Abstract
Glycerol-3-phosphate acyltransferase (GPAT) genes encode enzymes involved in the biosynthesis of plant oils. Rapeseed has four BnGPAT9 genes, but the expression patterns and functions of these four homologous copies in rapeseed for seed oil accumulation are not well understood. In this study, we cloned the four BnGPAT9 genes and their promoters from Brassica napus and found significant differences in the expression of BnGPAT9 genes among different rapeseed varieties. We confirmed that BnGPAT9-A01/C01 are highly conserved in rapeseed, with high expression levels in various tissues, especially during the late stages of silique development and seed maturation. All four BnGPAT9 genes (BnGPAT9-A01/C01/A10/C09) can promote seed oil accumulation, but BnGPAT9-A01/C01 have a greater effect. Overexpression in Arabidopsis and rapeseed increased seed oil content and altered fatty acid composition, significantly increasing linolenic acid content. Transcriptome analysis revealed that BnGPAT9 genes promote the upregulation of genes related to oil synthesis, particularly those in the Plant–pathogen interaction, alpha-Linolenic acid metabolism, MAPK signaling pathway—plant, and Glutathione metabolism pathways. In summary, these results indicate that the four BnGPAT9 genes in rapeseed have different expression patterns and roles in regulating seed oil accumulation, with BnGPAT9-A01/C01 contributing the most to promoting oil accumulation.
1. Introduction
As one of the world’s three major oil crops, rapeseed holds a very important position in the global oil crop production structure. Increasing the oil content in rapeseed seeds and improving the composition of different fatty acids in the seeds are important goals for the high-quality breeding of oil crops. Triacylglycerol (TAG) is the main storage form of oil in oil crop seeds. It usually combines with oleosin to form a stable oil body structure, providing carbon sources and energy support for seedlings from germination to photosynthetic autotrophy [1,2]. It is also closely related to processes such as cell division, stomatal opening and closing, membrane lipid remodeling, and pollen development [3,4,5].
There are two pathways for the synthesis of TAG. One is the acyl-CoA-dependent pathway. Glycerol-3-phosphate acyltransferase (GPAT) transfers an acyl group from acyl-CoA or an acyl carrier protein (ACP) to the sn-1 position of glycerol-3-phosphate, forming lysophosphatidic acid (LPA). This is the first step in the synthesis of various glycerolipids, including membrane lipids and triacylglycerol [6,7,8]. Finally, diacylglycerol acyltransferase (DGAT) catalyzes the acylation at the sn-3 position to produce TAG. DGAT is also considered a rate-limiting enzyme for oil storage in plants [9,10,11]. The other pathway is the acyl-CoA-independent pathway [12], where TAG can also be synthesized by phospholipids: diacylglycerol acyltransferase (PDAT), transferring an acyl group from the sn-2 position of phospholipids to the sn-3 position of sn-1,2-diacylglycerol [13,14]. In rapeseed, oil exists in the form of TAG, and GPAT plays an initiating role in the catalysis of triacylglycerol formation [7,8]. GPAT is the rate-limiting enzyme in this pathway [15]. Thus, GPAT plays a very important role in the formation of oil in rapeseed.
In Arabidopsis thaliana, GPAT9 (AT5G60620) is localized on the endoplasmic reticulum membrane and participates in the acylation reaction at the sn-1 position of the glycerol backbone during TAG biosynthesis, playing an important role in the Kennedy pathway of triglyceride biosynthesis [8,16]. A reduced expression of the AtGPAT9 gene affects the content and composition of TAG in seeds. Knocking out the AtGPAT9 gene reduces the oil content in seeds by 26–44% [8]. With the function of the GPAT9 gene being confirmed in Arabidopsis, the functions of GPAT9 genes in other plants are gradually being revealed. There are two GPAT9 genes in sunflowers, HaGPAT9-1 and HaGPAT9-2 [17]. Both sunflower genes are expressed during seed development and in vegetative tissues, but HaGPAT9-1 is the functional gene during seed development and exhibits GPAT enzyme activity. PpGPAT9 is a gene encoding GPAT9 in Physcomitrella patens. An overexpression of PpGPAT9 in Arabidopsis can increase seed oil content by approximately 10%, and the levels of polyunsaturated fatty acids (PUFAs) and saturated fatty acids (FAs) in transgenic seed oil increase by 60% and 43%, respectively [18]. In B. napus, four GPAT9 genes have been cloned and studied. GPAT9 genes located on the A1 and C1 chromosomes (BnGPAT9-A01/C01) encode functional GPAT enzymes, while those on the A10 and C9 chromosomes (BnGPAT9-A10/C09) do not encode functional enzymes [19]. Recent studies have shown that the overexpression of BnGPAT9-C01 in B. napus can promote the production of phosphatidic acid (PA), increase oil accumulation in seeds, and lead to an increase in C18:2 fatty acids [20]. However, the expression patterns of the four GPAT9 genes in B. napus during silique development and their regulatory roles in oil synthesis still need further elucidation.
Our initial research was based on the Arabidopsis AtGPAT9 (AT5G60620) sequence. Using the AtGPAT9 gene number, we retrieved the GPAT9 gene sequences of Brassica campestris and Brassica oleracea from the Brassica database (http://brassicadb.cn/#/ (accessed on 4 November 2015)). Based on the alignment results of the Arabidopsis, B. campestris, and B. oleracea GPAT9 sequences, we designed primers and cloned BnGPAT9-C01 (GenBank: KX344511.1) from Xiangyou 15 (Brassica napus L., XY15) [21,22]. We then conducted bioinformatics analysis and gene expression pattern analysis on it. Later, we searched for four BnGPAT9 genes in the B. napus varieties ZS11, Westar, Tapidor, No2127, and Darmor through BnTIR (https://yanglab.hzau.edu.cn/BnTIR (accessed on 15 October 2019)). Starting from gene cloning, we cloned the four BnGPAT9 genes and their promoters from XY 15. Using promoter–GUS fusion, we identified the expression patterns of the four BnGPAT9 genes in the plant. Through qPCR, we revealed the expression characteristics of BnGPAT9 genes in the roots, stems, leaves, flowers, and siliques of B. napus, dynamically identifying the relationship between the expression of the four BnGPAT9 genes and oil formation during seed development. By constructing overexpression vectors for the four BnGPAT9 genes and transforming them into Arabidopsis, we separately identified the roles of the four genes in seed oil accumulation. This study aims to clone, identify, and analyze the functions of the BnGPAT9 genes in B. napus, further elucidating the regulatory roles of the four homologous BnGPAT9 genes in oil synthesis. We also hope to improve the quality of rapeseed and increase the oil content in seeds through transgenic technology.
2. Materials and Methods
2.1. Materials
The experimental material used in this study is B. napus XY15, provided by the Oil Crops Research Institute of Hunan Agricultural University. XY15 and transgenic rapeseed were planted in the Yunyuan experimental field of Hunan Agricultural University, with net isolation planting. Other conditions were consistent with field planting. In March 2023, we collected roots, stems, leaves, and flowers of XY15 during the flowering period. The roots were cleaned thoroughly, immediately frozen in liquid nitrogen, and then stored at −80 °C for future use. For XY15 and transgenic B. napus, manual pollination was performed on freshly blooming flowers. Siliques were collected at 1 week, 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, and 7 weeks post-pollination. Seeds and silique walls were separated using tweezers, immediately frozen in liquid nitrogen, and then stored at −80 °C for future use. The materials used for subsequent promoter analysis and overexpression experiments in Arabidopsis thaliana were the Columbia wild-type strain, preserved by our laboratory.
