Validation of Molecular Markers Significant for Flowering Time, Plant Lodging, Stem Geometry Properties, and Raffinose Family Oligosaccharides in Pea (Pisum sativum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genotyping
2.3. Sanger Sequencing
2.4. Phenotyping
3. Results
3.1. Days to Flowering, Lodging, and Stem Geometry Parameters in Pea Cultivars
3.2. Soluble Carbohydrates in Seeds of Pea Cultivars
3.3. Trait Correlation
3.4. Closely Linked Markers
4. Discussion
4.1. The Variability of the Agronomic Traits
4.2. Evaluation of Closely Linked Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bourion, V.; Rizvi, S.; Fournier, S.; Larambergue, H.; Galmiche, F.; Marget, P.; Duc, G.; Burstin, J. Genetic Dissection of Nitrogen Nutrition in Pea through a Qtl Approach of Root, Nodule, and Shoot Variability. Theor. Appl. Genet. 2010, 121, 71–86. [Google Scholar] [CrossRef]
- Robinson, G.H.J.; Domoney, C. Perspectives on the Genetic Improvement of Health- and Nutrition-Related Traits in Pea. Plant Physiol. Biochem. 2021, 158, 353–362. [Google Scholar] [CrossRef]
- Gawłowska, M.; Knopkiewicz, M.; Święcicki, W.; Boros, L.; Wawer, A. Quantitative Trait Loci for Stem Strength Properties and Lodging in Two Pea Bi-Parental Mapping Populations (Pisum sativum L.). Crop Sci. 2021, 61, 1682–1697. [Google Scholar] [CrossRef]
- Mohan, M.; Nair, S.; Bhagwat, A.; Krishna, T.; Yano, M.; Bhatia, C.; Sasaki, T. Genome Mapping, Molecular Markers and Marker-Assisted Selection in Crop Plants. Mol. Breed. 1997, 3, 87–103. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-Assisted Selection: An Approach for Precision Plant Breeding in the Twenty-First Century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Renshaw, D.; Yang, H.; Yan, G. Development of a Co-Dominant DNA Marker Tightly Linked to Gene Tardus Conferring Reduced Pod Shattering in Narrow-Leafed Lupin (Lupinus Angustifolius L.). Euphytica 2010, 176, 49–58. [Google Scholar] [CrossRef]
- Masouleh, A.K.; Waters, D.L.; Reinke, R.F.; Henry, R.J. A High-Throughput Assay for Rapid and Simultaneous Analysis of Perfect Markers for Important Quality and Agronomic Traits in Rice Using Multiplexed Maldi-Tof Mass Spectrometry. Plant Biotechnol. J. 2009, 7, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Kroc, M.; Czepiel, K.; Wilczura, P.; Mokrzycka, M.; Święcicki, W. Development and Validation of a Gene-Targeted Dcaps Marker for Marker-Assisted Selection of Low-Alkaloid Content in Seeds of Narrow-Leafed Lupin (Lupinus angustifolius L.). Genes 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Mc Phee, K.E.; Inglis, D.A.; Gundersen, B.; Coyne, C.J. Mapping Qtl for Fusarium Wilt Race 2 Partial Resistance in Pea (Pisum sativum). Plant Breed. 2012, 131, 300–306. [Google Scholar] [CrossRef]
- Smýkal, P.; Šafářová, D.; Navrátil, M.; Dostalová, R. Marker Assisted Pea Breeding: Eif4e Allele Specific Markers to Pea Seed-Borne Mosaic Virus (Psbmv) Resistance. Mol. Breed. 2010, 26, 425–438. [Google Scholar] [CrossRef]
- Jha, A.B.; Gali, K.K.; Banniza, S.; Warkentin, T.D. Validation of Snp Markers Associated with Ascochyta Blight Resistance in Pea. Can. J. Plant Sci. 2019, 99, 243–249. [Google Scholar] [CrossRef]
- Zhang, C.; Tar’an, B.; Warkentin’, T.; Tullu, A.; Bett, K.E.; Vandenberg, B.; Somers, D.J. Selection for Lodging Resistance in Early Generations of Field Pea by Molecular Markers. Crop Sci. 2006, 46, 321–329. [Google Scholar] [CrossRef]
