Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Activity of the Copolymers
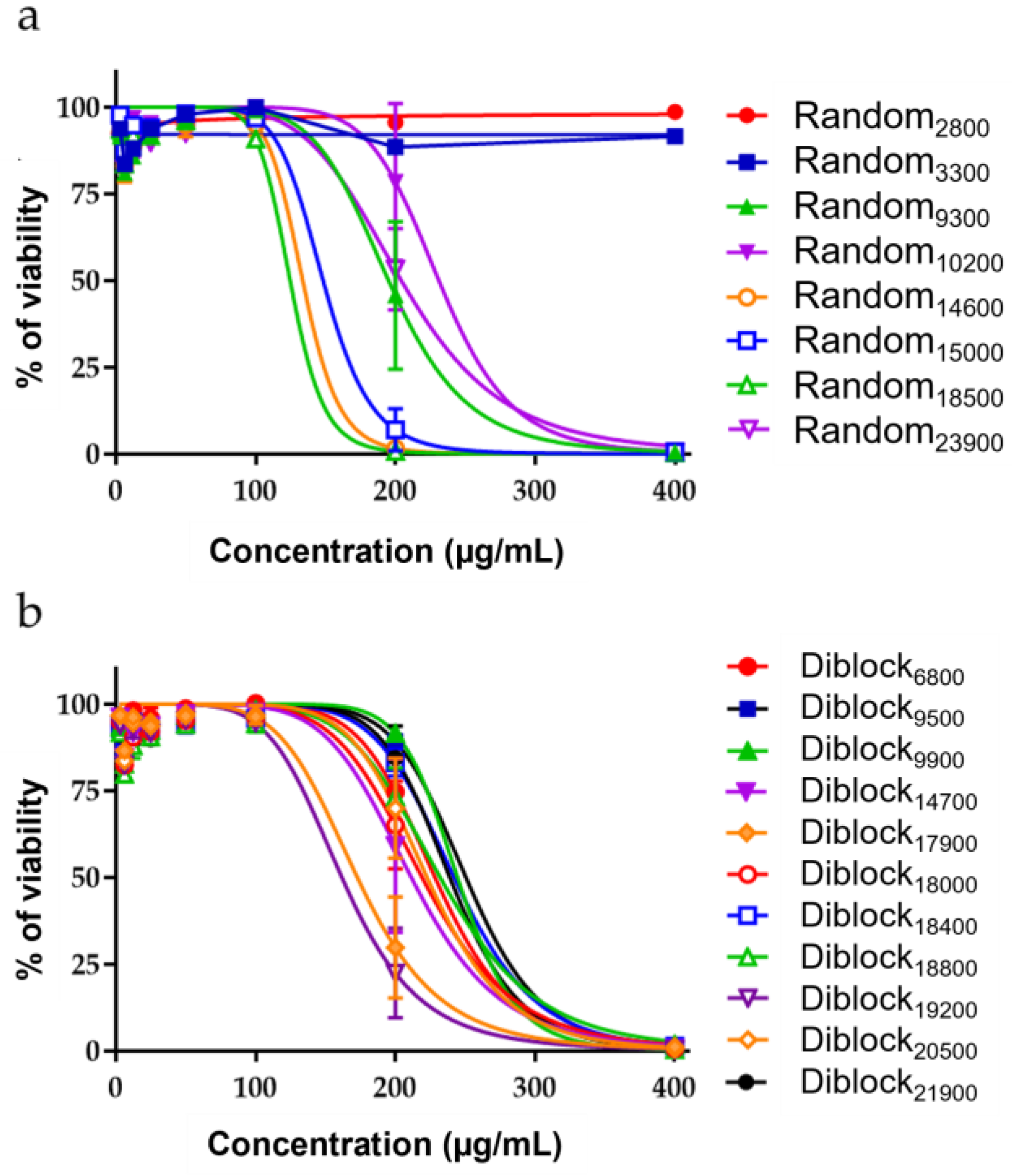
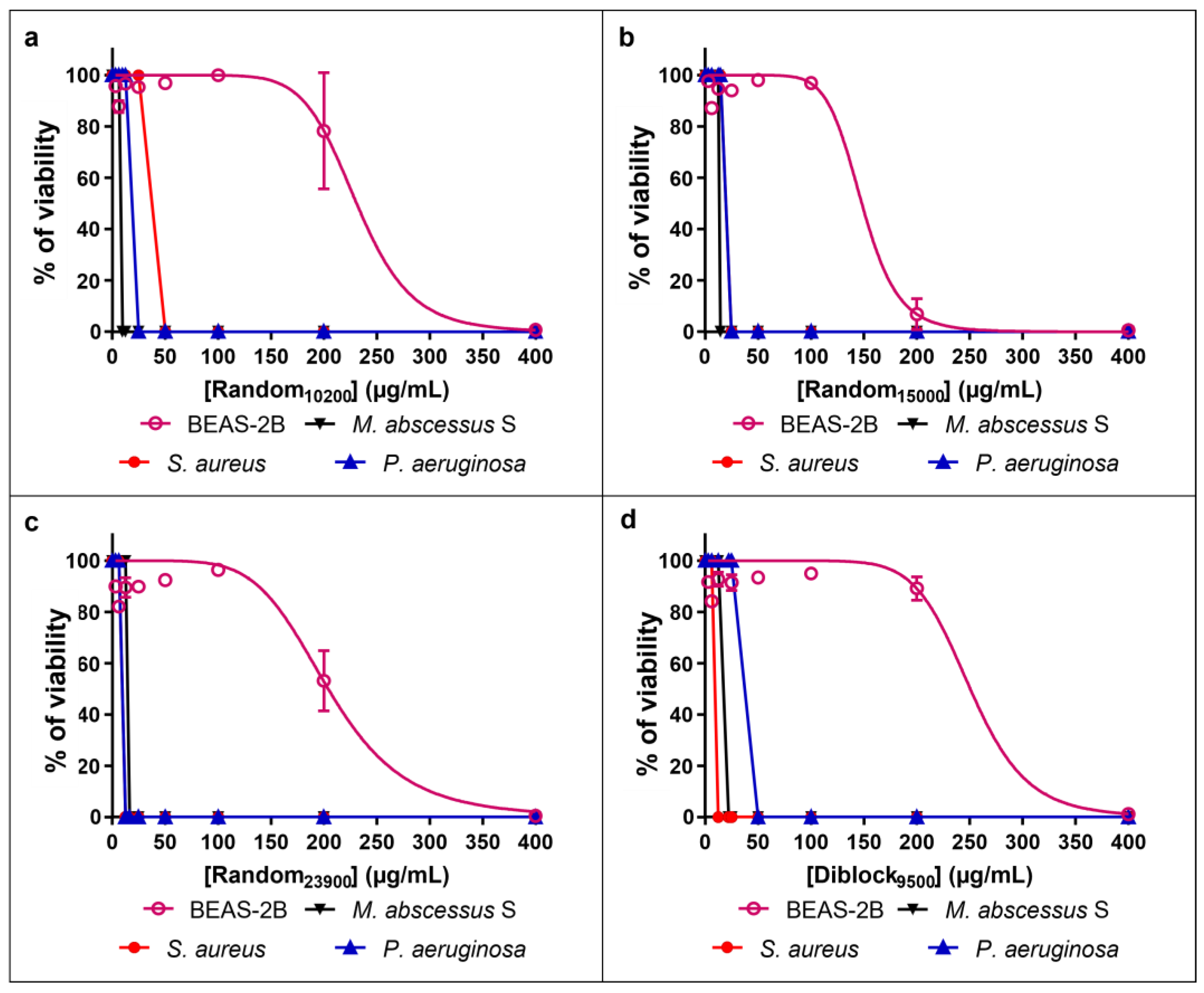