2.2. Cloning and Bioinformatics Analysis of BnGPAT9 Genes and Promoters
Using the AtGPAT9 gene sequence as a reference, we searched for the four BnGPAT9 genes in B. napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, and ZS11 in the BnTIR database (https://yanglab.hzau.edu.cn/BnTIR (accessed on 15 October 2019)) and downloaded their nucleotide and protein sequences. Using the ZS11 genome as a reference, we designed primers for gene and promoter cloning: BnGPAT9-A01 (Fw: TTGGCTCTGACTCTTACC; Rv: TGCATTCACTTGTCTTCC), BnGPAT9-A10 (Fw: GAACAATGAGCAGTGGGG; Rv: TTTTTATCTAATGGAAGG), BnGPAT9-C01 (Fw: TTCACCACCAAAAGAACCCAAT; Rv: TGAAACACCACAAGGAACAAGG), BnGPAT9-C09 (Fw: ATGAGCAGTACGGCGGGGAAGC; Rv: GAGGGTGCTTTATCATTACATT), pBnGPAT9-A01 (Fw: GTTTTCCTTCGTGTTTCTCTCT; Rv: TGTGTATTTCTAAGTGTTCGTC), pBnGPAT9-A10 (Fw: TACAGCTCGTGGCTACAACCGG; Rv: TTGTGGTGGTGGTGAAGAGAAG), pBnGPAT9-C01 (Fw: GTATGGAGGTCGATGTTGAGGT; Rv: CGAGTTTCTGACCTTGTGATGT), and pBnGPAT9-C09 (Fw: CTCATAAACAAGGAGAGGAAGT; Rv: TGAAGAGGAGAAGCGAGGGAAG). Using cDNA from rapeseed flowers as a template, we cloned the BnGPAT9 genes and their promoters using PCR technology.
Using the ORF Finder and CD-search functions of the NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 6 February 2024)), we analyzed the open reading frame and protein domains of the BnGPAT9 gene in rapeseed. We used the ExPASy tool (https://web.expasy.org/protparam/ (accessed on 9 March 2024)) to analyze basic physicochemical properties of the protein such as the molecular weight and isoelectric point. We utilized PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 16 March 2024)) and TBtools software v2.096 to analyze cis-acting elements. We also used the MEME program (http://meme-suite.org/tools/meme (accessed on 21 March 2024)) to analyze conserved domains in the protein sequence.
2.3. Plant Expression Vector Construction and Genetic Transformation
The vector PC1300S was digested with SacⅠ and BamHⅠ, and digested fragments were recovered. We recombined it with the PCR amplification products BnGPAT9-A01, BnGPAT9-A10, BnGPAT9-C01, and BnGPAT9-C09 from Section 2.2. We transformed the recombinant products into E. coli and verified by sequencing. Similarly, the pCAMBIA1301:GUS vector was digested with Hind III and BamH Ⅰ, and digested fragments were recovered. We recombined it with the PCR amplification products pBnGPAT9-A01, pBnGPAT9-A10, pBnGPAT9-C01, and pBnGPAT9-C09 from Section 2.2. We transformed the recombinant products into E. coli and verified by sequencing. We transferred the recombinant plasmids into Agrobacterium tumefaciens GV3101 and then transformed them into wild-type Arabidopsis using the floral dip method [23]. Using XY15 as the receptor material, we obtained transgenic lines through the Agrobacterium-mediated transformation of rapeseed. The Agrobacterium-mediated transformation process for B. napus refers to the method described by Dai [24].
2.4. GUS Staining of Plant Tissues
GUS staining was performed according to the method described by previous researchers [25]. The specific experimental procedure was as follows:
- Immerse the plant tissue samples in 1.5 mL centrifuge tubes containing 90% pre-chilled acetone and place them on ice for 10 min to fix the tissues.
- Remove the acetone and rinse 2–3 times with a staining buffer (rinse solution: 50 mM NaPO4, pH = 7.2; 0.5 mM K3Fe(CN)6; 0.5 mM K4Fe(CN)6), with each rinse lasting 2 min.
- Remove the rinse solution and add 2 mM X-Gluc staining solution (X-Gluc diluted with rinse solution, stored at −20 °C in the dark) to immerse the plant tissues. Apply a vacuum for 10 min to ensure thorough contact with the X-Gluc staining solution.
- Incubate at 37 °C for 10–20 h for adequate staining.
- Remove the staining solution and add 75% ethanol for 1 h to decolorize (if decolorization is insufficient, this step can be repeated).
- Add 95% ethanol and store at 4 °C for long-term preservation or observe and photograph directly under a stereomicroscope.
2.5. Determination of Oil Content and Fatty Acid Composition of Seeds
The oil content and fatty acid composition in Arabidopsis and rapeseed seeds were determined using gas chromatography. The specific steps for extracting fatty acids from the seeds were as follows:
- Weigh 10 mg of seeds.
- Add 2 mL of 2.5% H2SO4 in methanol (0.01% BHT).
- Add 100 μL of 16.2 μmol/mL (C17:0) fatty acid (FA) as an internal standard.
- For rapeseed, gently crush the seeds with a glass rod (not necessary for Arabidopsis).
- Incubate in a water bath at 85 °C for 2 h, checking every 10–20 min to ensure the lid is not loose or leaking.
- After cooling to room temperature, add 2 mL of ddH2O and 2 mL of n-hexane, and shake well.
- Centrifuge at 1000 rpm for 10 min.
- Take approximately 1 mL of the supernatant into the sample vial.
- Inject 1 μL of the sample into the gas chromatograph (GC), using a split ratio of 2:1 to 10:1.
To calculate the oil content in seeds using gas chromatography, the following formula is used: seed oil content (%) = (mass of internal standard FA (C17:0)/proportion of internal standard FA (C17:0)—mass of internal standard FA (C17:0))/seed mass.
2.6. RNA Extraction and qRT-PCR Analysis
Root, stem, leaf, flower, seed, and silique tissues were collected from XY15 and transgenic lines. RNA was extracted from plant tissues using the RNA Prep Pure Plant kit (Cat. No. LS1040, Promega, Madison, WI, USA). cDNA was synthesized using the Transcript RT kit (Cat. No. AE311, TransGen Biotech, Beijing, China), and the reverse transcription product was diluted 100-fold as the template. B. napus Actin2.1 was used as the internal reference gene. The qRT-PCR reaction system consisted of 5 μL of the 2×SYBR Green Master Mix, 0.4 μL each of the forward and reverse primers, and 4.2 μL of the cDNA template. The qRT-PCR primers used were qRT-PCR-BnGPAT9-A01 (Fw: TGGCTCTGACTCTTACCATCGC; Rv: GGAACCGGAAGGAAGGTAATCT), qRT-PCR-BnGPAT9-A10 (Fw: GAACAATGAGCAGTGGGGCGGG; Rv: TTCAGTGAGAGTTGGCGAGATG), qRT-PCR-BnGPAT9-C01 (Fw: GCATAGCGAACGCAAGCAACAG; Rv: GAAACACCACAAGGAACAAGG), qRT-PCR-BnGPAT9-C09 (Fw: ATGAGCAGTACGGCGGGGAAGC; Rv: CAGTCAGAGTTGGCGAGATGTC), and qRT-PCR-Actin (Fw: GGTTGGGATGGACCAGAAGG; Rv: TCAGGAGCAATACGGAGC). The qRT-PCR reaction program was as follows: 95 °C for 5 min; 95 °C for 30 s, 60 °C for 1 min, 72 °C for 10 min, 30 cycles. Each treatment had 3 biological replicates, and gene relative expression levels were analyzed using the 2–ΔΔCt method [26].