- Page, D.; Aubert, G.; Duc, G.; Welham, T.; Domoney, C. Combinatorial Variation in Coding and Promoter Sequences of Genes at the Tri Locus in Pisum sativum Accounts for Variation in Trypsin Inhibitor Activity in Seeds. Mol. Genet. Genom. 2002, 267, 359–369. [Google Scholar] [CrossRef]
- Javid, M.; Rosewarne, G.M.; Sudheesh, S.; Kant, P.; Leonforte, A.; Lombardi, M.; Kennedy, P.R.; Cogan, N.O.; Slater, A.T.; Kaur, S. Validation of Molecular Markers Associated with Boron Tolerance, Powdery Mildew Resistance and Salinity Tolerance in Field Peas. Front. Plant Sci. 2015, 6, 917. [Google Scholar] [CrossRef]
- Burstin, J.; Salloignon, P.; Chabert-Martinello, M.; Magnin-Robert, J.-B.; Siol, M.; Jacquin, F.; Chauveau, A.; Pont, C.; Aubert, G.; Delaitre, C.; et al. Genetic Diversity and Trait Genomic Prediction in a Pea Diversity Panel. BMC Genom. 2015, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Gawlowska, M.; Lahuta, L.; Święcicki, W.; Krajewski, P. Variability in the Oligosaccharide Concentration in Seeds of the Mapping Population of Pea (Pisum sativum L). Czech. J. Genet. Plant Breed. 2014, 50, 157–162. [Google Scholar] [CrossRef]
- Loridon, K.; Mc Phee, K.; Morin, J.; Dubreuil, P.; Pilet-Nayel, M.; Aubert, G.; Rameau, C.; Baranger, A.; Coyne, C.; Lejeune-Henaut, I. Microsatellite Marker Polymorphism and Mapping in Pea (Pisum sativum L.). Theor. Appl. Genet. 2005, 111, 1022–1031. [Google Scholar] [CrossRef]
- Fredslund, J.; Schauser, L.; Madsen, L.H.; Sandal, N.; Stougaard, J. Prifi: Using a Multiple Alignment of Related Sequences to Find Primers for Amplification of Homologs. Nucleic Acids Res. 2005, 33, W516–W520. [Google Scholar] [CrossRef]
- Tayeh, N.; Aluome, C.; Falque, M.; Jacquin, F.; Klein, A.; Chauveau, A.; Berard, A.; Houtin, H.; Rond, C.; Kreplak, J.; et al. Development of Two Major Resources for Pea Genomics: The Genopea 13.2k Snp Array and a High-Density, High-Resolution Consensus Genetic Map. Plant J. 2015, 84, 1257–1273. [Google Scholar] [CrossRef]
- Święcicki, W.; Gawłowska, M.; Bednarowicz, M.; Knopkiewicz, M. Localization of the Common Markers on the Pea Maps Wt10245 X Wt11238, Carneval X Mp1401 and P665 X Messire (Pisum sativum L.). Sci. Med. 2012, 3, 229–234. [Google Scholar]
- Gilpin, B.J.; McCallum, J.A.; Frew, T.J.; Timmerman-Vaughan, G.M. A Linkage Map of the Pea (Pisum sativum L.) Genome Containing Cloned Sequences of Known Function and Expressed Sequence Tags (Ests). Theor. Appl. Genet. 1997, 95, 1289–1299. [Google Scholar] [CrossRef]
- Tar’an, B.; Warkentin, T.; Somers, D.J.; Miranda, D.; Vandenberg, A.; Blade, S.; Woods, S.; Bing, D.; Xue, A.; DeKoeyer, D.; et al. Quantitative Trait Loci for Lodging Resistance, Plant Height and Partial Resistance to Mycosphaerella Blight in Field Pea (Pisum sativum L.). Theor. Appl. Genet. 2003, 107, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Joinmap Version 3.0. Software for the Calculation of Genetic Linkage Maps; Plant Research International B.V.: Wageningen, The Netherlands, 2001.
- Gottlieb, L.D. Enzyme Differentiation and Phylogeny in Clarkia franciscana, C. rubicunda and C. amoena. Evolution 1973, 27, 205–214. [Google Scholar] [CrossRef]
- Cardy, B.; Stuber, C.; Goodman, M. Techniques for Starch Gel Electrophoresis of Enzymes from Maize (Zea mays L.); North Carolina State University: Raleigh, NC, USA, 1980. [Google Scholar]
