Food Authentication: The Detection of Arbutus unedo and Olea europaea Leaves as an Admixture of Oregano Using LAMP- and Duplex LAMP-Based Test Systems with Lateral-Flow Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Material
2.2. DNA Isolation
2.2.1. High-Quality DNA Isolation
2.2.2. Point-of-Care (POC)-Suitable DNA Isolation
2.3. Primer Design
2.4. LAMP Assay
2.5. Optimization of LAMP Assay and Isolation Protocol with DoE
2.6. Lateral-Flow-Assay
3. Results
3.1. Primer Design and Selectivity
3.2. Optimization of Isolation Protocol and Reaction Conditions
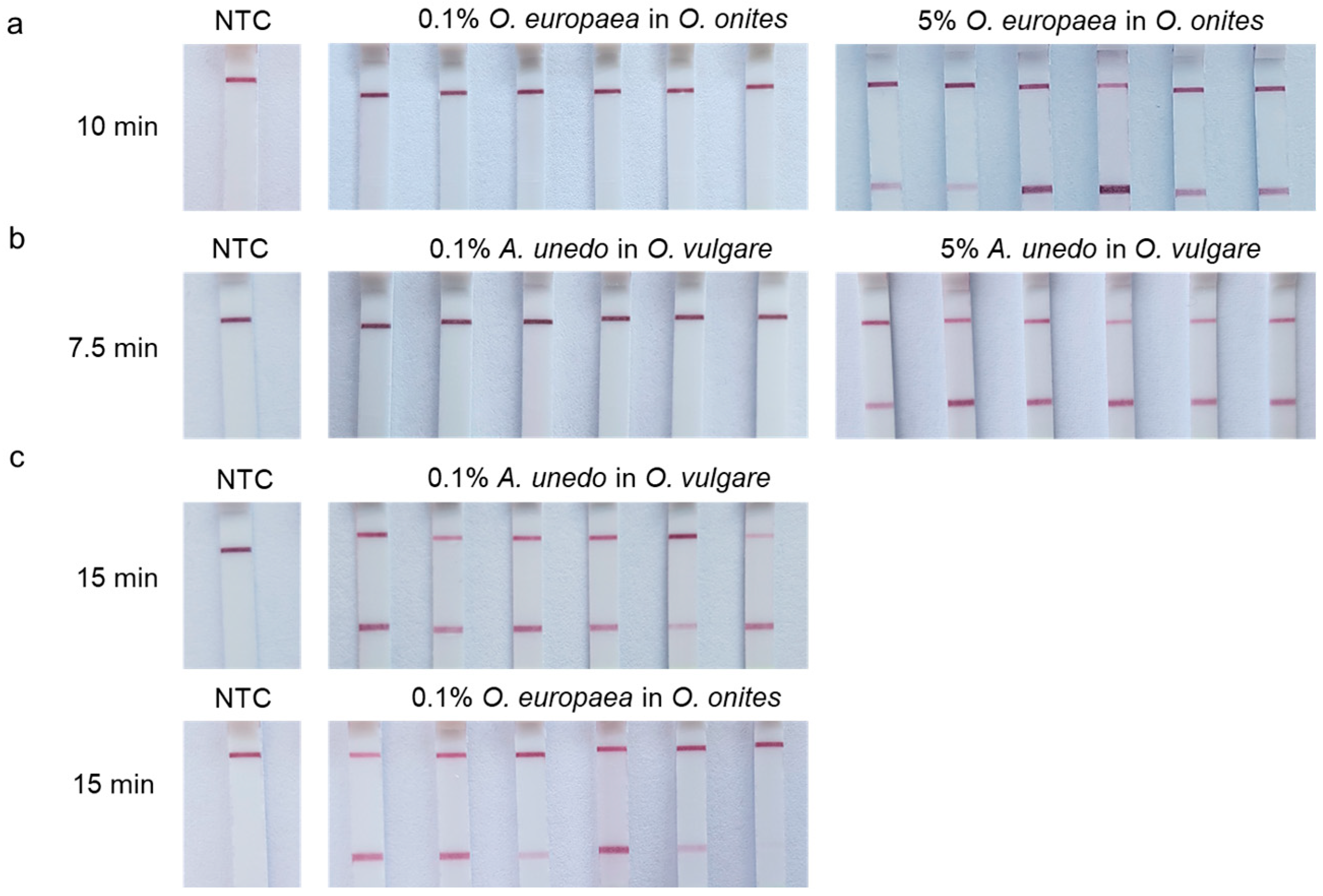
3.3. Lateral-Flow Assays
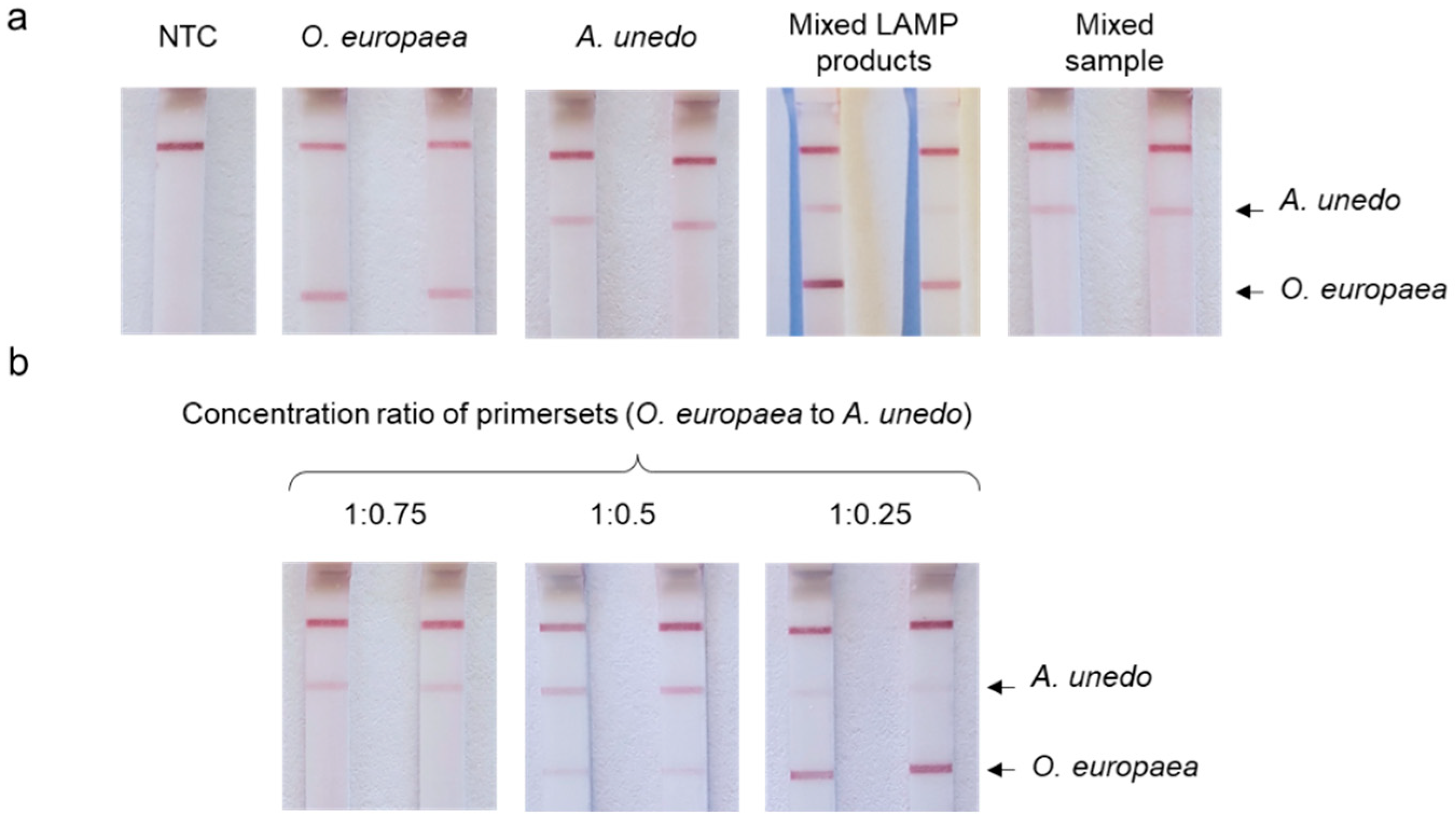
3.4. Duplex LAMP with LFA Detection
4. Discussion
4.1. Primer Design and Uniplex LAMP/LFA Assays
4.2. Duplex LAMP/LFA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ISO 7925:1999; Dried Oregano (Origanum vulgare L.)—Whole or Ground Leaves—Specification. ISO: Geneva, Switzerland, 2015. Available online: https://www.iso.org/obp/ui/#iso:std:iso:7925:ed-2:v1:en (accessed on 20 February 2024).