2.7. Transcriptome Sequencing and Differential Expression Genes Screening
Based on the oil content detection results of seeds from different transgenic Arabidopsis lines, siliques of 14-day-old transgenic Arabidopsis stored at −80 °C were used for transcriptome sequencing analysis. During the entire seed development process in Arabidopsis, the oil accumulation in seeds follows an “S” curve, rapidly increasing from the 7th day after fertilization and reaching a peak on the 18th day after fertilization [27,28]. Total RNA was extracted from tissue samples, and the concentration and purity of the extracted RNA were measured using a Nanodrop 2000 (Thermo Fisher Scientific, Shanghai, China). The integrity of the RNA was assessed by agarose gel electrophoresis, and the RQN value was determined using an Agilent 5300 (Agilent Technologies, Inc., Shanghai, China). For library construction, the total RNA required per sample was 1 μg, with a concentration of ≥30 ng/μL, an RQN > 6.5, and an OD 260/280 ratio between 1.8 and 2.2. The cDNA library was sequenced using the Illumina HiSeq2000 (Illumina, Shanghai, China) high-throughput sequencing platform. The raw data were filtered to obtain high-quality clean data, which were then aligned to the specified reference genome (TAIR 10_arabidopsis, download link: https://www.arabidopsis.org/, (accessed on 22 July 2024)) to obtain the mapped data. Differential expression analysis was conducted based on gene expression levels in different sample groups, using FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) as the metric for transcript or gene expression levels. Genes with a log2 fold change (|log2(FC)|) ≥ 1 and a p-value < 0.05 were considered differentially expressed genes (DEGs). KEGG enrichment analyses were performed on the identified DEGs.
3. Results
3.1. Characteristic Analysis of BnGPAT9 in B. napus
Using the AtGPAT9 gene sequence as a reference, we searched for four BnGPAT9 genes in the BnTIR database (https://yanglab.hzau.edu.cn/BnTIR (accessed on 15 October 2019)) across B. napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, and ZS11. Based on their chromosomal locations, the BnGPAT9 genes on chromosomes A01, A10, C01, and C09 were named BnGPAT9-A01, BnGPAT9-A10, BnGPAT9-C01, and BnGPAT9-C09, respectively. Primers were designed based on the gene sequences from ZS11, and using cDNA from XY15 flowers as a template, we cloned BnGPAT9-A01 (CDS length 1131 bp), BnGPAT9-A10 (CDS length 1116 bp), BnGPAT9-C01 (CDS length 1131 bp), and BnGPAT9-C09 (CDS length 1116 bp). BnGPAT9-A01 encodes 376 amino acids with a molecular weight of 43.25 kD. BnGPAT9-A10 encodes 371 amino acids with a molecular weight of 42.48 kD. BnGPAT9-C01 encodes 376 amino acids with a molecular weight of 43.25 kD. BnGPAT9-C09 encodes 371 amino acids with a molecular weight of 42.51 kD. The conserved motifs of the four BnGPAT9 proteins and the AtGPAT9 protein are highly conserved, each containing 10 motifs in the same order (Figure 1A). The protein sequence identities between BnGPAT9-A01, BnGPAT9-A10, BnGPAT9-C01, BnGPAT9-C09, and AtGPAT9 are 93%, 93%, 94%, and 94%, respectively. BnGPAT9-A01 and BnGPAT9-C09 have 98% protein sequence identity, while BnGPAT9-A01 and BnGPAT9-C01 have identical protein sequences (Figure 1B). We found some differences in the CDS and protein sequences of BnGPAT9 among different B. napus varieties (Supplementary Figure S1). Notably, the CDS and amino acid sequences of BnGPAT9-C09 are identical across all varieties (Supplementary Figures S1, S4, and S8), reflecting the high conservation of the BnGPAT9-C09 protein in rapeseed. Although there are nucleotide differences in the CDS sequences of BnGPAT9-A01 among different rapeseed varieties, these do not result in changes to the amino acid sequences, and the BnGPAT9-A01 protein sequences are identical across all varieties (Supplementary Figures S1 and S5). The BnGPAT9-C01 protein sequence in XY15 is identical to those in No2127 and Quinta, although there are nucleotide differences in the CDS sequences (Supplementary Figures S1, S3, and S7). The CDS and protein sequences of BnGPAT9-A10 show greater variability among different varieties (Supplementary Figures S1, S2, and S6).

Figure 1.
Characterization of the BnGPAT9 protein sequences. (A) The analysis of conserved motifs in BnGPAT9 and AtGPAT9 proteins; (B) the multiple sequence alignment of plant BnGPAT9 proteins.
3.2. Analysis of Promoter Cis-Acting Elements of BnGPAT9
Using DNA from XY15 flowers as a template, we cloned promoters from the upstream 2 kb regions of the four BnGPAT9 genes in XY15, obtaining the promoter sequences pBnGPAT9-A01 (1239 bp), pBnGPAT9-A10 (900 bp), pBnGPAT9-C01 (1509 bp), and pBnGPAT9-C09 (1174 bp). Using the PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 26 March 2024)), we conducted a cis-regulatory element analysis of the four promoter sequences. As shown in Figure 2, a total of 37 types of functional cis-regulatory elements were identified in the promoter sequences of the BnGPAT9 genes in rapeseed, which were categorized into five main classes based on their functions: stress response, light response, plant hormone response, metabolic regulation, and other elements. The elements involved in stress response were the most numerous and diverse, primarily including MYB and ARE elements. Light response elements were the next most common, including elements such as G-box, BOX 4, and MRE. Plant hormone response elements included those responsive to abscisic acid (ABRE), methyl jasmonate (CGTCA motif and TGACG motif), gibberellin (GARE motif), and salicylic acid (SARE and TCA). Additionally, there were elements involved in metabolic regulation, such as carbon metabolism (AAGAA motif), among others. The rich variety of regulatory elements suggests that BnGPAT9 may play an important regulatory role in the growth and development of rapeseed and be influenced by external environmental factors such as temperature, moisture, and various hormones.
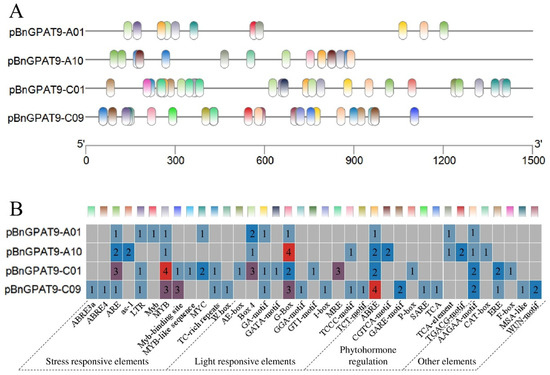
Figure 2.