- Wolko, B.; Święcicki, W.K. The Application of Electrophoretic Methods of Isozymes Separation for Genetical Characterization of Pea Cultivars. Genet. Pol. 1987, 19, 89–99. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA Sequencing with Chain-Terminating Inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Capal, P.; Novak, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A Reference Genome for Pea Provides Insight into Legume Genome Evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Strażyński, P.; Mrówczyński, M.; Horoszkiewicz-Janka, J.; Kozłowski, J.; Księżak, J.; Matyjaszczyk, E.; Osiecka, A.; Kierzek, R.; Krawczyk, R.; Korbas, M.; et al. Metodyka Integrowanej Ochrony Grochu Siewnego Dla Producentów; Strażyński, P., Mrówczyński, M., Eds.; Instytut Ochrony Roślin–Państwowy Instytut Badawczy: Poznań, Poland, 2014; ISBN 978-83-64655-01-2. (In Polish) [Google Scholar]
- Gawłowska, M.; Święcicki, W.; Lahuta, L.; Kaczmarek, Z. Raffinose Family Oligosaccharides in Seeds of Pisum Wild Taxa, Type Lines for Seed Genes, Domesticated and Advanced Breeding Materials. Genet. Resour. Crop Evol. 2017, 64, 569–579. [Google Scholar] [CrossRef]
- Lahuta, L.B. Biosynthesis of Raffinose Family Oligosaccharides and Galactosyl Pinitols in Developing and Maturing Seeds of Winter Vetch (Vicia villosa Roth.). Acta Soc. Bot. Pol. 2006, 75, 219–227. [Google Scholar] [CrossRef]
- Gabriel, K.R. A Procedure for Testing the Homogeneity of All Sets of Means in Analysis of Variance. Biometrics 1964, 20, 459–477. [Google Scholar] [CrossRef]
- Gali, K.K.; Liu, Y.; Sindhu, A.; Diapari, M.; Shunmugam, A.S.K.; Arganosa, G.; Daba, K.; Caron, C.; Lachagari, R.V.; Tar’an, B.; et al. Construction of High-Density Linkage Maps for Mapping Quantitative Trait Loci for Multiple Traits in Field Pea (Pisum sativum L.). BMC Plant Biol. 2018, 18, 172. [Google Scholar] [CrossRef]
- Huang, S.; Gali, K.K.; Tar2019an, B.; Warkentin, T.D.; Bueckert, R.A. Pea Phenology: Crop Potential in a Warming Environment. Crop Sci. 2017, 57, 1540–1551. [Google Scholar] [CrossRef]
- Niu, L.; Feng, S.; Ding, W.; Li, G. Influence of Speed and Rainfall on Large-Scale Wheat Lodging from 2007 to 2014 in China. PLoS ONE 2016, 11, e0157677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C. Implementation of Marker-Assisted Selection for Lodging Resistance in Pea Breeding. Ph.D. Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2004. [Google Scholar]
- Obraztsov, A.S.; Amelin, A.V. Ideotype of Pea Plants in Relation to Their Resistance to Lodging in the South of the Non-Chernozem Zone of the Rsfsr. Sel’skokhozyaĭstvennaya Biol. 1990, 1, 83–89. [Google Scholar]
- Pattee, H.E.; Isleib, T.G.; Giesbrecht, F.G.; Mc Feeters, R.F. Investigations into Genotypic Variations of Peanut Carbohydrates. J. Agric. Food Chem. 2000, 48, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Jaureguy, L.M.; Chen, P.; Scaboo, A.M. Heritability and Correlations among Food-Grade Traits in Soybean. Plant Breed. 2011, 130, 647–652. [Google Scholar] [CrossRef]
- Tahir, M.; Vandenberg, A.; Chibbar, R.N. Influence of Environment on Seed Soluble Carbohydrates in Selected Lentil Cultivars. J. Food Compos. Anal. 2011, 24, 596–602. [Google Scholar] [CrossRef]
- Gangola, M.P.; Khedikar, Y.P.; Gaur, P.M.; Båga, M.; Chibbar, R.N. Genotype and Growing Environment Interaction Shows a Positive Correlation between Substrates of Raffinose Family Oligosaccharides (Rfo) Biosynthesis and Their Accumulation in Chickpea (Cicer arietinum L.) Seeds. J. Agric. Food Chem. 2013, 61, 4943–4952. [Google Scholar] [CrossRef]
