Molecular Characteristics and Antimicrobial Resistance of Salmonella enterica Serovar Schwarzengrund from Chicken Meat in Japan
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Susceptibility
2.2. Antimicrobial Resistance Genes
2.3. Class 1 Integron Detection and Characterization
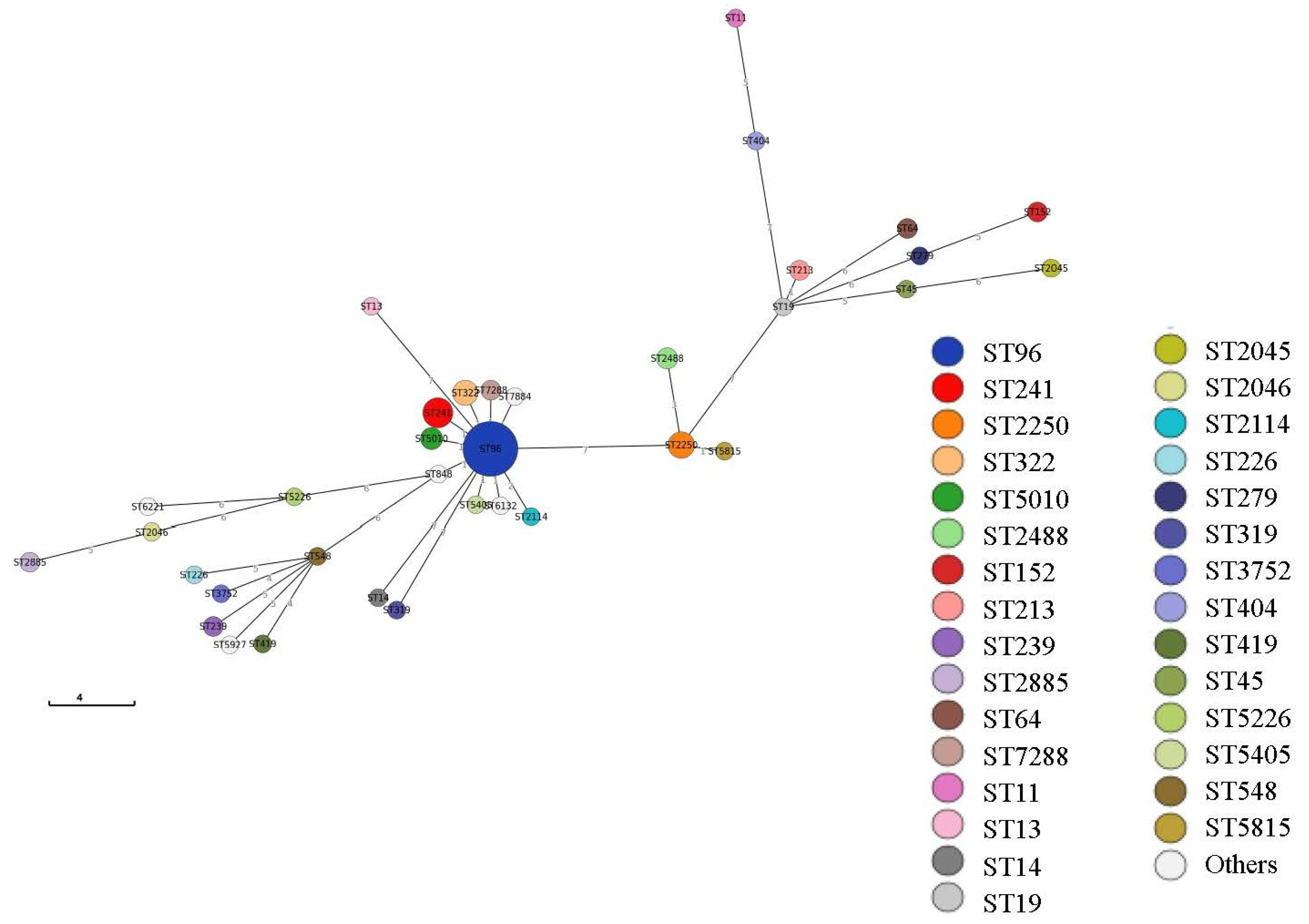
2.4. Multilocus Sequence Type (MLST) Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Antimicrobial Susceptibility Test
4.3. Antimicrobial Resistance Gene Detection
4.4. Integron Analysis
4.5. MLST Analysis
4.6. WGS
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanchez, S.; Hofacre, C.L.; Lee, M.D.; Maurer, J.J.; Doyle, M.R. Animal sources of salmonellosis in humans. J. Am. Vet. Med. Assoc. 2002, 221, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Food Safety Authority. The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J. 2007, 130, 1–352. [Google Scholar]
- Pires, S.M.; Vieira, A.R.; Hald, T.; Cole, D. Source attribution of human salmonellosis: An overview of methods and estimates. Foodborne Pathog. Dis. 2014, 11, 667–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, P.; Mourao, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Infect. Genet. Evol. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furukawa, I.; Ishihara, T.; Teranishi, H.; Saito, S.; Yatsuyanagi, J.; Wada, E.; Kumagai, Y.; Takahashi, S.; Konno, T.; Kashio, H.; et al. Prevalence and characteristics of Salmonella and Campylobacter in retail poultry meat in Japan. Jpn. J. Infect. Dis. 2017, 70, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Shigemura, H.; Matsui, M.; Sekizuka, T.; Onozuka, D.; Noda, T.; Yamashita, A.; Kuroda, M.; Suzuki, S.; Kimura, H.; Fujimoto, S.; et al. Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int. J. Food Microbiol. 2018, 274, 45–51. [Google Scholar] [CrossRef]
- Japanese Ministry of Agriculture, Forestry and Fisheries. Food Supply and Demand Table. Available online: https://www.maff.go.jp/j/zyukyu/fbs/ (accessed on 8 October 2020).
- Bangtrakulnonth, A.; Pornreongwong, S.; Pulsrikarn, C.; Sawanpanyalert, P.; Hendriksen, R.S.; Wong, D.; Aarestrup, F.M. Salmonella serovars from humans and other sources in Thailand, 1993–2002. Emerg. Infect. Dis. 2004, 10, 131–136. [Google Scholar] [CrossRef]