Cis elements detected in the promoter of the BnGPAT9 genes. (A) Promoter element distribution, where different colors correspond to different elements in the figure below; (B) the heat map shows the number of promoter elements, and the gray square indicates that the elements could not be detected.
3.3. Analysis of Expression Pattern of BnGPAT9
To gain a more comprehensive understanding of the spatiotemporal expression patterns of BnGPAT9, we transformed the pCAMBIA1301:GUS vector and the constructed promoter vectors pBnGPAT9-A01:GUS, pBnGPAT9-A10:GUS, pBnGPAT9-C01:GUS, and pBnGPAT9-C09:GUS into Arabidopsis thaliana wild-type Col-0 using the floral dip method mediated by Agrobacterium tumefaciens. Transgenic plant lines were successfully screened and obtained. As shown in Figure 3A, GUS staining results revealed that BnGPAT9-A01/C01 are highly expressed in stems, leaves, flowers, and siliques. BnGPAT9-C09 is expressed in leaves and flowers, but its expression level is significantly lower than that of BnGPAT9-A01/C01. BnGPAT9-A10 exhibits weak expression at the leaf margins, flowers, and siliques. These results indicate that BnGPAT9-A01/C01 are widely present in both the vegetative and reproductive organs of rapeseed and may be the primary genes performing the GPAT9 function. By examining the expression of BnGPAT9 in ZS11 through the BnTIR database, we found that the expression levels of BnGPAT9-A10 during the growth and development of rapeseed are significantly lower than those of BnGPAT9-A01/C01 (Figure 3B). BnGPAT9-A01/C01 are abundantly expressed in both vegetative and reproductive organs of ZS11, especially in flowers and developing seeds (Figure 3B). The GUS staining observations are consistent with the expression characteristics of BnGPAT9 in ZS11
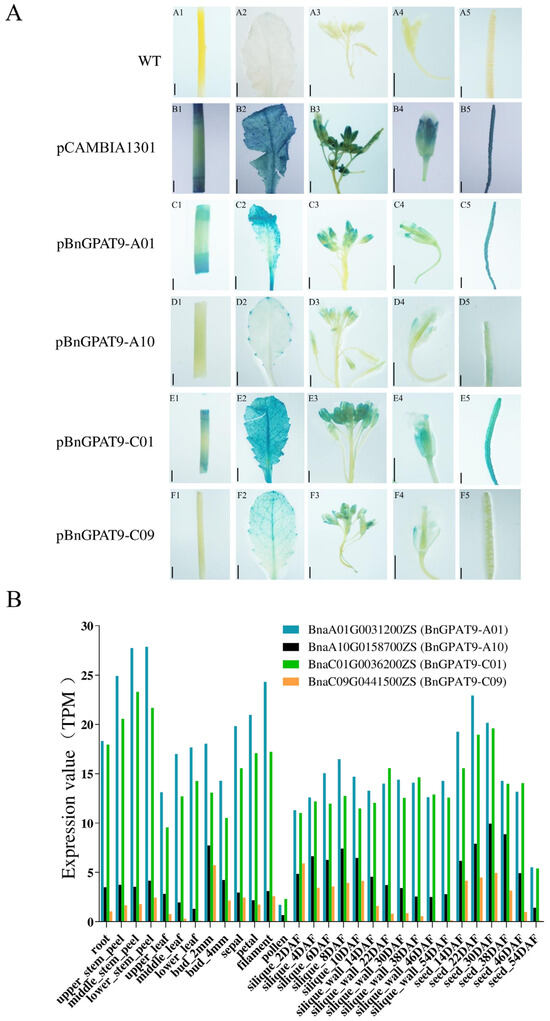
Figure 3.
Tissue expression patterns of BnGPAT9 genes. (A) The GUS staining results of Arabidopsis thaliana. (A1,B1,C1,D1,E1,F1): GUS staining in stems; (A2,B2,C2,D2,E2,F2): GUS staining in leaves; (A3,B3,C3,D3,E3,F3): GUS staining in inflorescences; (A4,B4,C4,D4,E4,F4): GUS staining in flowers; (A5,B5,C5,D5,E5,F5): GUS staining in siliques. Bar = 1 mm. (B) Expression profiles of BnGPAT9 genes in ZS11. Data sourced from BnTIR (https://yanglab.hzau.edu.cn/BnTIR (accessed on 15 October 2019)).
3.4. Tissue-Specific Expression Analysis of BnGPAT9 Genes and Oil Content Analysis in Seeds
To elucidate the regulatory roles of BnGPAT9-A01/A10/C01/C09 during the growth and development of rapeseed, this study employed qRT-PCR technology to analyze the expression patterns of these four BnGPAT9 genes in different tissues and at various developmental stages of seed development. Interestingly, we found that the expression level of BnGPAT9-C01 in XY15 was very low, but relatively high levels could be detected during the later stages of seed development (Figure 4A). As shown in Figure 4A, the genes BnGPAT9-A01/A10/C09 are expressed in the roots, stems, leaves, flowers, seeds, and silique walls of rapeseed, with high expression mainly observed during the late stages of seed development. Among them, BnGPAT9-A01 shows higher expression levels in seeds at the 6th and 7th weeks of development compared to BnGPAT9-A10/C09.
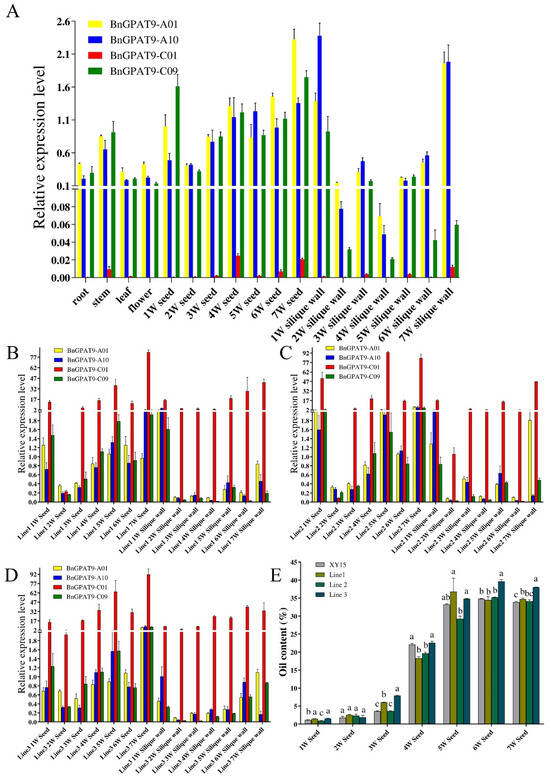
Figure 4.
Analysis of tissue-specific expression patterns of BnGPAT9 genes and seed oil accumulation. (A) qRT-PCR detection of BnGPAT9 expression in roots, stems, leaves, flowers, and seeds at 1–7 weeks of development, as well as siliques of XY15. (B–D) qRT-PCR detection of BnGPAT9 expression in seeds and siliques at 1–7 weeks of development in transgenic lines overexpressing the BnGPAT9-C01 gene. (E) Oil content in the seeds of XY15 and the three transgenic lines.