- Górecki, R.; Lahuta, L.; Jones, A.; Hedley, C. Soluble Sugars in Maturing Pea Seeds of Different Lines in Relation to Desiccation Tolerance. In Seed Biology: Advances and Applications; Black, M., Bradford, K., Vásquez-Ramos, J., Eds.; CAB International: Wallingford, UK, 2000; pp. 67–74. [Google Scholar]
- Górecki, R.J.; Fordonski, G.; Halmajan, H.; Horbowicz, M.; Jones, R.G.; Lahuta, L. Seed Physiology and Biochemistry. In Carbohydrates in Grain Legume Seeds: Improving Nutritional Quality and Agronomic Characteristics; Hedley, C.L., Ed.; CAB International: Norwich, UK, 2001; pp. 117–143. ISBN 08519946. [Google Scholar]
- Lahuta, L.B.; Łogin, A.; Rejowski, A.; Socha, A.; Zalewski, K. Influence of Water Deficit on the Accumulation of Sugars in Developing Field Bean (Vicia faba var. minor.) Seeds. Seed Sci. Technol. 2000, 28, 93–100. [Google Scholar]
- Lahuta, L.; Górecki, R.J.; Gojło, E.; Horbowicz, M. Effect of Exogenous Abscisic Acid on Accumulation of Raffinose Family Oligosaccharides and Galactosyl Cyclitols in Tiny Vetch Seeds (Vicia hirsuta [L.] S.F. Gray). Acta Soc. Bot. Pol. 2004, 73, 277–283. [Google Scholar] [CrossRef]
- Mądry, W.; Talbot, M.B.; Ukalski, K.; Drzazga, T.; Iwanska, M. Podstawy Teoretyczne Znaczenia Efektów Genotypowych I Interakcyjnych W Hodowli Roślin Na Przykładzie Pszenicy Ozimej. Biuletyn Instytutu Hodowli i Aklimatyzacji Roślin 2006, 240/241, 13–32. [Google Scholar]
- Piotrowicz-Cieślak, A.I. Contents of Soluble Carbohydrates in Yellow Lupin Seeds Maturated at Various Temperatures. Acta Physiol. Plant 2006, 28, 349–356. [Google Scholar] [CrossRef]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dedaldechamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thevenot, P.; Maurousset, L.; et al. Source-to-Sink Transport of Sugar and Regulation by Environmental Factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Sari, H.; Sari, D.; Eker, T.; Toker, C. De Novo Super-Early Progeny in Interspecific Crosses Pisum sativum L. × P. fulvum Sibth. Et Sm. Sci. Rep. 2021, 11, 19706. [Google Scholar] [CrossRef] [PubMed]
- Fondevilla, S.; Almeida, N.F.; Satovic, Z.; Rubiales, D.; Vaz Patto, M.C.; Cubero, J.I.; Torres, A.M. Identification of Common Genomic Regions Controlling Resistance to Mycosphaerella Pinodes, Earliness and Architectural Traits in Different Pea Genetic Backgrounds. Euphytica 2011, 182, 43–52. [Google Scholar] [CrossRef]
- Prioul, S.; Frankewitz, A.; Deniot, G.; Morin, G.; Baranger, A. Mapping of Quantitative Trait Loci for Partial Resistance to Mycosphaerella Pinodes in Pea (Pisum Sativum L.), at the Seedling and Adult Plant Stages. Theor. Appl. Genet. 2004, 108, 1322–1334. [Google Scholar] [CrossRef] [PubMed]
- Weller, J.L.; Ortega, R. Genetic Control of Flowering Time in Legumes. Front. Plant. Sci. 2015, 6, 207. [Google Scholar] [CrossRef]
- Weller, J.L.; Liew, L.C.; Hecht, V.F.G.; Rajandran, V.; Laurie, R.E.; Ridge, S.; Wenden, B.; Schoor, J.K.V.; Jaminon, O.; Blassiau, C.; et al. A Conserved Molecular Basis for Photoperiod Adaptation in Two Temperate Legumes. Proc. Natl. Acad. Sci. USA 2012, 109, 21158–21163. [Google Scholar] [CrossRef]
- Tafesse, E.G.; Gali, K.K.; Lachagari, V.B.R.; Bueckert, R.; Warkentin, T.D. Genome-Wide Association Mapping for Heat and Drought Adaptive Traits in Pea. Genes 2021, 12, 1897. [Google Scholar] [CrossRef]