- Marieschi, M.; Torelli, A.; Bianchi, A.; Bruni, R. Detecting Satureja montana L. and Origanum majorana L. by means of SCAR–PCR in commercial samples of Mediterranean oregano. Food Control 2011, 22, 542–548. [Google Scholar] [CrossRef]
- Kintzios, S.E. Oregano: The Genera Origanum and Lippia, 1st ed.; Medicinal and Aromatic Plants-Industrial Profiles; v. 25; Taylor and Francis: London, UK, 2002. [Google Scholar]
- Marieschi, M.; Torelli, A.; Poli, F.; Sacchetti, G.; Bruni, R. RAPD-based method for the quality control of Mediterranean oregano and its contribution to pharmacognostic techniques. J. Agric. Food Chem. 2009, 57, 1835–1840. [Google Scholar] [CrossRef]
- Maquet, A.; Lievens, A.; Paracchini, V.; Kaklamanos, G.; de La Calle, B.; Garlant, L.; Papoci, S.; Pietretti, D.; Zdiniakova, T.; Breidbach, A.; et al. Results of an EU Wide Coordinated Control Plan to Establish the Prevalence of Fraudulent Practices in the Marketing of Herbs and Spices; EUR30877EN; JRC126785; Publications Office of the European Union: Luxembourg, 2021. [Google Scholar]
- Besnard, G.; Garcia-Verdugo, C.; de Casas, R.R.; Treier, U.A.; Galland, N.; Vargas, P. Polyploidy in the olive complex (Olea europaea): Evidence from flow cytometry and nuclear microsatellite analyses. Ann. Bot. 2008, 101, 25–30. [Google Scholar] [CrossRef]
- Vannozzi, A.; Lucchin, M.; Barcaccia, G. cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches. Diversity 2018, 10, 98. [Google Scholar] [CrossRef]
- Mandrone, M.; Marincich, L.; Chiocchio, I.; Petroli, A.; Gođevac, D.; Maresca, I.; Poli, F. NMR-based metabolomics for frauds detection and quality control of oregano samples. Food Control 2021, 127, 108141. [Google Scholar] [CrossRef]
- Sasikumar, B.; Swetha, V.P.; Parvathy, V.A.; Sheeja, T.E. Advances in Adulteration and Authenticity Testing of Herbs and Spices. Adv. Food Authent. Test. 2016, 585–624. [Google Scholar] [CrossRef]
- Marieschi, M.; Torelli, A.; Bianchi, A.; Bruni, R. Development of a SCAR marker for the identification of Olea europaea L.: A newly detected adulterant in commercial Mediterranean oregano. Food Chem. 2011, 126, 705–709. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Meth. 2007, 70, 499–501. [Google Scholar] [CrossRef]
- Kiddle, G.; Hardinge, P.; Buttigieg, N.; Gandelman, O.; Pereira, C.; McElgunn, C.J.; Rizzoli, M.; Jackson, R.; Appleton, N.; Moore, C.; et al. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 2012, 12, 15. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef]
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Y.; Ding, L.; Huang, X.; Xiong, Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. TrAC Trends Anal. Chem. 2021, 145, 116452. [Google Scholar] [CrossRef]
- Kampeera, J.; Pasakon, P.; Karuwan, C.; Arunrut, N.; Sappat, A.; Sirithammajak, S.; Dechokiattawan, N.; Sumranwanich, T.; Chaivisuthangkura, P.; Ounjai, P.; et al. Point-of-care rapid detection of Vibrio parahaemolyticus in seafood using loop-mediated isothermal amplification and graphene-based screen-printed electrochemical sensor. Biosens. Bioelectron. 2019, 132, 271–278. [Google Scholar] [CrossRef]
- Lalle, M.; Possenti, A.; Dubey, J.P.; Pozio, E. Loop-Mediated Isothermal Amplification-Lateral-Flow Dipstick (LAMP-LFD) to detect Toxoplasma gondii oocyst in ready-to-eat salad. Food Microbiol. 2018, 70, 137–142. [Google Scholar] [CrossRef]
- Focke, F.; Haase, I.; Fischer, M. Loop-mediated isothermal amplification (LAMP): Methods for plant species identification in food. J. Agric. Food Chem. 2013, 61, 2943–2949. [Google Scholar] [CrossRef]
