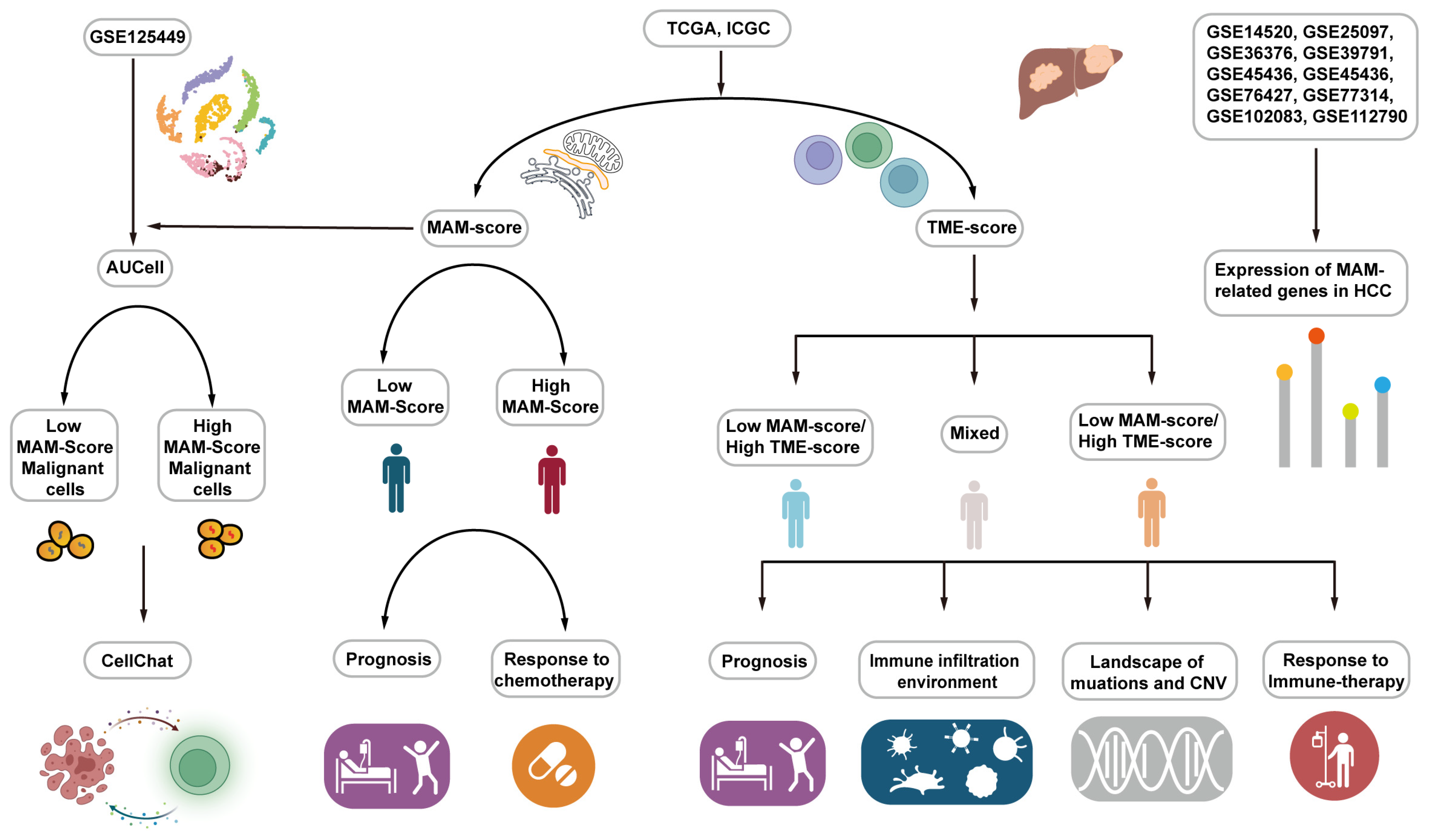
2.15. Work Flow
3. Results
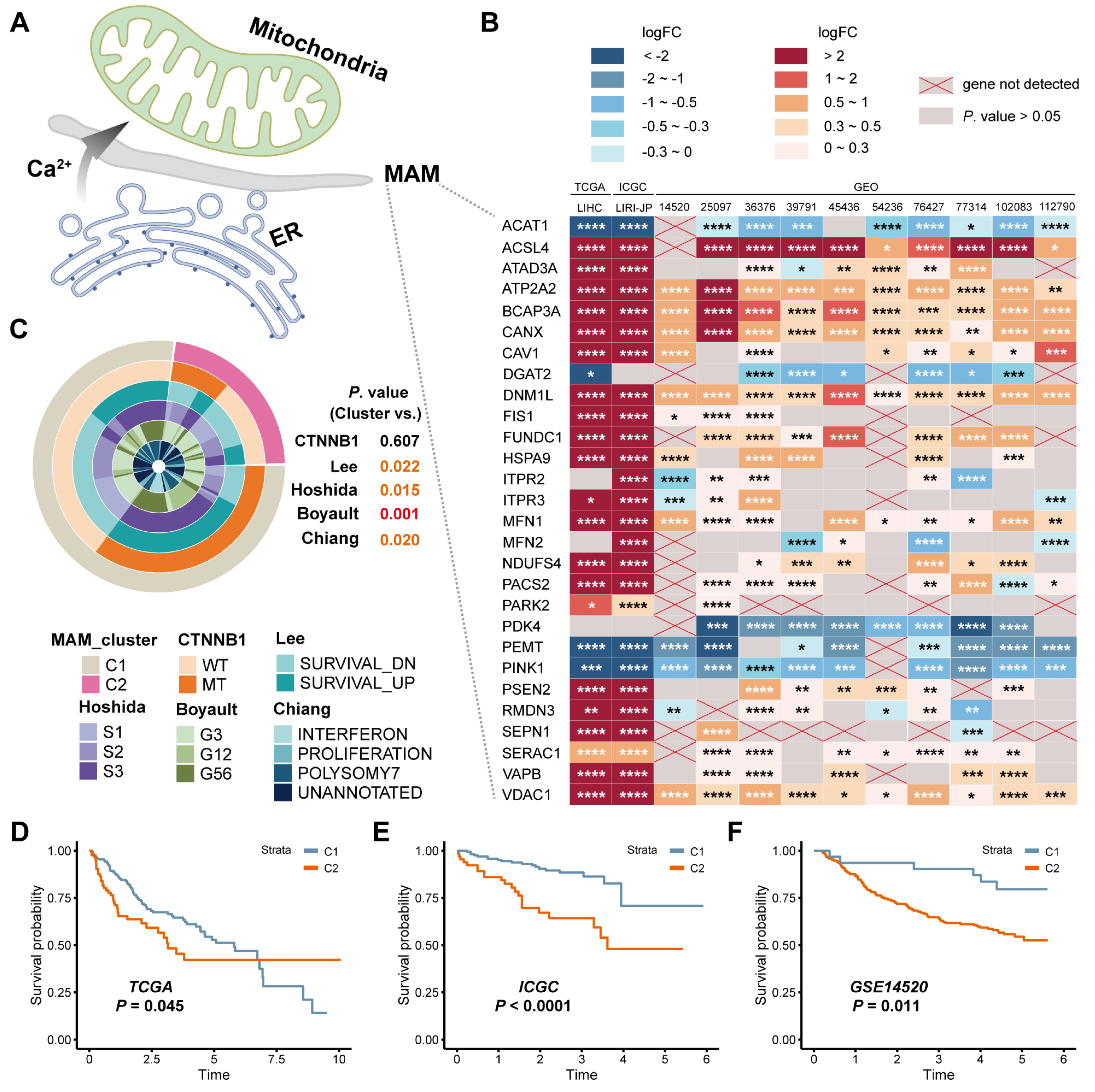
3.1. Identification of the Expression and Prognostic Value of MAM in HCC
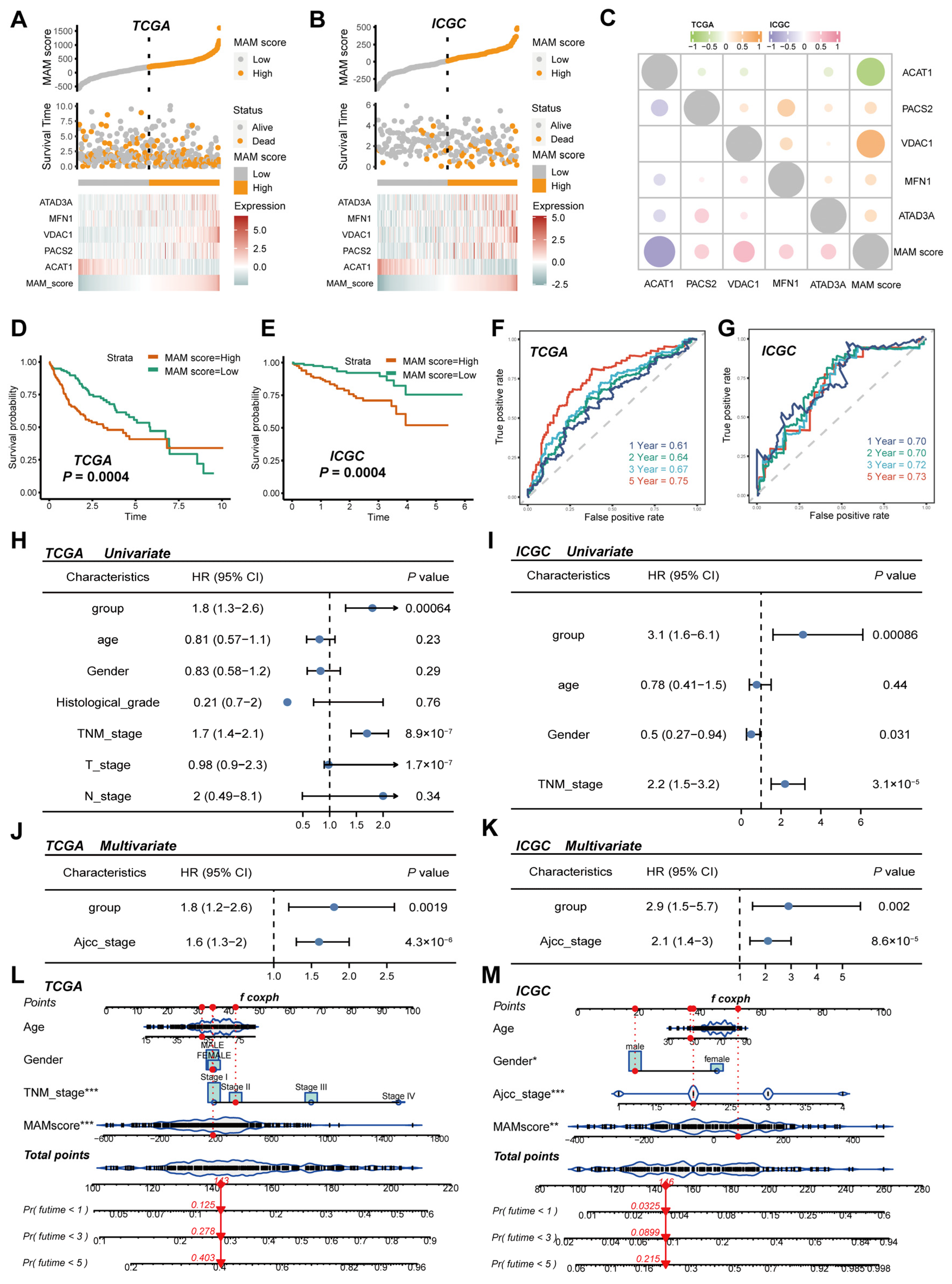
3.2. Construction and Verification of the MAM-Based Score in HCC
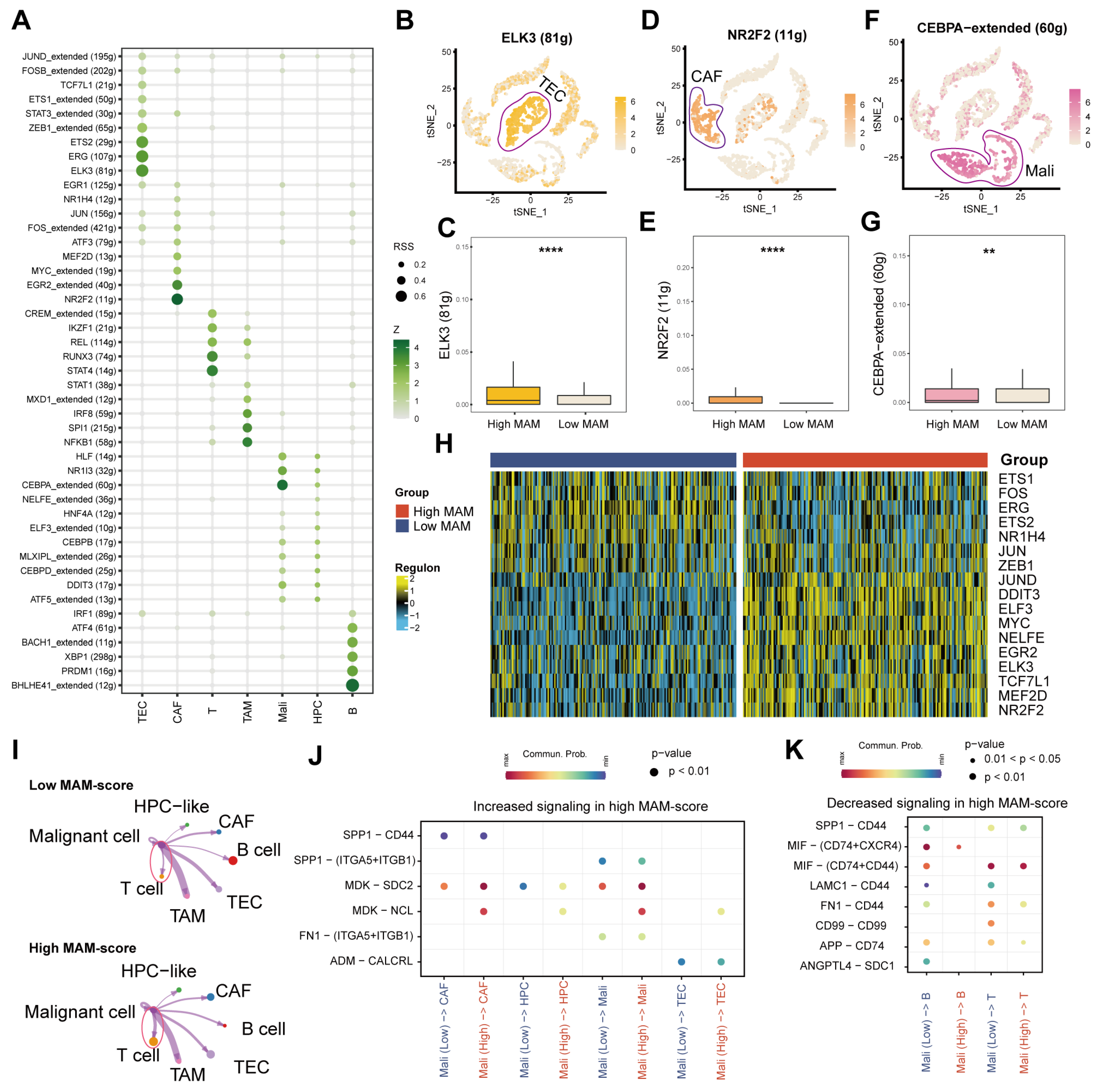
3.3. Identification of the MAM Score and Its Potential Enriched Pathways at the Single-Cell Level
3.4. Identification of the Underlying Mechanisms of MAM
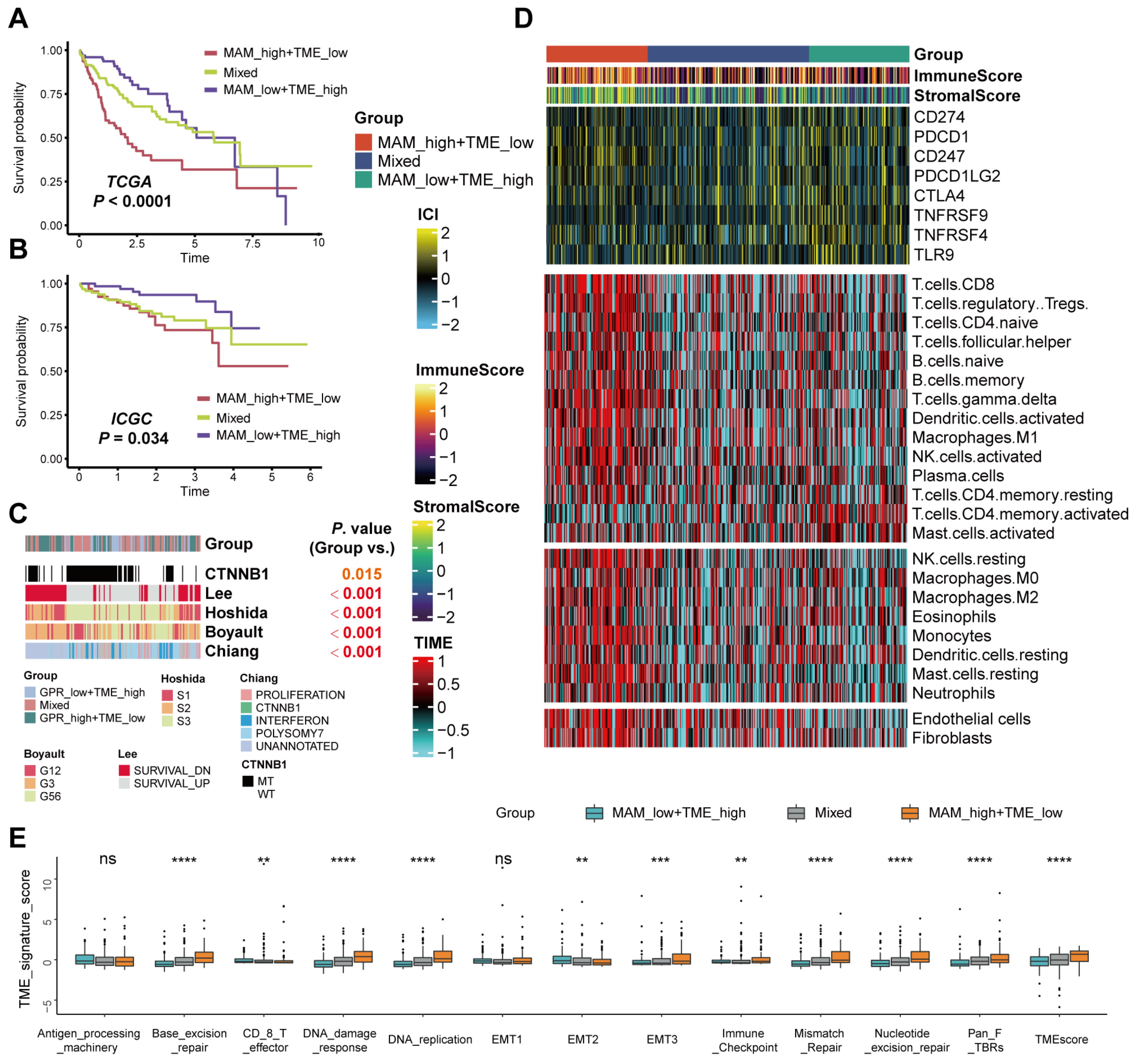
3.5. Differences in the Tumor Microenvironment Infiltration between Different MAM—Subgroups
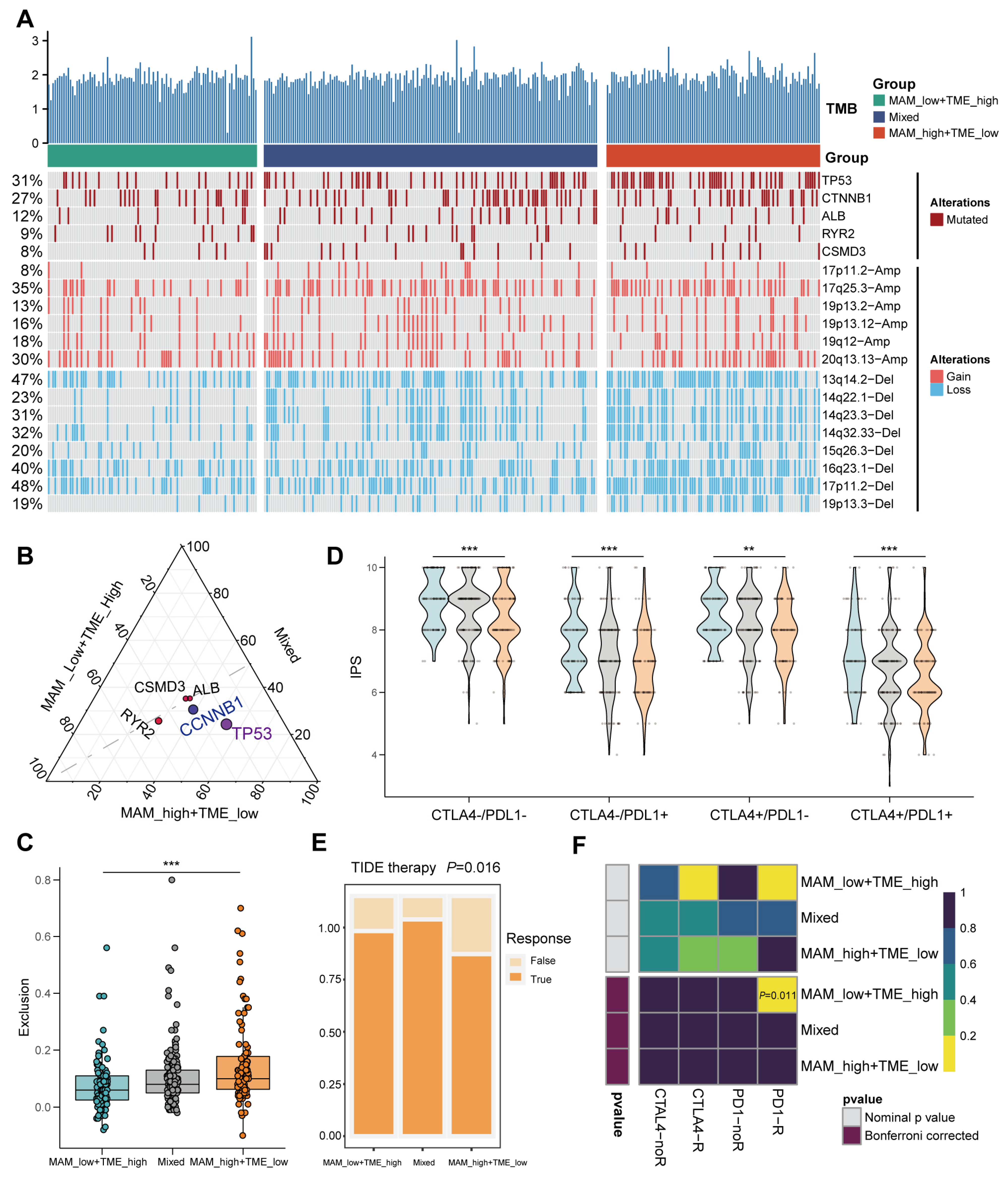
3.6. Mutation Landscape and Response to the Immune Checkpoint Block (ICB) Therapy
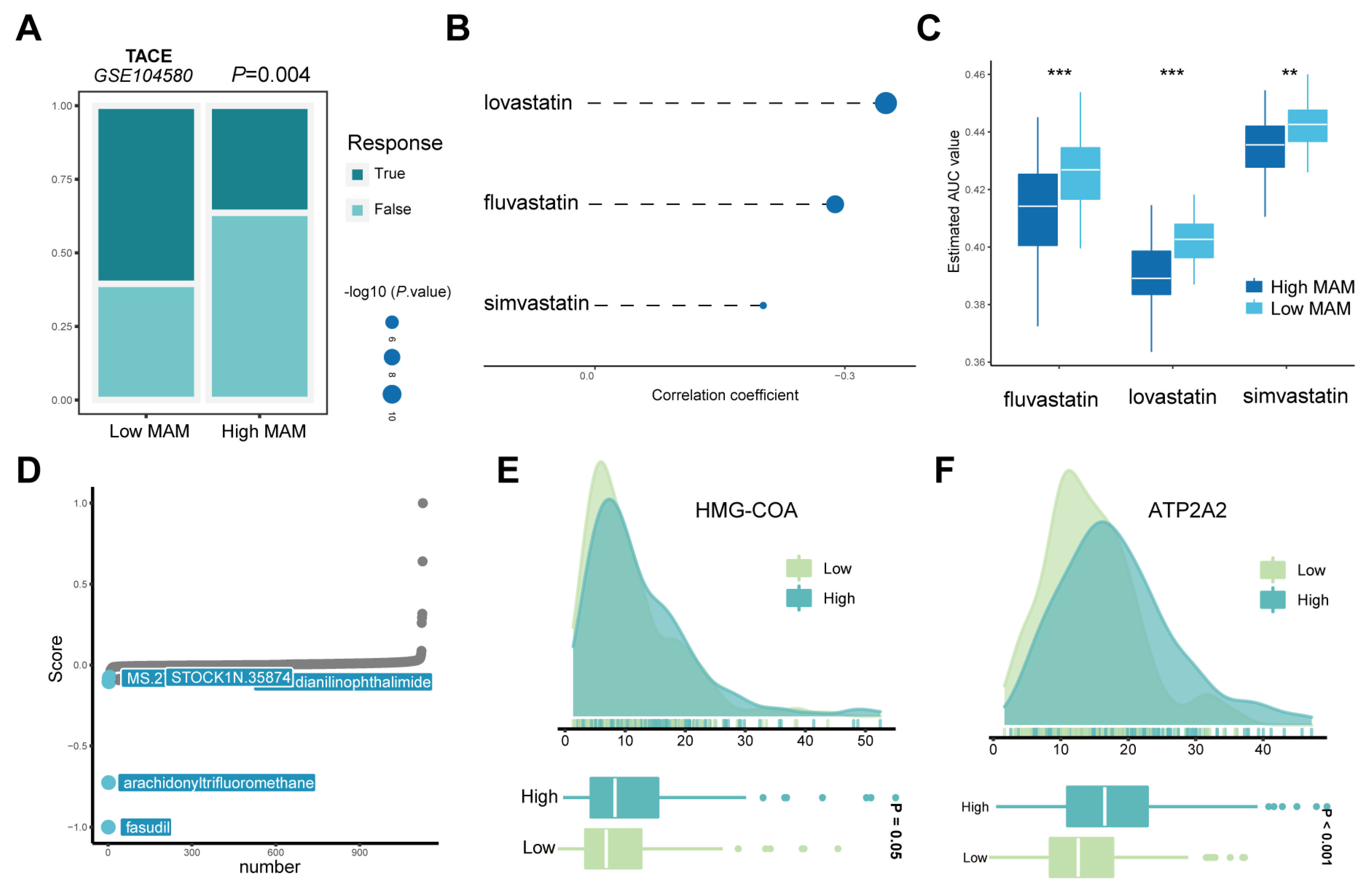
3.7. Response of the High- and Low-MAM-Score Groups to Chemotherapy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MAM | mitochondria-associated endoplasmic reticulum membrane |
| HCC | hepatocellular carcinoma |
| ER | endoplasmic reticulum |
| TME | tumor microenvironment |
| CNV | copy number variations |
| TACE | transcatheter arterial chemoembolization |
| ICGC | International Cancer Genome Consortium |
| CCLE | Cancer Cell Line Encyclopedia |
| KM | Kaplan–Meier |
| ROC | receiver operating characteristic |
| PCA | principal component analysis |
| T-SNE | t-distributed stochastic neighbor embedding |
| CAF | cancer-associated fibroblasts |
| TEC | tumor-associated endothelial cells |
| TAM | tumor-associated macrophages |
| HPC-like | hepatic progenitor cells |
| TF | transcriptional factors |
| TCIA | Cancer Immunome Atlas |
| CTRP | Cancer Therapeutics Response Portal |
| AUC | area under the curve |
Appendix A