Studies have reported that BnGPAT9-C01 is a dominant homologous gene in B. napus that can promote oil accumulation in seeds [20]. However, our study found that the expression level of BnGPAT9-C01 in XY15 is not high. To understand the impact of BnGPAT9-C01 on oil synthesis in XY15 seeds, we constructed an overexpression vector PC1300S::BnGPAT9-C01 and transformed it into rapeseed, obtaining three transgenic lines of XY15 (line 1, line 2, line 3). In the siliques of transgenic lines at 1–7 weeks of development, the expression level of BnGPAT9-C01 was significantly higher than that of BnGPAT9-A01/A10/C09 (Figure 4B–D). As shown in Figure 4E, the oil content in seeds at 1 week, 3 weeks, 5 weeks, 6 weeks, and 7 weeks of the transgenic lines was significantly higher than that of XY15, while the oil content in 2-week seeds showed no significant difference compared to XY15. The oil content in seeds of line 1 and line 2 at 4 weeks was even significantly lower than that of XY15. Overall, we found that the mature seeds (7 weeks) of the three lines overexpressing BnGPAT9-C01 had the highest increase in oil content by 4.2%, indicating that this gene has a positive regulatory function in seed oil content. We observed significant changes in the fatty acid composition of seeds from the transgenic lines (Table 1). Compared to the control, the content of oleic acid (C18:1) significantly decreased, while the content of Linoleic acid (C18:2) and linolenic acid (C18:3) significantly increased from early seed development (3 weeks) to seed maturity (7 weeks). The results indicate that BnGPAT9-C01 can promote oil accumulation in seeds and increase the content of Linoleic acid (C18:2) and linolenic acid (C18:3).

Table 1.
Brassica napus seeds FA composition.
3.5. The Role of the BnGPAT9 Gene in Seed Oil Accumulation
To elucidate the functions of BnGPAT9-A01/A10/C01/C09 in seed oil accumulation, we transformed the constructed overexpression vectors of PC1300S::BnGPAT9-A01/A10/C01/C09 into Arabidopsis thaliana wild-type Col-0 using the Agrobacterium-mediated floral dip method. We harvested T2 homozygous transgenic lines’ seeds to measure thousand-seed weight, oil content, and fatty acid composition. We found that the thousand-seed weight significantly decreased in BnGPAT9-C01 transgenic line 2 and significantly increased in BnGPAT9-C09 transgenic line 2 (Figure 5A). However, overall, the four BnGPAT9 genes did not affect the thousand-seed weight, indicating that BnGPAT9 does not influence seed weight. The seed oil content measurement results are shown in Figure 5B. An overexpression of BnGPAT9-A01 increased seed oil content by 0.29–9.51%, with line 3 being significantly higher than the control. An overexpression of BnGPAT9-A10 increased seed oil content by 1.58–2.54%, but seed oil content in line 2 decreased by 2.58% compared to the control. An overexpression of BnGPAT9-C01 increased seed oil content by 0.19–3.81%, with line 1 being significantly higher than the control. An overexpression of BnGPAT9-C09 significantly increased seed oil content by 0.88–1.04%, but seed oil content in line 1 decreased by 1.86% compared to the control. According to the significance analysis results, an overexpression of BnGPAT9-A01/C01/C09 significantly increased seed oil content, while an overexpression of BnGPAT9-A10 also increased seed oil content but did not reach a significant level. To further analyze the reasons for the increase in seed oil content, we examined the fatty acid composition of seeds from lines with significantly increased oil content. We found that the contents of Linoleic acid (C18:2) and linolenic acid (C18:3) in seeds were significantly increased compared to the control, accompanied by a decrease in oleic acid (C18:1) content (Table 2). This indicates that BnGPAT9 has the function of altering fatty acid composition, promoting the increase in Linoleic acid (C18:2) and linolenic acid (C18:3) content in seeds.
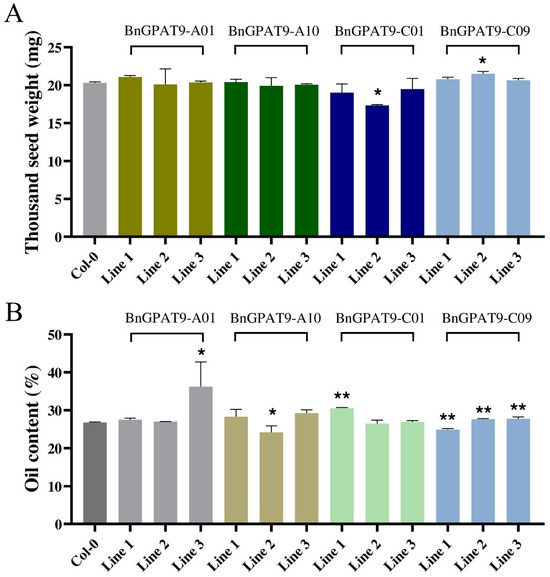
Figure 5.
Analysis of the thousand-seed weight and oil content in BnGPAT9 transgenic Arabidopsis seeds. (A) Thousand-seed weight of seeds from four BnGPAT9 transgenic lines. (B) Oil content of seeds from four BnGPAT9 transgenic lines. “*” indicates highly significant differences, p < 0.05. “**” indicates highly significant differences, p < 0.01.

Table 2.
Arabidopsis thaliana seeds FA composition.
3.6. Transcriptome Sequencing and DEG Analysis of Developing Siliques in BnGPAT9 Transgenic Arabidopsis
To understand the regulatory function of the BnGPAT9 gene in promoting seed oil accumulation, we performed transcriptome sequencing analysis on siliques from transgenic Arabidopsis at 14 days post-fertilization. As shown in Figure 6A, compared to the wild type, the BnGPAT9-A01 transgenic plants had 281 upregulated DEGs and 83 downregulated DEGs. The BnGPAT9-A10 transgenic plants had 747 upregulated DEGs and 271 downregulated DEGs. The BnGPAT9-C01 transgenic plants had 654 upregulated DEGs and 41 downregulated DEGs. The BnGPAT9-C09 transgenic plants had 681 upregulated DEGs and 123 downregulated DEGs. The number of upregulated DEGs in transgenic plants was significantly higher than the number of downregulated DEGs, with the BnGPAT9-C01 transgenic plants having the fewest downregulated DEGs. Among the BnGPAT9 transgenic plants, there were 168 commonly upregulated DEGs and only 7 commonly downregulated DEGs (Figure 6B,C). The heatmap of gene expression levels showed that the gene expression patterns in the four BnGPAT9 transgenic lines were significantly different from those in the wild type (Figure 6D). Compared to the wild type, many genes in the transgenic plants exhibited increased expression levels, with the BnGPAT9-C01 transgenic plants having the highest number of genes with elevated expression levels. This suggests that the BnGPAT9 gene may promote oil content accumulation during seed development by upregulating the expression of related genes.