- Chen, M.H.; Wang, S.W.; Hwang, W.Z.; Tsai, S.J.; Hsih, Y.C.; Chiou, C.S.; Tsen, H.Y. Contamination of Salmonella Schwarzengrund cells in chicken meat from traditional marketplaces in Taiwan and comparison of their antibiograms with those of the human isolates. Poult. Sci. 2010, 89, 359–365. [Google Scholar] [CrossRef]
- Tejada, T.S.; Silva, C.S.J.; Lopes, N.A.; Silva, D.T.; Agostinetto, A.; Silva, E.F.; Menezes, D.B.; Timm, C.D. DNA profiles of Salmonella spp. isolated from chicken products and from broiler and human feces. Braz. J. Poult. Sci. 2016, 18, 693–699. [Google Scholar] [CrossRef]
- Mechesso, A.F.; Moon, D.C.; Kim, S.J.; Song, H.J.; Kang, H.Y.; Na, S.H.; Choi, J.H.; Kim, H.Y.; Yoon, S.S.; Lim, S.K. Nationwide surveillance on serotype distribution and antimicrobial resistance profiles of non-typhoidal Salmonella serovars isolated from food-producing animals in South Korea. Int. J. Food Microbiol. 2020, 335, 108893. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Hendriksen, R.S.; Lockett, J.; Gay, K.; Teates, K.; McDermott, P.F.; White, D.G.; Hasman, H.; Sorensen, G.; Bangtrakulnonth, A.; et al. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg. Infect. Dis. 2007, 13, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Lauderdale, T.L.; Lo, D.Y.; Chen, C.L.; Chen, P.C.; Liang, S.Y.; Kuo, J.C.; Liao, Y.S.; Liao, C.H.; Tsao, C.S.; et al. An association of genotypes and antimicrobial resistance patterns among Salmonella isolates from pigs and humans in Taiwan. PLoS ONE 2014, 9, e95772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihara, K.; Takahashi, T.; Morioka, A.; Kojima, A.; Kijima, M.; Asai, T.; Tamura, Y. National surveillance of Salmonella enterica in food-producing animals in Japan. Acta Vet. Scand. 2009, 51, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, T.; Esaki, H.; Kojima, A.; Ishihara, K.; Tamura, Y.; Takahashi, T. Antimicrobial resistance in Salmonella isolates from apparently healthy food-producing animal from 2000 to 2003: The first stage of Japanese Veterinary Antimicrobial Resistance Monitoring (JVARM). J. Vet. Med. Sci. 2006, 68, 881–884. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Murakami, K.; Ozawa, M.; Koike, R.; Ishikawa, H. Relationships between multidrug-resistant Salmonella enterica serovar Schwarzengrund and both broiler chickens and retail chicken meats in Japan. Jpn. J. Infect. Dis. 2009, 62, 198–200. [Google Scholar] [PubMed]
- Duc, V.M.; Nakamoto, Y.; Fujiwara, A.; Toyofuku, H.; Obi, T.; Chuma, T. Prevalence of Salmonella in broiler chickens in Kagoshima, Japan in 2009 to 2012 and the relationship between serovars changing and antimicrobial resistance. BMC Vet. Res. 2019, 15, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duc, V.M.; Shin, J.; Nagamatsu, Y.; Fuhiwara, A.; Toyofuku, H.; Obi, T.; Chuma, T. Increased Salmonella Schwarzengrund prevalence and antimicrobial susceptibility of Salmonella enterica isolated from broiler chickens in Kagoshima prefecture in Japan between 2013 and 2016. J. Vet. Med. Sci. 2020, 82, 585–589. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, K.; Nakazawa, C.; Nomura, S.; Elahi, S.; Yamashita, M.; Fujikawa, H. Effects of climatic elements on Salmonella contamination in broiler chicken meat in Japan. J. Vet. Med. Sci. 2020, 82, 646–652. [Google Scholar] [CrossRef] [Green Version]
- National Institute of Infectious Diseases. Infectious Agents Surveillance Report. 2012. Available online: https://www.niid.go.jp/niid/ja/typhi-m/iasr-reference/230-iasr-data/3037-iasr-table-b-pm.html (accessed on 9 March 2021).
- National Institute of Infectious Diseases. Infectious Agents Surveillance Report. 2021. Available online: https://www.niid.go.jp/niid/ja/typhi-m/iasr-reference/510-graphs/1524-iasrgb.html (accessed on 9 March 2021).
- Iwabuchi, E.; Yamamoto, S.; Endo, Y.; Ochiai, T.; Hirai, K. Prevalence of Salmonella isolates and antimicrobial resistance patterns in chicken meat throughout Japan. J. Food Prot. 2011, 74, 270–273. [Google Scholar] [CrossRef]
- Salmonella Genome Databases. Available online: https://pubmlst.org/salmonella/ (accessed on 12 May 2020).
- EnteroBase. Available online: https://enterobase.warwick.ac.uk/ (accessed on 12 May 2020).