References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bayard, Q.; Meunier, L.; Peneau, C.; Renault, V.; Shinde, J.; Nault, J.C.; Mami, I.; Couchy, G.; Amaddeo, G.; Tubacher, E.; et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat. Commun. 2018, 9, 5235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Shimizu, S.; Sato, T.; Morimoto, M.; Kojima, Y.; Inaba, Y.; Hagihara, A.; Kudo, M.; Nakamori, S.; Kaneko, S.; et al. Sorafenib plus hepatic arterial infusion chemotherapy with cisplatin versus sorafenib for advanced hepatocellular carcinoma: Randomized phase II trial. Ann. Oncol. 2016, 27, 2090–2096. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Haas, N.B.; Donskov, F.; Gross-Goupil, M.; Varlamov, S.; Kopyltsov, E.; Lee, J.L.; Melichar, B.; Rini, B.I.; Choueiri, T.K.; et al. Randomized Phase III Trial of Adjuvant Pazopanib Versus Placebo After Nephrectomy in Patients with Localized or Locally Advanced Renal Cell Carcinoma. J. Clin. Oncol. 2017, 35, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Satala, C.B.; Jung, I.; Kobori, L.; Kovacs, Z.; Fodor, D.; Szodorai, R.; Gurzu, S. Benefits of the 8th American Joint Committee on Cancer System for Hepatocellular Carcinoma Staging. J. Gastrointest. Cancer 2021, 52, 243–248. [Google Scholar] [CrossRef]
- Hung, M.H.; Wang, X.W. Molecular Alterations and Heterogeneity in Hepatocellular Carcinoma. In Hepatocellular Carcinoma: Translational Precision Medicine Approaches; Cham, H.Y., Ed.; Human Press: Totowa, NJ, USA, 2019; pp. 293–316. [Google Scholar]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Li, B.; Cui, Y.; Nambiar, D.K.; Sunwoo, J.B.; Li, R. The Immune Subtypes and Landscape of Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 3528–3537. [Google Scholar] [CrossRef] [Green Version]
- Ruella, M.; Klichinsky, M.; Kenderian, S.S.; Shestova, O.; Ziober, A.; Kraft, D.O.; Feldman, M.; Wasik, M.A.; June, C.H.; Gill, S. Overcoming the Immunosuppressive Tumor Microenvironment of Hodgkin Lymphoma Using Chimeric Antigen Receptor T Cells. Cancer Discov. 2017, 7, 1154–1167. [Google Scholar] [CrossRef] [Green Version]