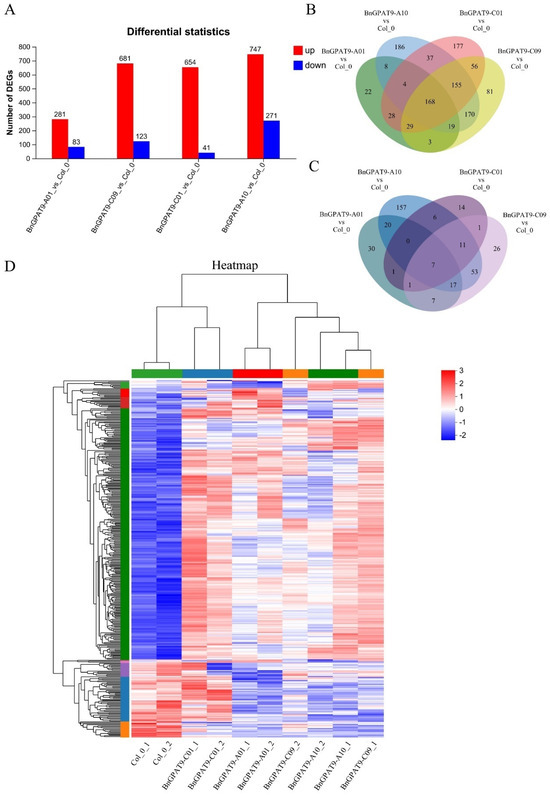
Figure 6.
An analysis of differential gene expression in siliques of BnGPAT9 transgenic Arabidopsis. (A) A statistical analysis of the number of DEGs. The red bars represent upregulated genes, while the blue bars represent downregulated genes. (B) A Venn diagram analysis of upregulated DEGs. This diagram illustrates the common and unique upregulated genes across the different BnGPAT9 transgenic lines. (C) A Venn diagram analysis of downregulated DEGs. This diagram shows the common and unique downregulated genes among the various BnGPAT9 transgenic lines. (D) A clustering heatmap of DEGs. Each column represents a sample, and each row represents a gene. The color in the heatmap indicates the normalized expression level of the gene in each sample, with red representing higher expression levels and blue representing lower expression levels.
3.7. KEGG Enrichment Analysis of Differentially Expressed Genes
We used KEGG enrichment analysis to understand the main metabolic and signal transduction pathways involved in seed oil accumulation for DEGs. We found that DEGs in BnGPAT9-A01 transgenic plants were significantly enriched in the Plant–pathogen interaction, alpha-Linolenic acid metabolism, MAPK signaling pathway—plant, Glutathione metabolism, Glucosinolate biosynthesis, Linoleic acid metabolism, and Glycerophospholipid metabolism pathways (Figure 7A). The DEGs in BnGPAT9-A10 transgenic plants were significantly enriched in the Plant–pathogen interaction, Protein processing in the endoplasmic reticulum, MAPK signaling pathway—plant, Glutathione metabolism, Tryptophan metabolism, and alpha-Linolenic acid metabolism pathways (Figure 7B). The DEGs in BnGPAT9-C01 transgenic plants were significantly enriched in the Plant–pathogen interaction, MAPK signaling pathway—plant, Photosynthesis—antenna proteins, alpha-Linolenic acid metabolism, Glutathione metabolism, and Taurine and hypotaurine metabolism pathways (Figure 7C). The DEGs in BnGPAT9-C09 transgenic plants were significantly enriched in the Plant–pathogen interaction, alpha-Linolenic acid metabolism, Glutathione metabolism, MAPK signaling pathway—plant, and Glucosinolate biosynthesis pathways (Figure 7D). We found that the Plant–pathogen interaction, alpha-Linolenic acid metabolism, MAPK signaling pathway—plant, and Glutathione metabolism are common pathways for the differential genes in the four BnGPAT9 transgenic plants, with alpha-Linolenic acid metabolism being the most directly related pathway to fatty acid metabolism. Importantly, the differential genes enriched in the alpha-Linolenic acid metabolism pathway were all significantly upregulated (Figure 7E). This suggests that the BnGPAT9 gene may influence the expression of genes related to linolenic acid synthesis during the process of promoting oil accumulation, thereby affecting the fatty acid composition and promoting linolenic acid synthesis.
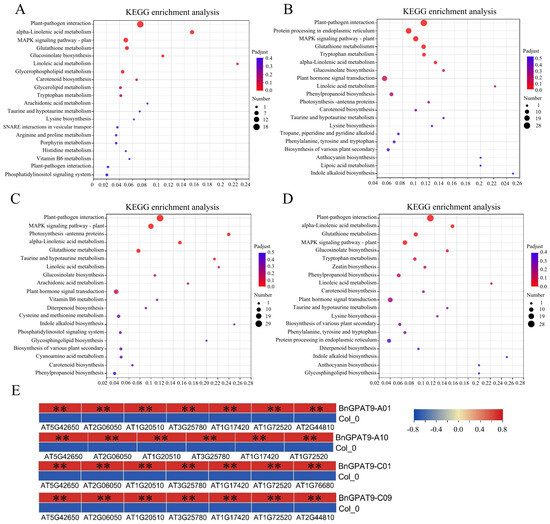
Figure 7.
KEGG enrichment analysis of DEGs in four BnGPAT9 transgenic Arabidopsis thaliana. (A) KEGG enrichment analysis of DEGs between BnGPAT9-A01 transgenic Arabidopsis thaliana and wild type. (B) KEGG enrichment analysis of DEGs between BnGPAT9-A10 transgenic Arabidopsis thaliana and wild type. (C) KEGG enrichment analysis of DEGs between BnGPAT9-C01 transgenic Arabidopsis thaliana and wild type. (D) KEGG enrichment analysis of DEGs between BnGPAT9-C09 transgenic Arabidopsis thaliana and wild type. (E) Heatmap of expression of DEGs enriched in alpha-Linolenic acid metabolism pathway. “**” indicates highly significant differences, p < 0.01.
4. Discussion
Glycerol-3-phosphate acyltransferase (GPAT) catalyzes the first acylation step in the synthesis of nearly all glycerolipids. Specifically, GPAT catalyzes the transfer of a fatty acid from an acyl donor (acyl-CoA or acyl carrier protein) to the hydroxyl group of glycerol-3-phosphate (sn-1 or sn-2) to form lysophosphatidic acid. This reaction is considered a key rate-limiting step, linking the pathways of fatty acid and glycerolipid synthesis [29]. In this study, we obtained four BnGPAT9 genes from the B. napus cultivar XY15. BnGPAT9-A01/C01 encode 376 amino acids, while BnGPAT9-A10/C09 encode 371 amino acids. The sequences of the four BnGPAT9 proteins include 10 motifs, which are consistent with the motif arrangement of the Arabidopsis thaliana AtGPAT9 protein. This indicates that BnGPAT9 in B. napus possesses the essential structural elements of the GPAT family proteins, conferring the catalytic function of plant glycerol-3-phosphate acyltransferase, and thereby participating in the regulation of oil accumulation and other processes in B. napus.