2.2. Innocuity of the Copolymers
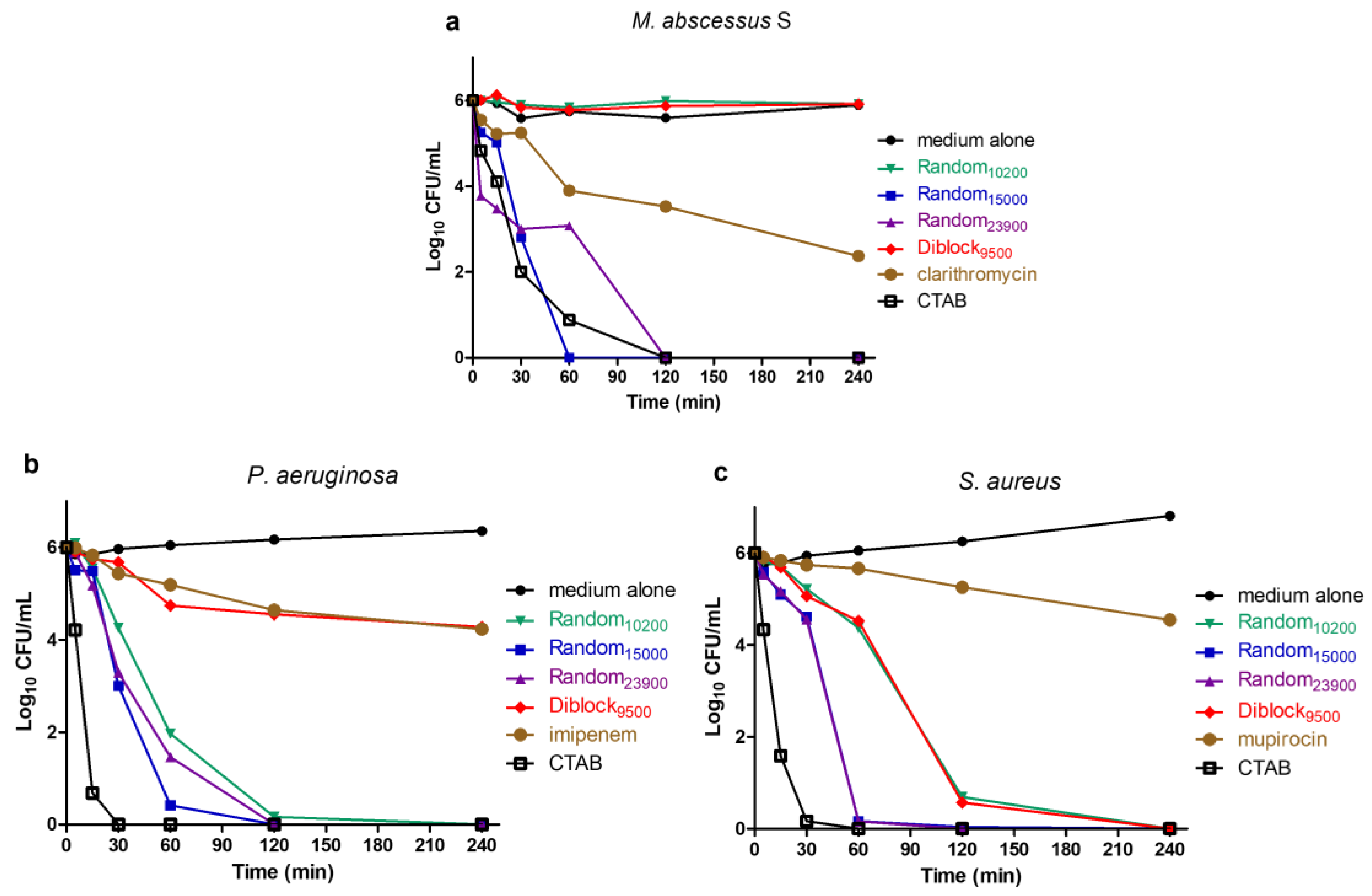
2.3. Determination of