- Holz, N.; Illarionov, B.; Wax, N.; Schmidt, C.; Fischer, M. Point-of-care suitable identification of the adulterants Carthamus tinctorius and Curcuma longa in Crocus sativus based on loop-mediated isothermal amplification (LAMP) and lateral-flow-assay (LFA). Food Control 2023, 148, 109637. [Google Scholar] [CrossRef]
- Wax, N.; Pförtner, L.S.; Holz, N.; Sterzl, S.; Melnik, M.; Kappel, K.; Bade, P.; Schröder, U.; Haase, I.; Fritsche, J.; et al. Fast and User-Friendly Detection of Flatfish Species (Pleuronectes platessa and Solea solea) via Loop-Mediated Isothermal Amplification (LAMP). J. Agric. Food Chem. 2023, 71, 14795–14805. [Google Scholar] [CrossRef]
- Zahradnik, C.; Martzy, R.; Mach, R.L.; Krska, R.; Farnleitner, A.H.; Brunner, K. Detection of the food allergen celery via loop-mediated isothermal amplification technique. Anal. Bioanal. Chem. 2014, 406, 6827–6833. [Google Scholar] [CrossRef]
- Doria, G.; Clemente, C.; Coelho, E.; Colaço, J.; Crespo, R.; Semikhodskii, A.; Mansinho, H.; Dinis, M.; Carvalho, M.F.; Casmarrinha, M.; et al. An isothermal lab-on-phone test for easy molecular diagnosis of SARS-CoV-2 near patients and in less than 1 hour. Int. J. Infect. Dis. 2022, 123, 1–8. [Google Scholar] [CrossRef]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef]
- Allgöwer, S.M.; Hartmann, C.A.; Holzhauser, T. The Development of Highly Specific and Sensitive Primers for the Detection of Potentially Allergenic Soybean (Glycine max) Using Loop-Mediated Isothermal Amplification Combined with Lateral Flow Dipstick (LAMP-LFD). Foods 2020, 9, 423. [Google Scholar] [CrossRef]
- Ge, Y.; Wu, B.; Qi, X.; Zhao, K.; Guo, X.; Zhu, Y.; Qi, Y.; Shi, Z.; Zhou, M.; Wang, H.; et al. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS ONE 2013, 8, e69941. [Google Scholar] [CrossRef]
- Yu, J.; Wang, F.; Zhan, X.; Wang, X.; Zuo, F.; Wei, Y.; Qi, J.; Liu, Y. Improvement and evaluation of loop-mediated isothermal amplification combined with a chromatographic flow dipstick assay and utilization in detection of Vibrio cholerae. Anal. Bioanal. Chem. 2019, 411, 647–658. [Google Scholar] [CrossRef]
- Rolando, J.C.; Jue, E.; Barlow, J.T.; Ismagilov, R.F. Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020, 48, e42. [Google Scholar] [CrossRef]
- Wong, Y.-P.; Othman, S.; Lau, Y.-L.; Radu, S.; Chee, H.-Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef]
- Yin, H.Y.; Fang, T.J.; Wen, H.W. Combined multiplex loop-mediated isothermal amplification with lateral flow assay to detect sea and seb genes of enterotoxic Staphylococcus aureus. Lett. Appl. Microbiol. 2016, 63, 16–24. [Google Scholar] [CrossRef]
- Liang, C.; Chu, Y.; Cheng, S.; Wu, H.; Kajiyama, T.; Kambara, H.; Zhou, G. Multiplex loop-mediated isothermal amplification detection by sequence-based barcodes coupled with nicking endonuclease-mediated pyrosequencing. Anal. Chem. 2012, 84, 3758–3763. [Google Scholar] [CrossRef]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. BioTechniques 2012, 53, 81–89. [Google Scholar] [CrossRef]
- Karthik, K.; Rathore, R.; Thomas, P.; Arun, T.R.; Viswas, K.N.; Dhama, K.; Agarwal, R.K. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX 2014, 1, 137–143. [Google Scholar] [CrossRef]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques 2015, 58, 59–68. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Chen, L.; Quan, S.; Jiang, S.; Zhang, D.; Yang, L. One simple DNA extraction device and its combination with modified visual loop-mediated isothermal amplification for rapid on-field detection of genetically modified organisms. Anal. Chem. 2013, 85, 75–82. [Google Scholar] [CrossRef]