- Slaney, C.Y.; von Scheidt, B.; Davenport, A.J.; Beavis, P.A.; Westwood, J.A.; Mardiana, S.; Tscharke, D.C.; Ellis, S.; Prince, H.M.; Trapani, J.A.; et al. Dual-specific Chimeric Antigen Receptor T Cells and an Indirect Vaccine Eradicate a Variety of Large Solid Tumors in an Immunocompetent, Self-antigen Setting. Clin. Cancer Res. 2017, 23, 2478–2490. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Lopez-Moyado, I.F.; Seo, H.; Lio, C.J.; Hempleman, L.J.; Sekiya, T.; Yoshimura, A.; Scott-Browne, J.P.; Rao, A. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 2019, 567, 530–534. [Google Scholar] [CrossRef]
- Hato, T.; Goyal, L.; Greten, T.F.; Duda, D.G.; Zhu, A.X. Immune checkpoint blockade in hepatocellular carcinoma: Current progress and future directions. Hepatology 2014, 60, 1776–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vance, J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990, 265, 7248–7256. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wen, Y.; Dong, J.; Cao, C.; Yuan, S. Systematic In-Depth Proteomic Analysis of Mitochondria-Associated Endoplasmic Reticulum Membranes in Mouse and Human Testes. Proteomics 2018, 18, e1700478. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Varnai, P.; Golenar, T.; Roy, S.; Purkins, G.; Schneider, T.G.; Balla, T.; Hajnoczky, G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell 2010, 39, 121–132. [Google Scholar] [CrossRef]
- Doghman-Bouguerra, M.; Lalli, E. ER-mitochondria interactions: Both strength and weakness within cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dai, X.; Wu, S.; Xu, W.; Song, P.; Huang, K. FUNDC1-dependent mitochondria-associated endoplasmic reticulum membranes are involved in angiogenesis and neoangiogenesis. Nat. Commun. 2021, 12, 2616. [Google Scholar] [CrossRef]