- Liew, L.C.; Hecht, V.; Laurie, R.E.; Knowles, C.L.; Vander Schoor, J.K.; Macknight, R.C.; Weller, J.L. Die Neutralis and Late Bloomer 1 Contribute to Regulation of the Pea Circadian Clock. Plant Cell 2009, 21, 3198–3211. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Diapari, M.; Bueckert, R.A.; Tar’an, B.; Warkentin, T.D. Population Structure and Association Mapping of Traits Related to Reproductive Development in Field Pea. Euphytica 2017, 213, 215. [Google Scholar] [CrossRef]
- Weeden, N.; Ellis, T.; Timmerman-Vaughan, G.; Swiecicki, W.; Rozov, S.; Berdnikov, V. A Consensus Linkage Map for Pisum sativum. Pisum Genet. 1998, 30, 1–4. [Google Scholar]
- Keller, M.; Karutz, C.; Schmid, E.J.; Stamp, P.; Winzeler, M.; Keller, B.; Messmer, M.M. Quantitative Trait Loci for Lodging Resistance in a Segregating Wheat×Spelt Population. Theor. Appl. Genet. 1999, 98, 1171–1182. [Google Scholar] [CrossRef]
- Dumont, E.; Fontaine, V.; Vuylsteker, C.; Sellier, H.; Bodele, S.; Voedts, N.; Devaux, R.; Frise, M.; Avia, K.; Hilbert, J.-L. Association of Sugar Content Qtl and Pql with Physiological Traits Relevant to Frost Damage Resistance in Pea under Field and Controlled Conditions. Theor. Appl. Genet. 2009, 118, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Aubert, G.; Morin, J.; Jacquin, F.; Loridon, K.; Quillet, M.; Petit, A.; Rameau, C.; Lejeune-Henaut, I.; Huguet, T.; Burstin, J. Functional Mapping in Pea, as an Aid to the Candidate Gene Selection and for Investigating Synteny with the Model Legume Medicago Truncatula. Theor. Appl. Genet. 2006, 112, 1024–1041. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.B.; Breton, G.; Limin, A.E.; Mahfoozi, S.; Sarhan, F. Photoperiod and Temperature Interactions Regulate Low-Temperature-Induced Gene Expression in Barley. Plant. Physiol. 2001, 127, 1676–1681. [Google Scholar] [CrossRef] [PubMed]
- Gawłowska, M.; Święcicki, W. The fa2 gene and molecular markers mapping in the gp segment of the Pisum linkage group V. J. Appl. Genet. 2016, 57, 317–322. [Google Scholar] [CrossRef]
- Berbel, A.; Ferrándiz, C.; Hecht, V.; Dalmais, M.; Lund, O.S.; Sussmilch, F.C.; Taylor, S.A.; Bendahmane, A.; Ellis, T.H.N.; Beltrán, J.P.; et al. VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 2012, 3, 797. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2015, 137, 1420–1434. [Google Scholar] [CrossRef]
- Knopkiewicz, M.; Gawłowska, M.; Święcicki, W. Poszukiwanie polimorficznych markerów zdefiniowanych sekwencyjnie w populacji grochu Carneval x MP1401. Searching for polymorphic sequence-defined markers in the pea. Fragm. Agron. 2012, 29, 87–94. [Google Scholar]
- Sindhu, A.; Ramsay, L.; Sanderson, L.A.; Stonehouse, R.; Li, R.; Condie, J.; Shunmugam, A.S.; Liu, Y.; Jha, A.B.; Diapari, M.; et al. Gene-based SNP discovery and genetic mapping in pea. Theor. Appl. Genet. 2014, 127, 2225–2241. [Google Scholar] [CrossRef]

| No. | Genotype Name Polish Register | Leaf Type | Pedigree | No. | Genotype Name German Register | Delivered by | Leaf Type |
|---|---|---|---|---|---|---|---|
| 1 | Akord | afila | (A × B) × B | 1 | Alvesta | KWS Lochow GmbH | afila |
| 2 | Baryton | afila | ua | 2 | Casablanca | KWS Lochow GmbH | afila |
| 3 | Batuta | afila | C × D/95 | 3 | La Mancha | KWS Lochow GmbH | afila |
| 4 | Bohun | afila | E × F | 4 | Santana | KWS Lochow GmbH | afila |
| 5 | Boruta | afila | G × H | 5 | Gregor | Pflanzenzucht Hans-Georg Lembke | afila |
| 6 | Brutus | afila | ua | 6 | Starter | Pflanzenzucht Hans-Georg Lembke | afila |
| 7 | Brylant | afila | ua | 7 | Salamanca | Pflanzenzucht Hans-Georg Lembke | afila |