- Kidgell, C.; Reichard, U.; Wain, J.; Linz, B.; Torpdahl, M.; Dougan, G.; Achtman, M. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2002, 2, 39–45. [Google Scholar] [CrossRef]
- Bell, R.L.; Gonzalez-Escalona, N.; Stones, R.; Brown, E.W. Phylogenetic evaluation of the ‘Typhimurium’ complex of Salmonella strains using a seven-gene multi-locus sequence analysis. Infect. Genet. Evol. 2011, 11, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Achtman, M.; Hale, J.; Murphy, R.A.; Boyd, E.F.; Porwollik, S. Population structures in the SARA and SARB reference collections of Salmonella enterica according to MLST, MLEE and microarray hybridization. Infect. Genet. Evol. 2013, 16, 314–325. [Google Scholar] [CrossRef] [Green Version]
- Krauland, M.G.; Marsh, J.W.; Paterson, D.L.; Harrison, L.H. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg. Infect. Dis. 2009, 15, 388–396. [Google Scholar] [CrossRef]
- Yamazaki, W.; Uemura, R.; Sekiguchi, S.; Dong, J.B.; Watanabe, S.; Kirino, Y.; Mekata, H.; Nonaka, N.; Norimine, J.; Sueyoshi, M.; et al. Campylobacter and Salmonella are prevalent in broiler farms in Kyushu, Japan: Results of a 2-year distribution and circulation dynamics audit. J. Appl. Microbiol. 2016, 120, 1711–1722. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ikeda, A.; Ishikawa, K.; Murakami, M.; Kusukawa, M.; Asai, T.; Yamada, Y. Prevalence and antimicrobial susceptibility of Salmonella in Japanese broiler flocks. Epidemiol. Infect. 2012, 140, 2074–2081. [Google Scholar] [CrossRef] [Green Version]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.C.; Chang, Y.C.; Chuang, H.L.; Chiu, C.C.; Yeh, K.S.; Chang, C.C.; Hsuan, S.L.; Chen, T.H. Antibiotic resistance, integrons and Salmonella genomic island 1 among Salmonella Schwarzengrund in broiler chicken and pig. Afr. J. Microbiol. Res. 2010, 4, 677–681. [Google Scholar]
- Khemtong, S.; Chuanchuen, R. Class 1 integrons and Salmonella genomic island 1 among Salmonella enterica isolated from poultry and swine. Microb. Drug Resis. 2008, 14, 65–70. [Google Scholar] [CrossRef]
- Miko, A.; Pries, K.; Schroeter, A.; Helmuth, R. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 2005, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, T.; Kojima, A.; Harada, K.; Ishihara, K.; Takahashi, T.; Tamura, Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn. J. Infect. Dis. 2005, 58, 369–372. [Google Scholar]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Veterinary Assay Laboratory. Monitoring of AMR. Available online: https://www.maff.go.jp/nval/yakuzai/yakuzai_p3.html (accessed on 26 August 2020).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; M100-29; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, L.K.; Mulvey, M.R.; Martin, I.; Peters, G.A.; Johnson, W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar typhimurium DT104. Antimicrob. Agents Chemother. 1999, 43, 3018–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepan, R.M.; Sherwood, J.S.; Petermann, S.R.; Logue, C.M. Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE, MLST and NARMS. BMC Microbiol. 2011, 11, 153. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carrico, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Sekizuka, T.; Yatsu, K.; Inamine, Y.; Segawa, T.; Nishio, M.; Kishi, N.; Kuroda, M. Complete genome sequence of a blaKPC-2-positive Klebsiella pneumonias strain isolated from the effluent of an urban sewage treatment plant in Japan. mSphere 2018, 19, e00314-18. [Google Scholar]
- ResFinder. Available online: https://cge.cbs.dtu.dk/services/ResFinder/ (accessed on 29 October 2021).