- Li, J.; Qi, F.; Su, H.; Zhang, C.; Zhang, Q.; Chen, Y.; Chen, P.; Su, L.; Chen, Y.; Yang, Y.; et al. GRP75-faciliated Mitochondria-associated ER Membrane (MAM) Integrity controls Cisplatin-resistance in Ovarian Cancer Patients. Int. J. Biol. Sci. 2022, 18, 2914–2931. [Google Scholar] [CrossRef]
- Shimada, S.; Mogushi, K.; Akiyama, Y.; Furuyama, T.; Watanabe, S.; Ogura, T.; Ogawa, K.; Ono, H.; Mitsunori, Y.; Ban, D.; et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. eBioMedicine 2019, 40, 457–470. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar]
- Park, S.Y. Nomogram: An analogue tool to deliver digital knowledge. J. Thorac. Cardiovasc. Surg. 2018, 155, 1793. [Google Scholar] [CrossRef] [Green Version]
- Aibar, S.; Gonzalez-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Yang, S.; Ma, J.; Chen, Z.; Song, G.; Rao, D.; Cheng, Y.; Huang, S.; Liu, Y.; Jiang, S.; et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022, 12, 134–153. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, D.; Li, M.; Zhou, R.; Zhang, J.; Sun, H.; Shi, M.; Bin, J.; Liao, Y.; Rao, J.; Liao, W. Tumor Microenvironment Characterization in Gastric Cancer Identifies Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol. Res. 2019, 7, 737–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, K.; Murphy, D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Hernandez, M.O.; Zhao, Y.; Mehta, M.; Tran, B.; Kelly, M.; Rae, Z.; Hernandez, J.M.; Davis, J.L.; Martin, S.P.; et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 2019, 36, 418–430.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Raoul, J.L.; Forner, A.; Bolondi, L.; Cheung, T.T.; Kloeckner, R.; de Baere, T. Updated use of TACE for hepatocellular carcinoma treatment: How and when to use it based on clinical evidence. Cancer Treat. Rev. 2019, 72, 28–36. [Google Scholar] [CrossRef]