| 8 | Cysterski | afila | ua | 8 | Navarro | Pflanzenzucht Hans-Georg Lembke | afila |
| 9 | Dymek | afila | I × J | 9 | Madonna | Pflanzenzucht Hans-Georg Lembke | afila |
| 10 | Ezop | afila | K ×L | ||||
| 11 | Goplik | afila | Ł/82 × M | ||||
| 12 | Kavalir | afila | ua | ||||
| 13 | Lasso | afila | ua | ||||
| 14 | Mecenas | afila | N × O | ||||
| 15 | Medal | afila | MERLIN × PIAST | ||||
| 16 | Mentor | afila | R × S | ||||
| 17 | Merlin | afila | T × U | ||||
| 18 | Profi | afila | ua | ||||
| 19 | Ramrod (Piast) | afila | Bred by Łagiewniki Breeding Station | ||||
| 20 | Set | normal | Line from map. pop. × LP1/90 | ||||
| 21 | Tarchalska | afila | ((W × Y) × Z) × Ż | ||||
| 22 | Terno | afila | ua | ||||
| 23 | Turkus | afila | Bred by Sobótka Breeding Station | ||||
| 24 | Wenus | afila | Ź × Ż | ||||
| 25 | Zekon | afila | ua |
| Character | 2014 | 2015 | GxY Interaction | ||||
|---|---|---|---|---|---|---|---|
| Mean | Range | CV% | Mean | Range | CV% | F Statistic | |
| DTF (days) | 58 | 53–64 | 1.4 | 68 | 68–71 | 0.6 | 6.3 ** |
| Plant height (cm) | 93.8 | 72–113 | 7.6 | 67.4 | 53–87 | 10.5 | 2.4 ** |
| Lodging 2nd (the end of flowering) | 7.6 | 6.3–8.7 | 9.0 | 8.1 | 4.7–9.0 | 7.5 | 2.9 ** |
| Lodging 3rd (maturity) | 5.3 | 5.3–5.5 | 13.8 | 6.8 | 3.7–8.5 | 13.5 | 2.7 ** |
| Stem wall thickness in the bottom of the stem (mm) | 0.5 | 0.39–0.53 | 8.9 | 0.4 | 0.33–0.50 | 10.1 | - |
| Stem wall thickness in the middle part of the stem (mm) | 0.4 | 0.37–0.54 | 9.4 | 0.4 | 0.37–0.53 | 10.4 | 1.9 ** |
| Stem wall thickness in the upper part of the stem (mm) | 0.4 | 0.35–0.50 | 9.8 | 0.4 | 0.35–0.57 | 14.0 | - |
| Stem diameter in the bottom of the stem (mm) | 3.7 | 3.17–4.19 | 5.3 | 2.9 | 2.52–3.38 | 8.8 | - |
| Stem diameter in the middle part of the stem (mm) | 4.9 | 4.23–5.36 | 5.9 | 4.0 | 3.19–4.94 | 10.6 | 2.0 ** |
| Stem diameter in the upper part of the stem (mm) | 4.7 | 3.95–5.35 | 6.6 | 3.7 | 2.77–4.70 | 11.7 | 2.03 ** |
| Character | Subgroup | 2014 | 2015 |
|---|---|---|---|
| DTF (days) | I | 60 | 68 |
| II | 56 | 66 | |
| Plant height (cm) | I | 103.8 | 73.6 |
| II | 87.9 | 59.2 | |
| Lodging 2nd (the end of flowering) | I | – | 8.5 |
| II | – | 6.6 | |
| Lodging 3rd (maturity) | I | – | 7.4 |
| II | – | 4.7 | |
| Stem wall thickness in the bottom part | I | – | 0.43 |
| II | – | 0.36 | |
| Stem wall thickness in the middle part | I | – | 0.47 |
| II | – | 0.40 | |
| Stem wall thickness in the upper part | I | – | 0.48 |
| II | – | 0.38 | |
| Stem diameter in the bottom part | I | – | 3.1 |
| II | – | 2.7 | |
| Stem diameter in the middle part | I | – | 4.4 |
| II | – | 3.7 | |
| Stem diameter in the upper part | I | – | 4.0 |
| II | – | 3.4 |
| Year | 2013 | 2014 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Character | G × Y Interaction | ||||||||||||
| (mg g−1) DW | Cultivar with min. concentration | Mean | CV% | Cultivar with max. concentration | Mean | CV% | Cultivar with min. concentration | Mean | CV% | Cultivar with max. concentration | Mean | CV% | F statistic |
| TSCs 1 | Turkus | 84.2 | 0.9 | Zekon | 118.0 | 4.2 | Batuta | 101.5 | 12.2 | Navarro | 135.8 | 7.9 | 4.19 ** |
| Fructose | Brylant | 0.2 | 9.2 | Mentor | 0.5 | 8.1 | Brylant | 0.3 | 15.9 | Merlin | 0.7 | 7.0 | 2.52 ** |
| Sucrose | La Mancha | 22.0 | 2.8 | Goplik | 39.5 | 3.2 | Salamanca | 21.9 | 3.2 | Bohun | 48.0 | 6.7 | 6.91 ** |