Ten GPAT genes have been identified in Arabidopsis thaliana. Among them, AtGPAT1-8 are sn-2 position acyltransferase genes specific to terrestrial plants, mainly involved in the synthesis of polar glycerolipids such as cutin and suberin, with no direct evidence linking them to TAG synthesis [30,31]. The GPAT9 gene has been the most extensively studied in Arabidopsis thaliana, confirming that this gene is a functional member of the GPAT gene family involved in TAG synthesis, regulating plant TAG biosynthesis and thereby affecting oil content [8,16]. B. napus has four homologous copies of the BnGPAT9 gene, located on chromosomes 1 and 10 of the A genome and chromosomes 1 and 9 of the C genome, respectively. Researchers have cloned the four BnGPAT9 genes from ZS11 and Westar, confirming that BnGPAT9 regulates oil accumulation in rapeseed seeds. They found that among the four homologous copies of the BnGPAT9 gene, only the homologs from BnGPAT9-A01/C01 encode functional GPAT enzymes, while the homologs from BnGPAT9-A10/C09 do not encode functional enzymes [19]. Interestingly, the BnGPAT9 genes exhibit different expression patterns in ZS11 and Westar. In ZS11, BnGPAT9-A01/C01 are highly expressed in all tissues, including roots, stems, leaves, flowers, and developing seeds [19]. In contrast, in Westar, only BnGPAT9-C01 is highly expressed in roots, stems, leaves, flowers, and developing seeds, while the other three homologous copies are almost not expressed in roots, stems, leaves, flowers, and early developing seeds [20]. Differential expression of homologous genes is widely observed in allopolyploids, where homologous genes often exhibit functional divergence after polyploidization, particularly in terms of gene expression [32,33,34]. Promoter GUS staining also confirmed that BnGPAT9-A01/C01 are highly expressed in stems, leaves, flowers, and siliques, consistent with the expression patterns observed in qRT-PCR analysis. This differential expression pattern of BnGPAT9 homologous genes may reflect the adaptability of different rapeseed cultivars during domestication.
Not only do the expression patterns of the genes differ, but the functions of the BnGPAT9-C01 gene in ZS11 and Westar also have distinct differences. Although the overexpression of BnGPAT9-C01 can promote oil accumulation in seeds in both cases, the overexpression of BnGPAT9-C01 from ZS11 leads to an increase in C18:2 fatty acids, whereas the overexpression of BnGPAT9-C01 from Westar results in an increase in erucic acid [19,20]. We found that the expression pattern of the BnGPAT9 gene in XY15 is more similar to that in ZS11 and significantly different from that in Westar. BnGPAT9-A01 can increase the oil content in seeds by up to 9.51%, and BnGPAT9-C01 can increase it by up to 3.81%. Following this, BnGPAT9-A01 (increasing by 2.54%) and BnGPAT9-C01 (increasing by 1.04%) also contribute to the increase in seed oil content. This indicates that all four BnGPAT9 genes can regulate oil accumulation in seeds, but BnGPAT9-A01/C01 contribute more significantly to promoting oil accumulation. Notably, BnGPAT9 from XY15 has the function of reducing oleic acid content and promoting the accumulation of Linoleic and linolenic acids. This functional difference in the gene may be closely related to variations in gene sequences and protein structures. Through sequence alignment analysis of the four BnGPAT9 genes in various B. napus varieties such as Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15, we found that the CDS sequence of BnGPAT9-C09 is completely identical across different varieties, showing a high degree of conservation. Although the CDS sequence of BnGPAT9-A01 varies among different varieties, the protein sequence remains completely consistent, also reflecting high conservation. The protein sequence of BnGPAT9-C01 shows changes at specific amino acid sites, which may cause functional differences in promoting oil synthesis. However, both the CDS and protein sequences of BnGPAT9-A10 exhibit significant variations among different varieties. From a crop improvement perspective, identifying homologous genes that are potentially functionally conserved is crucial for elucidating the genetic basis of targeted trait design [35]. The conservation of the homologous genes BnGPAT9-A01/C01/C09, along with the significant structural and sequence differences in the BnGPAT9-A10 homologous gene, may indicate major gene variation events and strong selective pressures during the coordinated evolution of the subgenomes in allopolyploid B. napus, which is of great significance for the process of allopolyploidization in rapeseed.
The GPAT9 gene is a positive regulator of oil accumulation in seeds, with its function validated in Arabidopsis thaliana [8], rapeseed [19,20], peanut [36], and sunflower [17]. These studies primarily focus on the correlation analysis between gene expression levels and oil content, as well as functional validation at the cellular level, while the regulation at the transcriptional level remains unclear. We found that the four BnGPAT9 genes in rapeseed mainly promote oil accumulation by upregulating gene expression, particularly BnGPAT9-A01/C01. These differentially upregulated genes are significantly enriched in pathways such as the Plant–pathogen interaction, alpha-Linolenic acid metabolism, MAPK signaling pathway—plant, Glutathione metabolism, Glucosinolate biosynthesis, Linoleic acid metabolism, Glycerophospholipid metabolism, Protein processing in endoplasmic reticulum, Tryptophan metabolism, Photosynthesis—antenna proteins, and Taurine and hypotaurine metabolism. Importantly, alpha-Linolenic acid metabolism is a commonly significantly enriched pathway in the four BnGPAT9 transgenic plants, and the significant increase in linolenic acid content in seeds may be related to the upregulation of genes in the alpha-Linolenic acid metabolism pathway. Alpha-Linolenic acid (ALA, C18:3) is an important omega-3 polyunsaturated fatty acid and also a precursor for the synthesis of eicosapentaenoic acid (EPA, C20:5) and docosahexaenoic acid (DHA, C22:6) [37]. DHA and EPA are essential lipid nutrients for the human body, and when supplementation of DHA and EPA is needed, it can also be achieved through the intake of alpha-Linolenic acid [38].
5. Conclusions
In this study, we cloned four BnGPAT9 genes from the B. napus variety XY15. By comparing these genes and their proteins with those from other rapeseed varieties such as Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, and ZS11, we found that the homologous genes BnGPAT9-A01/C01/C09 are relatively conserved. Different rapeseed varieties exhibited significant differences in gene expression patterns. BnGPAT9-A01/C01 showed higher expression levels across various tissues in rapeseed, especially during the late stages of seed development, playing a major role in seed oil accumulation. We confirmed that overexpression of the rapeseed BnGPAT9 genes can promote oil accumulation in seeds by upregulating the expression of many differentially expressed genes, particularly those in the alpha-Linolenic acid metabolism pathway, which promotes an increase in linolenic acid content. This has significant implications for improving the fatty acid composition of rapeseed oil. Therefore, our work provides an important reference value for enriching the functional research of BnGPAT9 genes in rapeseed and for enhancing the yield and quality of rapeseed oil.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agriculture14081334/s1, Figure S1: Sequence alignment of the BnGPAT9-A01 gene in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S2: Sequence alignment of the BnGPAT9-A10 gene in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S3: Sequence alignment of the BnGPAT9-C01 gene in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S4: Sequence alignment of the BnGPAT9-C09 gene in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S5: Amino acid sequence alignment of the BnGPAT9-A01 in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S6: Amino acid sequence alignment of the BnGPAT9-A10 in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S7: Amino acid sequence alignment of the BnGPAT9-C01 in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15. Figure S8: Amino acid sequence alignment of the BnGPAT9-C09 in Brassica napus varieties Gangan, No2127, Quinta, Shengli, Tapidor, Westar, Zheyou, ZS11, and XY15.