- Zhong, S.; Li, L.; Liang, N.; Zhang, L.; Xu, X.; Chen, S.; Yin, H. Acetaldehyde Dehydrogenase 2 regulates HMG-CoA reductase stability and cholesterol synthesis in the liver. Redox Biol. 2021, 41, 101919. [Google Scholar] [CrossRef]
- Chen, Y.C.; Yuan, T.Y.; Zhang, H.F.; Wang, D.S.; Niu, Z.R.; Li, L.; Fang, L.H.; Du, G.H. Fasudil evokes vasodilatation of rat mesenteric vascular bed via Ca(2+) channels and Rho/ROCK pathway. Eur. J. Pharmacol. 2016, 788, 226–233. [Google Scholar] [CrossRef]
- Nakajima, K.; Ishiwata, M.; Weitemier, A.Z.; Shoji, H.; Monai, H.; Miyamoto, H.; Yamakawa, K.; Miyakawa, T.; McHugh, T.J.; Kato, T. Brain-specific heterozygous loss-of-function of ATP2A2, endoplasmic reticulum Ca2+ pump responsible for Darier's disease, causes behavioral abnormalities and a hyper-dopaminergic state. Hum. Mol. Genet. 2021, 30, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Means, R.E.; Katz, S.G. Yes, MAM! Mol. Cell. Oncol. 2021, 8, 1919473. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, A. The recent insights into the function of ACAT1: A possible anti-cancer therapeutic target. Life Sci. 2019, 232, 116592. [Google Scholar] [CrossRef]
- Zhu, Y.; Gu, L.; Lin, X.; Liu, C.; Lu, B.; Cui, K.; Zhou, F.; Zhao, Q.; Prochownik, E.V.; Fan, C.; et al. Dynamic Regulation of ME1 Phosphorylation and Acetylation Affects Lipid Metabolism and Colorectal Tumorigenesis. Mol. Cell 2020, 77, 138–149.e5. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.J.; Li, X.; Zeng, Y.; Jiang, J.; Jiang, R. Icariin inhibits the formation of mitochondria-associated membranes (MAMs) and improves erectile function in rats treated with prostate radiation. Andrology 2022, 10, 1208–1216. [Google Scholar] [CrossRef]
- Thoudam, T.; Ha, C.M.; Leem, J.; Chanda, D.; Park, J.S.; Kim, H.J.; Jeon, J.H.; Choi, Y.K.; Liangpunsakul, S.; Huh, Y.H.; et al. PDK4 Augments ER-Mitochondria Contact to Dampen Skeletal Muscle Insulin Signaling During Obesity. Diabetes 2019, 68, 571–586. [Google Scholar] [CrossRef]