| Raffinose | Boruta | 7.4 | 2.9 | Zekon | 20.0 | 4.0 | Boruta | 8.9 | 6.3 | Zekon | 23.2 | 3.6 | 4.43 ** |
| Stachyose | Boruta | 20.6 | 2.9 | Zekon | 40.1 | 5.6 | Boruta | 24.0 | 6.7 | Lasso | 48.3 | 3.3 | 2.30 ** |
| Verbascose | Kavalier | 15.3 | 3.5 | Mecenas | 36.1 | 3.9 | Merlin | 20.8 | 6.9 | Wenus | 50.9 | 3.5 | 11.56 ** |
| Total RFOs | Turkus | 58.4 | 1.2 | Mecenas | 81.7 | 3.6 | Batuta | 69.1 | 13.6 | Navarro | 97.6 | 4.8 | 5.86 ** |
| RFOs content (% of TSCs) | Tarchalska | 61.0 | 0.4 | Starter | 75.0 | 0.4 | Goplik | 66.0 | 1.2 | Salamanca | 77.0 | 0.05 | 24.25 ** |
| Character (mg g−1 DW) | Genotype Group of High and Low Concentration | Mean Value | |
|---|---|---|---|
| 2013 (No. of Objects) | 2014 (No. of Objects) | ||
| TSCs | High | 106.6 (16) | 130.3 (08) |
| Low | 93.7 (18) | 112.6 (26) | |
| Fructose | High | 0.4 (20) | 0.6 (18) |
| Low | 0.3 (14) | 0.4 (16) | |
| Sucrose | High | 34.6 (11) | 38.3 (09) |
| Low | 26.3 (23) | 28.4 (25) | |
| Raffinose | High | 14.6 (24) | 19.1 (12) |
| Low | 9.6 (10) | 13.3 (22) | |
| Stachyose | High | 34.1 (16) | 40.2 (13) |
| Low | 24.1 (18) | 27.1 (21) | |
| Verbascose | High | 30.4 (19) | 41.7 (18) |
| Low | 20.5 (15) | 27.8 (16) | |
| Total RFOs | High | 75.2 (10) | 86.6 (23) |
| Low | 65.6 (24) | 74.4 (11) | |
| RFOs content (% of TSCs) | High | 71.2% (20) | 73.4% (19) |
| Low | 64.9% (14) | 67.7% (15) | |
| 2014 | DTF | H | Lodg2 | Lodg3 | BDiam | Mdiam | Tdiam | Bthic | Mthic | Tthic |
|---|---|---|---|---|---|---|---|---|---|---|
| DTF | 1 | |||||||||
| H | ns | 1 | ||||||||
| Lodg2 | ns | ns | 1 | |||||||
| Lodg3 | 0.26 (ns) | ns | 0.57 | 1 | ||||||
| BDiam | ns | ns | ns | ns | 1 | |||||
| Mdiam | ns | 0.48 | −0.40 | ns | 0.36 | 1 | ||||
| Tdiam | ns | ns | ns | ns | 0.34 | 0.70 | 1 | |||
| Bthic | ns | ns | ns | ns | ns | ns | ns | 1 | ||
| Mthic | ns | ns | ns | ns | ns | 0.34 | 0.31 (ns) | ns | 1 | |
| Tthic | ns | ns | ns | ns | ns | ns | 0.32 (ns) | ns | ns | 1 |
| 2015 | DTF | H | Lodg2 | Lodg3 | Bdiam | Mdiam | Tdiam | Bthic | Mthic | Tthic |
| DTF | 1 | |||||||||
| H | −0.32 (ns) | 1 | ||||||||
| Lodg2 | 0.35 | −0.64 | 1 | |||||||
| Lodg3 | 0.38 | −0.70 | 0.91 | 1 | ||||||
| Bdiam | ns | ns | ns | ns | 1 | |||||
| Mdiam | 0.37 | ns | ns | ns | 0.79 | 1 | ||||
| Tdiam | 0.35 | ns | ns | ns | 0.54 | 0.84 | 1 | |||
| Bthic | ns | ns | ns | ns | ns | ns | ns | 1 | ||
| Mthic | ns | ns | ns | ns | 0.47 | 0.48 | 0.37 | ns | 1 | |
| Tthic | 0.43 | ns | ns | ns | 0.53 | 0.73 | 0.62 | ns | 0.67 | 1 |
| Character | Year | Linkage Group | Marker (Distance from QTL) | Mean Value of the Character for A Cultivar Group | F Statistics | Sign | |
|---|---|---|---|---|---|---|---|
| Allele I (mg g−1 DW) (No. of Objects) | Allele II (mg g−1 DW) (No. of Objects) | ||||||
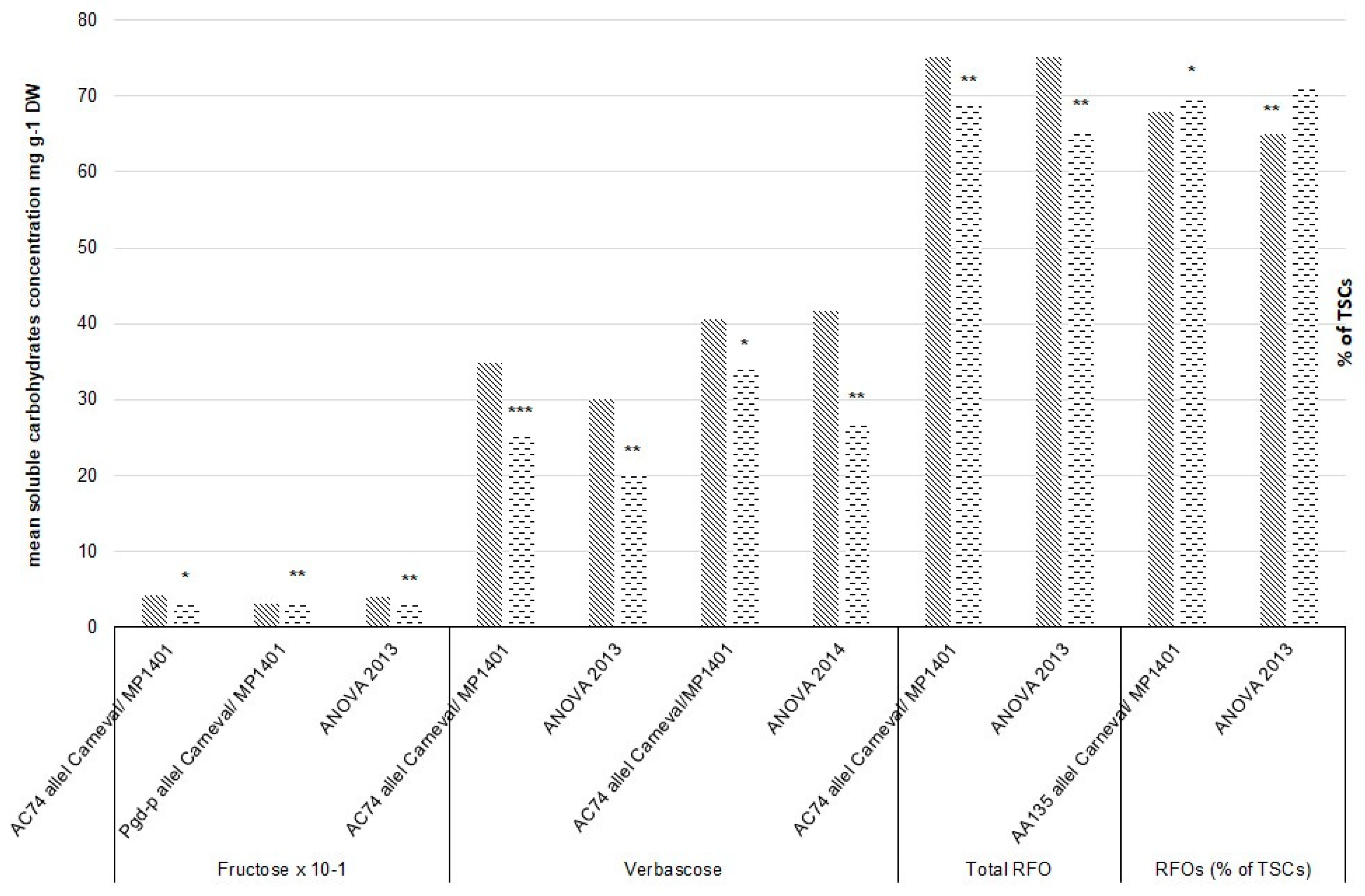
| Fructose | 2013 | chr1LG6 | AC74 | 0.42 (02) | 0.34 (29) | 0.05 | * |
| 2013 | chr7LG7 | Pgd-p | 0.30 (07) | 0.35 (24) | 0.033 | ** | |
| Verbascose | 2013 | chr1LG6 | AC74 (1 cM) | 34.85 (02) | 25.17 (29) | <0.001 | *** |
| 2014 | chr1LG6 | AC74 (1 cM) | 40.55 (02) | 33.98 (29) | 0.021 | * | |
| Total RFO | 2013 | chr1LG6 | AC74 (5 cM) | 75.09 (02) | 68.70 (29) | 0.01 | ** |
| RFOs (% of TSCs) | 2013 | chr7LG7 | AA135 (5 cM) | 0.68 (18) | 0.70 (13) | 0.012 | * |
| Character | Year | Linkage Group | Marker (Distance from QTL) | Mean Value of the Character for A Cultivar Group | F Statistics | Sign | |
| Allele I (No. of Objects) | Allele II (No. of Objects) | ||||||