Author Contributions
M.X., B.H. and L.H. designed the experiment. M.X., Z.P. and M.L. conducted the experiment. M.L., D.Z., X.L., S.Q., C.S. and B.H. helped and provided useful suggestions during the experiment. M.X., M.L. and L.H. processed and analyzed data and wrote the first draft. C.G., X.X., L.H. and M.X. revised and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the High-Tech Research and Development Program 863 (2011AA10A104) and a special fund for guiding the city and county science and technology development of Jiangxi Province.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Graham, I.A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef]
- Cui, S.; Hayashi, Y.; Otomo, M.; Mano, S.; Oikawa, K.; Hayashi, M.; Nishimura, M. Sucrose Production Mediated by Lipid Metabolism Suppresses the Physical Interaction of Peroxisomes and Oil Bodies during Germination of Arabidopsis thaliana. J. Biol. Chem. 2016, 291, 19734–19745. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 Acyltransferases have overlapping functions in Arabidopsis Triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Morimoto, R.; Tabeta, H.; Asaoka, M.; Ishida, M.; Maeshima, M.; Tsukaya, H.; Ferjani, A. Compensated Cell Enlargement in fugu5 is Specifically Triggered by Lowered Sucrose Production from Seed Storage Lipids. Plant Cell Physiol. 2017, 58, 668–678. [Google Scholar] [CrossRef]
- Yang, Y.; Benning, C. Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 2018, 49, 191–198. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA: Glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef] [PubMed]
- Gidda, S.K.; Shockey, J.M.; Rothstein, S.J.; Dyer, J.M.; Mullen, R.T. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 2009, 47, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.; Regmi, A.; Cotton, K.; Adhikari, N.; Browse, J.; Bates, P.D. Identification of Arabidopsis GPAT9 (AT5G60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 2015, 170, 163–179. [Google Scholar] [CrossRef]
- Lewin, T.M.; Wang, P.; Coleman, R.A. Analysis of amino acid motifs diagnostic for the SN-Glycerol-3-phosphate acyltransferase reaction. Biochemistry 1999, 38, 5764–5771. [Google Scholar] [CrossRef]
- Weselake, R.J.; Shah, S.; Tang, M.; Quant, P.A.; Snyder, C.L.; Furukawa-Stoffer, T.L.; Zhu, W.; Taylor, D.C.; Zou, J.; Kumar, A.; et al. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. J. Exp. Bot. 2008, 59, 3543–3549. [Google Scholar] [CrossRef]
- Li, R.; Yu, K.; Hildebrand, D.F. DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 2010, 45, 145–157. [Google Scholar] [CrossRef]
- Weiss, S.B.; Kennedy, E.P.; Kiyasu, J.Y. The enzymatic synthesis of triglycerides. J. Biol. Chem. 1960, 235, 40–44. [Google Scholar] [CrossRef]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef]
- Banaś, A.; Dahlqvist, A.; Ståhl, U.; Lenman, M.; Stymne, S. The involvement of phospholipid: Diacylglycerol acyltransferases in triacylglycerol production. Biochem. Soc. Trans. 2000, 28, 703–705. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Siloto, R.M.P.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a Pair of Phospholipid: Diacylglycerol Acyltransferases from Developing Flax (Linum usitatissimum L.) Seed Catalyzing the Selective Production of Trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Singer, S.D.; Chen, G.; Mietkiewska, E.; Tomasi, P.; Jayawardhane, K.; Dyer, J.M.; Weselake, R.J. Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 2016, 67, 4627–4638. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Aznar-Moreno, J.A.; Balbuena, T.S.; Haslam, R.P.; Gidda, S.K.; Pérez-Hormaeche, J.; Mullen, R.T.; Thelen, J.J.; Napier, J.A.; Salas, J.J.; et al. Sunflower HaGPAT9-1 is the predominant GPAT during seed development. Plant Sci. 2016, 252, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.U.; Kim, J.; Kim, H.; Suh, M.C. Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression. Plants 2019, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, J.; Zhang, B.; Li, Q.; Liu, C.; Huang, Q.; Cui, P. The functional divergence of homologous GPAT9 genes contributes to the erucic acid content of Brassica napus seeds. BMC Plant Biol. 2024, 24, 69. [Google Scholar] [CrossRef]
- Gong, W.; Chen, W.; Gao, Q.; Qian, L.; Yuan, X.; Tang, S.; Hong, Y. Glycerol-3-Phosphate Acyltransferase GPAT9 Enhanced Seed Oil Accumulation and Eukaryotic Galactolipid Synthesis in Brassica napus. Int. J. Mol. Sci. 2023, 24, 16111. [Google Scholar] [CrossRef]
- Xing, M.; Liu, S.F.; Wu, X.M.; Guan, C.Y.; Xiong, X.H. Cloning and Bioinformatics Analysis of GPAT9 Gene in Rapeseed. Mol. Plant Breed. 2016, 14, 3282–3288. [Google Scholar] [CrossRef]
- Xing, M.; Zhou, X.Q.; He, T.; Zhang, Z.R.; Yue, N.Y.; Zeng, X.G.; Wu, X.M.; Guan, C.Y.; Xiong, X.H. Expression pattern of BnGPAT9 gene in Brassica napus and its expression under abiotic stresses. Chin. J. Oil Crop Sci. 2017, 39, 454–461. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Dai, C.; Li, Y.; Li, L.; Du, Z.; Lin, S.; Tian, X.; Li, S.; Yang, B.; Yao, W.; Wang, J.; et al. An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol. Breed. 2020, 40, 96. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Dubreucq, B.; Miquel, M.; Rochat, C.; Lepiniec, L. Storage reserve accumulation in Arabidopsis: Metabolic and developmental control of seed filling. Arab. Book 2008, 6, e0113. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Lepiniec, L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 2009, 47, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Chen, X.; Snyder, C.L.; Truksa, M.; Shah, S.; Weselake, R.J. sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal. Behav. 2011, 6, 1695–1699. [Google Scholar] [CrossRef]
- Yang, W.; Simpson, J.P.; Li-Beisson, Y.; Beisson, F.; Pollard, M.R.; Ohlrogge, J.B. A Land-Plant-Specific Glycerol-3-Phosphate Acyltransferase Family in Arabidopsis: Substrate Specificity, sn-2 Preference, and Evolution. Plant Physiol. 2012, 160, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- VanBuren, R.; Wai, C.M.; Wang, X.; Pardo, J.; Yocca, A.E.; Wang, H.; Chaluvadi, S.R.; Han, G.; Bryant, D.; Edger, P.P.; et al. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Peng, P.F.; Li, Y.C.; Mei, D.S.; Colasanti, J.; Fu, L.; Liu, J.; Chen, Y.F.; Hu, Q. Expression divergence of FRUITFULL homeologs enhanced pod shatter resistance in Brassica napus. Genet. Mol. Res. 2015, 14, 871–885. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, X.; Luo, L.; Yang, H.; Li, P.; Zhang, K.; Liu, F.; Wan, Y. Characterization of glycerol-3-phosphate acyltransferase 9 (AhGPAT9) genes, their allelic polymorphism and association with oil content in peanut (Arachis hypogaea L.). Sci. Rep. 2020, 10, 14648. [Google Scholar] [CrossRef] [PubMed]
- Ouagueni, A.; Al-Zoubi, R.M.; Zarour, A.; Al-Ansari, A.; Bawadi, H. Effects of omega-3 polyunsaturated fatty acids, docosahexaenoic acid and eicosapentaenoic acid, on Post-Surgical Complications in Surgical Trauma Patients: Mechanisms, nutrition, and challenges. Mar. Drugs 2024, 22, 207. [Google Scholar] [CrossRef] [PubMed]
- Block, R.C.; Duff, R.; Lawrence, P.; Kakinami, L.; Brenna, J.T.; Shearer, G.C.; Meednu, N.; Mousa, S.; Friedman, A.; Harris, W.S.; et al. The effects of EPA, DHA, and aspirin ingestion on plasma lysophospholipids and autotaxin. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 87–95. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).