- Azeez, J.M.; Vini, R.; Remadevi, V.; Surendran, A.; Jaleel, A.; Santhosh Kumar, T.R.; Sreeja, S. VDAC1 and SERCA3 Mediate Progesterone-Triggered Ca2(+) Signaling in Breast Cancer Cells. J. Proteome Res. 2018, 17, 698–709. [Google Scholar] [CrossRef]
- Li, C.; Liu, J.; Hou, W.; Kang, R.; Tang, D. STING1 Promotes Ferroptosis Through MFN1/2-Dependent Mitochondrial Fusion. Front. Cell. Dev. Biol. 2021, 9, 698679. [Google Scholar] [CrossRef]
- Lang, L.; Loveless, R.; Teng, Y. Emerging Links between Control of Mitochondrial Protein ATAD3A and Cancer. Int. J. Mol. Sci. 2020, 21, 7917. [Google Scholar] [CrossRef]
- Sala-Vila, A.; Navarro-Lerida, I.; Sanchez-Alvarez, M.; Bosch, M.; Calvo, C.; Lopez, J.A.; Calvo, E.; Ferguson, C.; Giacomello, M.; Serafini, A.; et al. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 2016, 6, 27351. [Google Scholar] [CrossRef] [Green Version]
- Wiley, C.D.; Velarde, M.C.; Lecot, P.; Liu, S.; Sarnoski, E.A.; Freund, A.; Shirakawa, K.; Lim, H.W.; Davis, S.S.; Ramanathan, A.; et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016, 23, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Wang, X. Lysosome biogenesis: Regulation and functions. J. Cell Biol 2021, 220, e202102001. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Trere, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef] [PubMed]
- Sckisel, G.D.; Mirsoian, A.; Minnar, C.M.; Crittenden, M.; Curti, B.; Chen, J.Q.; Blazar, B.R.; Borowsky, A.D.; Monjazeb, A.M.; Murphy, W.J. Differential phenotypes of memory CD4 and CD8 T cells in the spleen and peripheral tissues following immunostimulatory therapy. J. Immunother. Cancer 2017, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Nagasaki, J.; Inozume, T.; Sax, N.; Ariyasu, R.; Ishikawa, M.; Yamashita, K.; Kawazu, M.; Ueno, T.; Irie, T.; Tanji, E.; et al. PD-1 blockade therapy promotes infiltration of tumor-attacking exhausted T cell clonotypes. Cell Rep. 2022, 38, 110331. [Google Scholar] [CrossRef]