| Stem wall thickness (in the middle part) | 2014 | chr3LG5 | PsCam962 | 0.42 (12) | 0.45 (23) | 0.014 | * |
| 2015 | chr5LG3 | AB141 | 0.45 (13) | 0.42 (20) | 0.019 | * | |
| 2015 | chr3LG5 | PisGen9_3_1 | 0.42 (10) | 0.44 (24) | 0.036 | * | |
| 2015 | chr5LG3 | A001 | 0.44 (12) | 0.42 (22) | 0.040 | * | |
| 2015 | chr3LG5 | PsCam962 | 0.42 (12) | 0.44 (22) | 0.043 | * | |
| Stem wall thickness (top) | 2014 | chr3LG5 | AA81 | 0.45 (13) | 0.41 (14) | 0.004 | ** |
| 2014 | chr3LG5 | mtmt_EST_03378 | 0.40 (14) | 0.44 (19) | 0.001 | *** | |
| Stem diameter (in the bottom part) | 2015 | chr4LG4 | P393 | 2.86 (29) | 3.09 (05) | 0.004 | ** |
| Stem diameter (in the middle part) | 2015 | chr5LG3 | A004 | 4.14 (18) | 3.93 (16) | 0.009 | ** |
| 2015 | chr5LG3 | A001 | 3.99 (28) | 4.28 (06) | 0.006 | ** | |
| Stem diameter (in the upper part) | 2014 | chr5LG3 | AB141 | 4.54 (13) | 4.76 (20) | 0.035 | * |
| 2014 | chr3LG5 | AB83 | 4.47 (20) | 4.73 (09) | 0.027 | * | |
| 2015 | chr5LG3 | A004 | 3.82 (18) | 3.65 (16) | 0.038 | * | |
| 2015 | chr5LG3 | A001 | 3.69 (28) | 4.02 (06) | 0.002 | ** | |
| DTF | 2014 | chr6LG2 | Leg_65 | 60.00 (20) | 67.4 (09) | 0.005 | *** |
| 2014 | chr5LG3 | AB141 | 58.00 (13) | 59.00 (20) | 0.027 | * | |
| 2014 | chr3LG5 | PisGen9_3_1 | 58.00 (10) | 59.00 (24) | 0.030 | * | |
| 2014 | chr1LG6 | PisGen16 | 58.00 (15) | 59.00 (19) | 0.010 | ** | |
| Plant height | 2014 | chr3LG5 | OPG9b | 89.2 (07) | 95.0 (27) | 0.036 | * |
| Lodging in 2nd term (the end of flowering) | 2014 | chr5LG3 | AB141 | 7.41 (13) | 7.76 (20) | 0.028 | * |
| 2015 | chr3LG5 | PisGen9_3_1 | 8.57 (10) | 7.95 (24) | 0.008 | ** | |
| 2015 | chr5LG3 | A001 | 8.28 (28) | 7.39 (6) | <.001 | *** | |
| Lodging in 3rd term (maturity) | 2015 | chr3LG5 | PisGen9_3_1 | 7.35 (10) | 6.64 (24) | 0.024 | * |
| 2015 | chr5LG3 | A001 | 7.05 (28) | 5.83 (6) | 0.001 | ** | |
| 2015 | chr4LG4 | P393 | 6.97 (29) | 6.07 (5) | 0.026 | * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawłowska, M.; Lahuta, L.; Boros, L.; Sawikowska, A.; Kumar, P.; Knopkiewicz, M.; Kaczmarek, Z.; Święcicki, W. Validation of Molecular Markers Significant for Flowering Time, Plant Lodging, Stem Geometry Properties, and Raffinose Family Oligosaccharides in Pea (Pisum sativum L.). Agriculture 2022, 12, 1125. https://doi.org/10.3390/agriculture12081125
Gawłowska M, Lahuta L, Boros L, Sawikowska A, Kumar P, Knopkiewicz M, Kaczmarek Z, Święcicki W. Validation of Molecular Markers Significant for Flowering Time, Plant Lodging, Stem Geometry Properties, and Raffinose Family Oligosaccharides in Pea (Pisum sativum L.). Agriculture. 2022; 12(8):1125. https://doi.org/10.3390/agriculture12081125
Chicago/Turabian StyleGawłowska, Magdalena, Lesław Lahuta, Lech Boros, Aneta Sawikowska, Pankaj Kumar, Michał Knopkiewicz, Zygmunt Kaczmarek, and Wojciech Święcicki. 2022. "Validation of Molecular Markers Significant for Flowering Time, Plant Lodging, Stem Geometry Properties, and Raffinose Family Oligosaccharides in Pea (Pisum sativum L.)" Agriculture 12, no. 8: 1125. https://doi.org/10.3390/agriculture12081125
APA StyleGawłowska, M., Lahuta, L., Boros, L., Sawikowska, A., Kumar, P., Knopkiewicz, M., Kaczmarek, Z., & Święcicki, W. (2022). Validation of Molecular Markers Significant for Flowering Time, Plant Lodging, Stem Geometry Properties, and Raffinose Family Oligosaccharides in Pea (Pisum sativum L.). Agriculture, 12(8), 1125. https://doi.org/10.3390/agriculture12081125