| Antimicrobial Agents | Breakpoint | No. of Resistant Isolates (%) | |
|---|---|---|---|
| (μg/mL) | 2008 (n = 37) | 2015–2019 (n = 87) | |
| Ampicillin | 32 | 0 | 0 |
| Cefazolin | 8 | 0 | 0 |
| Streptomycin or Dihydrostreptomycin (a) | 16 | 37 (100%) (b) | 41 (47.1%) (b) |
| Gentamicin | 16 | 0 | 0 |
| Kanamycin | 64 | 19 (51.4%) (c) | 78 (89.7%) (c) |
| Tetracycline | 16 | 37 (100%) (d) | 41 (47.1%) (d) |
| Chloramphenicol | 32 | 0 | 0 |
| Nalidixic acid | 32 | 0e) | 8 (9.2%) (e) |
| Ciprofloxacin | 1 | 0 | 0 |
| Trimethoprim | 16 | 21 (56.8%) (f) | 14 (16.1%) (f) |
| Trimethoprim-sulfamethoxazole | 4/76 | 26 (70.3%) (g) | 39 (44.8%) (g) |
| Antimicrobial Resistance Gene | Class 1 Integron (Gene Casettes) | Antimicrobial Resistance | 2008 (a) | 2015–2019 | |||
|---|---|---|---|---|---|---|---|
| West Japan (b) | West Japan | East Japan | Domestic (c) | Sub-Total | |||
| aac(6’)-Iaa-aadA1-tetA-sul1-dfrA14-aphA1 | 1.0 kb (aadA1) | STR-TET-KAN | 1 | 1 | |||
| STR-TET-KAN-NAL-SXT | 2 | 3 | 5 | ||||
| STR-TET-KAN-NAL-TMP-SXT | 1 | 1 | 2 | ||||
| STR-TET-KAN-SXT | 1 | 4 | 9 | 6 | 20 | ||
| STR-TET-KAN-TMP-SXT | 7 | 5 | 1 | 13 | |||
| aac(6’)-Iaa-aadA1-tetA-sul1-dfrA14 | 1.0 kb (aadA1) | STR-TET-SXT | 5 | 1 | 1 | 7 | |
| STR-TET-TMP-SXT | 13 | 13 | |||||
| aac(6’)-Iaa-aadA1-tetA-sul1-aphA1 | 1.0 kb (aadA1) | STR-TET-KAN | 9 | 1 | 3 | 13 | |
| STR-TET-KAN-TMP | 1 | 1 | |||||
| aac(6’)-Iaa-aadA1-tetA-sul1 | 1.0 kb (aadA1) | STR-TET | 2 | 1 | 3 | ||
| aac(6’)-Iaa-strA/strB-sul2-dfrA14-aphA1 | Not determined | KAN-TMP-SXT | 3 | 2 | 5 | ||
| aac(6’)-Iaa-aphA1 | Not determined | KAN | 1 | 24 | 8 | 33 | |
| KAN-NAL | 1 | 1 | |||||
| aac(6’)-Iaa-dfrA14-aphA1 | Not determined | KAN-TMP | 1 | 1 | |||
| KAN | 1 | 1 | 2 | ||||
| aac(6’)-Iaa | Not determined | susceptible | 1 | 2 | 1 | 4 | |
| Total | 37 | 10 | 49 | 28 | 124 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsui, K.; Nakazawa, C.; Thiri Maung Maung Khin, S.; Iwabuchi, E.; Asai, T.; Ishihara, K. Molecular Characteristics and Antimicrobial Resistance of Salmonella enterica Serovar Schwarzengrund from Chicken Meat in Japan. Antibiotics 2021, 10, 1336. https://doi.org/10.3390/antibiotics10111336
Matsui K, Nakazawa C, Thiri Maung Maung Khin S, Iwabuchi E, Asai T, Ishihara K. Molecular Characteristics and Antimicrobial Resistance of Salmonella enterica Serovar Schwarzengrund from Chicken Meat in Japan. Antibiotics. 2021; 10(11):1336. https://doi.org/10.3390/antibiotics10111336
Chicago/Turabian StyleMatsui, Kaoru, Chisato Nakazawa, Shwe Thiri Maung Maung Khin, Eriko Iwabuchi, Tetsuo Asai, and Kanako Ishihara. 2021. "Molecular Characteristics and Antimicrobial Resistance of Salmonella enterica Serovar Schwarzengrund from Chicken Meat in Japan" Antibiotics 10, no. 11: 1336. https://doi.org/10.3390/antibiotics10111336