- Ke, L.; Shen, J.; Feng, J.; Chen, J.; Shen, S.; Li, S.; Kuang, M.; Liang, L.; Lu, C.; Li, D.; et al. Somatic Mutation Profiles Revealed by Next Generation Sequencing (NGS) in 39 Chinese Hepatocellular Carcinoma Patients. Front. Mol. Biosci. 2021, 8, 800679. [Google Scholar] [CrossRef]
- Long, J.; Wang, A.; Bai, Y.; Lin, J.; Yang, X.; Wang, D.; Yang, X.; Jiang, Y.; Zhao, H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. eBioMedicine 2019, 42, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Pinyol, R.; Sia, D.; Llovet, J.M. Immune Exclusion-Wnt/CTNNB1 Class Predicts Resistance to Immunotherapies in HCC. Clin. Cancer Res. 2019, 25, 2021–2023. [Google Scholar] [CrossRef] [Green Version]
- Biton, J.; Mansuet-Lupo, A.; Pecuchet, N.; Alifano, M.; Ouakrim, H.; Arrondeau, J.; Boudou-Rouquette, P.; Goldwasser, F.; Leroy, K.; Goc, J.; et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin. Cancer Res. 2018, 24, 5710–5723. [Google Scholar] [CrossRef] [Green Version]
- Malekzadeh, P.; Pasetto, A.; Robbins, P.F.; Parkhurst, M.R.; Paria, B.C.; Jia, L.; Gartner, J.J.; Hill, V.; Yu, Z.; Restifo, N.P.; et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Investig. 2019, 129, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Edeline, J.; Meyer, T.; Blanc, J.F.; Raoul, J.L. New Challenges Facing Systemic Therapies of Advanced HCC in the Era of Different First-Line Immunotherapy-Based Combinations. Cancers 2022, 14, 5868. [Google Scholar] [CrossRef] [PubMed]