- Liu, N.; Zou, D.; Dong, D.; Yang, Z.; Ao, D.; Liu, W.; Huang, L. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci. Rep. 2017, 7, 45601. [Google Scholar] [CrossRef]
- Wagner, A.; Blackstone, N.; Cartwright, P.; Dick, M.; Misof, B.; Snow, P.; Wagner, G.P.; Bartels, J.; Murtha, M.; Pendleton, J. Surveys of Gene Families Using Polymerase Chain Reaction: PCR Selection and PCR Drift. Syst. Biol. 1994, 43, 250. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Varma, A.; Padh, H.; Shrivastava, N. Plant genomic DNA isolation: An art or a science. Biotechnol. J. 2007, 2, 386–392. [Google Scholar] [CrossRef]
- Sheu, S.-C.; Wang, Y.-J.; Huang, P.-C.; Lien, Y.-Y.; Lee, M.-S. Authentication of olive oil in commercial products using specific, sensitive, and rapid loop-mediated isothermal amplification. J. Food Sci. Technol. 2023, 60, 1834–1840. [Google Scholar] [CrossRef]
- Sakamoto, W.; Takami, T. Chloroplast DNA Dynamics: Copy Number, Quality Control and Degradation. Plant Cell Physiol. 2018, 59, 1120–1127. [Google Scholar] [CrossRef]
- Giannoulia, K.; Banilas, G.; Hatzopoulos, P. Oleosin gene expression in olive. J. Plant Physiol. 2007, 164, 104–107. [Google Scholar] [CrossRef]
- Longo, M.C.; Berninger, M.S.; Hartley, J.L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 1990, 93, 125–128. [Google Scholar] [CrossRef]
- He, L.; Xu, H. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of white spot syndrome virus and infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp. Aquaculture 2011, 311, 94–99. [Google Scholar] [CrossRef]
- Hsieh, K.; Mage, P.L.; Csordas, A.T.; Eisenstein, M.; Soh, H.T. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem. Commun. 2014, 50, 3747–3749. [Google Scholar] [CrossRef]
- Carter, C.; Akrami, K.; Hall, D.; Smith, D.; Aronoff-Spencer, E. Lyophilized visually readable loop-mediated isothermal reverse transcriptase nucleic acid amplification test for detection Ebola Zaire RNA. J. Virol. Methods 2017, 244, 32–38. [Google Scholar] [CrossRef]
- Song, X.; Coulter, F.J.; Yang, M.; Smith, J.L.; Tafesse, F.G.; Messer, W.B.; Reif, J.H. A lyophilized colorimetric RT-LAMP test kit for rapid, low-cost, at-home molecular testing of SARS-CoV-2 and other pathogens. Sci. Rep. 2022, 12, 7043. [Google Scholar] [CrossRef]
- Becherer, L.; Bakheit, M.; Frischmann, S.; Stinco, S.; Borst, N.; Zengerle, R.; von Stetten, F. Simplified Real-Time Multiplex Detection of Loop-Mediated Isothermal Amplification Using Novel Mediator Displacement Probes with Universal Reporters. Anal. Chem. 2018, 90, 4741–4748. [Google Scholar] [CrossRef]
- Crego-Vicente, B.; Fernández-Soto, P.; García-Bernalt Diego, J.; Febrer-Sendra, B.; Muro, A. Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp.—A Laboratory Approach to Point-Of-Care. Int. J. Mol. Sci. 2023, 24, 893. [Google Scholar] [CrossRef]


| No. | Species | Origin/Supplier | Processing Stage |
|---|---|---|---|
| 1 | Arbutus unedo | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 2 | Cistus creticus | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 3 | Olea europaea | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 4 | Olea europaea | Local drug store | Dried, rubbed |
| 5 | Origanum majorana | Husarich GmbH | Dried, rubbed, germinated |
| 6 | Origanum onites | Husarich GmbH | Dried, rubbed, germinated |
| 7 | Origanum onites | Hela GmbH | Dried, rubbed, germinated |