- Guven, D.C.; Erul, E.; Sahin, T.K.; Dizdar, O.; Yalcin, S.; Sahin, I.H. The benefit of immunotherapy in patients with hepatocellular carcinoma: A systematic review and meta-analysis. Future Med. 2022, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Han, J.; Yang, Y.; Chen, Y. PD-1/PD-L1 checkpoint inhibitors in advanced hepatocellular carcinoma immunotherapy. Front. Immunol. 2022, 13, 1070961. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.R.; Imrichova, H.; Wang, H.; Chao, T.; Xiao, Z.; Gao, M.; Rincon-Restrepo, M.; Franco, F.; Genolet, R.; Cheng, W.C.; et al. Disturbed mitochondrial dynamics in CD8(+) TILs reinforce T cell exhaustion. Nat. Immunol. 2020, 21, 1540–1551. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Xia, S.; Zhang, L.; Qin, X.; Zhu, Z.; Ma, T.; Lu, S.; Chen, J.; Shi, X.; Ren, H. Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Is a Promising Signature to Predict Prognosis and Therapies for Hepatocellular Carcinoma (HCC). J. Clin. Med. 2023, 12, 1830. https://doi.org/10.3390/jcm12051830
Chen Y, Xia S, Zhang L, Qin X, Zhu Z, Ma T, Lu S, Chen J, Shi X, Ren H. Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Is a Promising Signature to Predict Prognosis and Therapies for Hepatocellular Carcinoma (HCC). Journal of Clinical Medicine. 2023; 12(5):1830. https://doi.org/10.3390/jcm12051830
Chicago/Turabian StyleChen, Yuyan, Senzhe Xia, Lu Zhang, Xueqian Qin, Zhengyi Zhu, Tao Ma, Shushu Lu, Jing Chen, Xiaolei Shi, and Haozhen Ren. 2023. "Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Is a Promising Signature to Predict Prognosis and Therapies for Hepatocellular Carcinoma (HCC)" Journal of Clinical Medicine 12, no. 5: 1830. https://doi.org/10.3390/jcm12051830
APA StyleChen, Y., Xia, S., Zhang, L., Qin, X., Zhu, Z., Ma, T., Lu, S., Chen, J., Shi, X., & Ren, H. (2023). Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Is a Promising Signature to Predict Prognosis and Therapies for Hepatocellular Carcinoma (HCC). Journal of Clinical Medicine, 12(5), 1830. https://doi.org/10.3390/jcm12051830