| 8 | Origanum vulgare | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 9 | Thymus vulgaris | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 10 | Carthamus tinctorius | Botanical Garden, University of Hamburg | Freeze dried 24 h, ground |
| 11 | Ocimum basilicum | Local retail | Freeze dried 24 h, ground |
| 12 | Brassica oleracea var. italica | Local retail | Freeze dried 24 h, ground |
| 13 | Crocus sativus | Husarich GmbH | Dried, ground |
| 14 | Curcuma longa | Hela GmbH | Dried, rubbed, germinated |
| 15 | Apium graveolens | Local retail | Freeze dried 24 h, ground |
| 16 | Salvia officinalis | Local retail | Freeze dried 24 h, ground |
| Primer Set I: 5′-3′ Species: Olea europaea, Locus: trnL-trnF Intergenic Spacer | |
|---|---|
| F3 | CACATGTGATATATAATACACATCC |
| B3 | CATTCCCAATGTAACATTAACATC |
| FIP (F2 + F1C) | GGATCTTCAAAAAGACGACTTTGTC-TTAAGCAAGGAATCCCCAT |
| BIP (B2 + B1C) | ATTCCAGGACTTGGAGAAAACTTTG-CCATCCTCATTTTATTAGATGACT |
| loopB | CCCCCCTTGTCCTTTTAATTGACAT |
| 6-FAM/loopB | 6-FAM/CCCCCCTTGTCCTTTTAATTGACAT |
| Biotin/BIP | Biotin/ATTCCAGGACTTGGAGAAAACTTTG-CCATCCTCATTTTATTAGATGACT |
| Primer Set II: 5′-3′ Species: Arbutus unedo, Locus: Internal Transcribed Spacer | |
| F3 | CCTCCGGGAACAATTGAGC |
| B3 | AACACAGCCCACGAATGG |
| FIP (F2 + F1C) | GAACACGTTTCCCGAAGGACCG-CCAGTTGTCGCCTTCCATT |
| BIP (B2 + B1C) | GTGAAATAACGAAACCCGGCGC-TGGGAGACGTGCATCTGTT |
| loopF | ACCCGCTCGAGGAGGAA |
| loopB | AACCGCGCCAAGGAAACT |
| 6-FAM/loopF | 6-FAM/ACCCGCTCGAGGAGGAA |
| Biotin/FIP | Biotin/ATTCCAGGACTTGGAGAAAACTTTG-CCATCCTCATTTTATTAGATGACT |
| DIG/FIP | DIG/ATTCCAGGACTTGGAGAAAACTTTG-CCATCCTCATTTTATTAGATGACT |
| Parameter | New England Biolabs (NEB) | Value Range for the Parameter | Holz et al. 2023 [21] | POC-Optimized |
|---|---|---|---|---|
| Parameters of DNA isolation optimized with DoE | ||||
| Washing steps | - | one/two | two | one |
| Buffer conc. [mol/L] | - | 1–2.5 | 2.5 | 2.5 |
| Sample weight [mg] | - | 5–25 | 20 | 15 |
| Optimized with OFAT | ||||
| DNA sample per reaction [µL] | - | 0.1–5.0 | 2 | 5 |
| Parameters of LAMP reaction optimized with DoE | ||||
| Temperature [°C] | 65 | 62–72 | 72 | 67 |
| MgSO4 [mM] | 6 | 4–8 | 4 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holz, N.; Wax, N.; Illarionov, B.A.; Iskhakova, M.; Fischer, M. Food Authentication: The Detection of Arbutus unedo and Olea europaea Leaves as an Admixture of Oregano Using LAMP- and Duplex LAMP-Based Test Systems with Lateral-Flow Assays. Agriculture 2024, 14, 597. https://doi.org/10.3390/agriculture14040597
Holz N, Wax N, Illarionov BA, Iskhakova M, Fischer M. Food Authentication: The Detection of Arbutus unedo and Olea europaea Leaves as an Admixture of Oregano Using LAMP- and Duplex LAMP-Based Test Systems with Lateral-Flow Assays. Agriculture. 2024; 14(4):597. https://doi.org/10.3390/agriculture14040597
Chicago/Turabian StyleHolz, Nathalie, Nils Wax, Boris A. Illarionov, Margarita Iskhakova, and Markus Fischer. 2024. "Food Authentication: The Detection of Arbutus unedo and Olea europaea Leaves as an Admixture of Oregano Using LAMP- and Duplex LAMP-Based Test Systems with Lateral-Flow Assays" Agriculture 14, no. 4: 597. https://doi.org/10.3390/agriculture14040597
APA StyleHolz, N., Wax, N., Illarionov, B. A., Iskhakova, M., & Fischer, M. (2024). Food Authentication: The Detection of Arbutus unedo and Olea europaea Leaves as an Admixture of Oregano Using LAMP- and Duplex LAMP-Based Test Systems with Lateral-Flow Assays. Agriculture, 14(4), 597. https://doi.org/10.3390/agriculture14040597