Time-Kill Kinetics
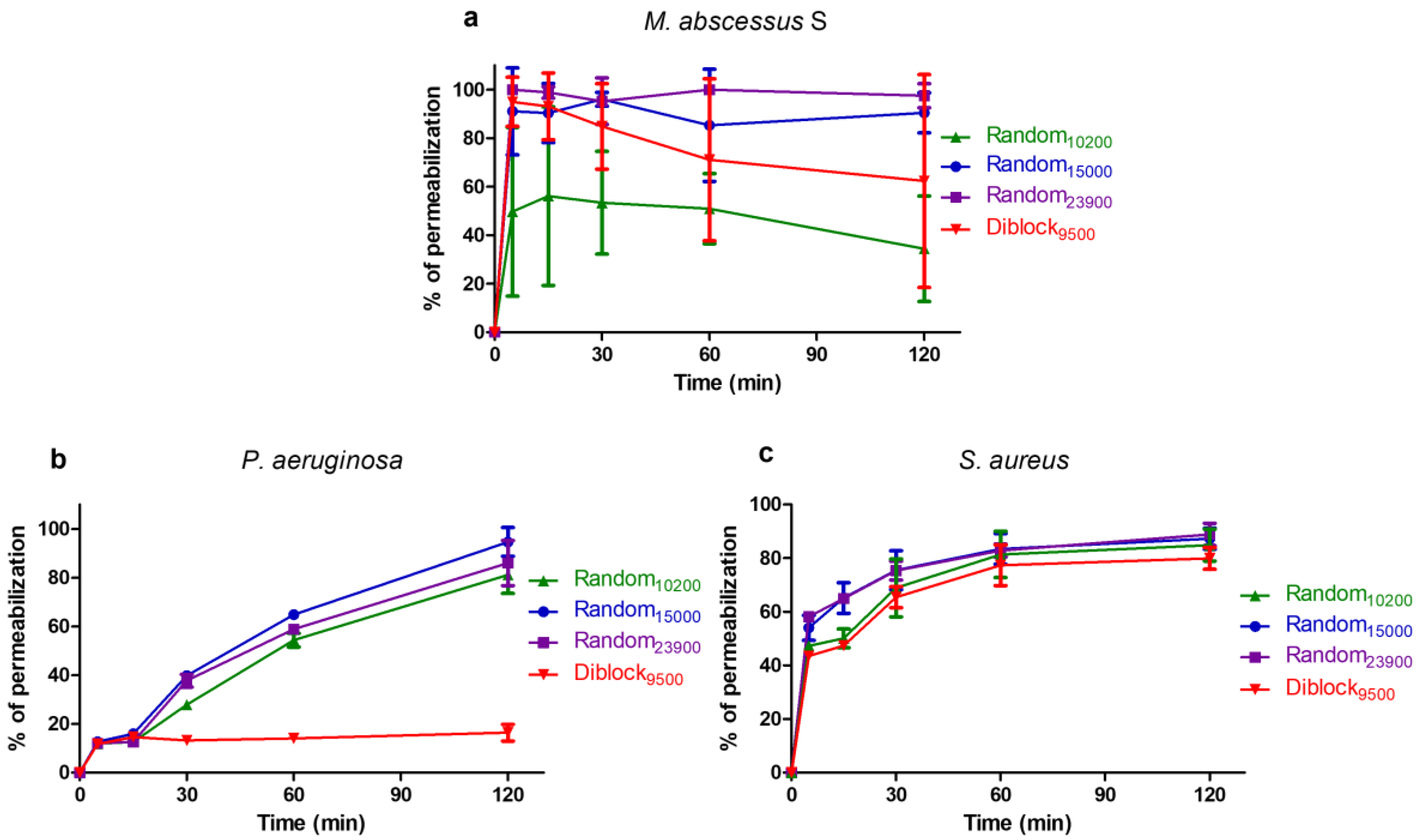
2.4. Mechanism of Action
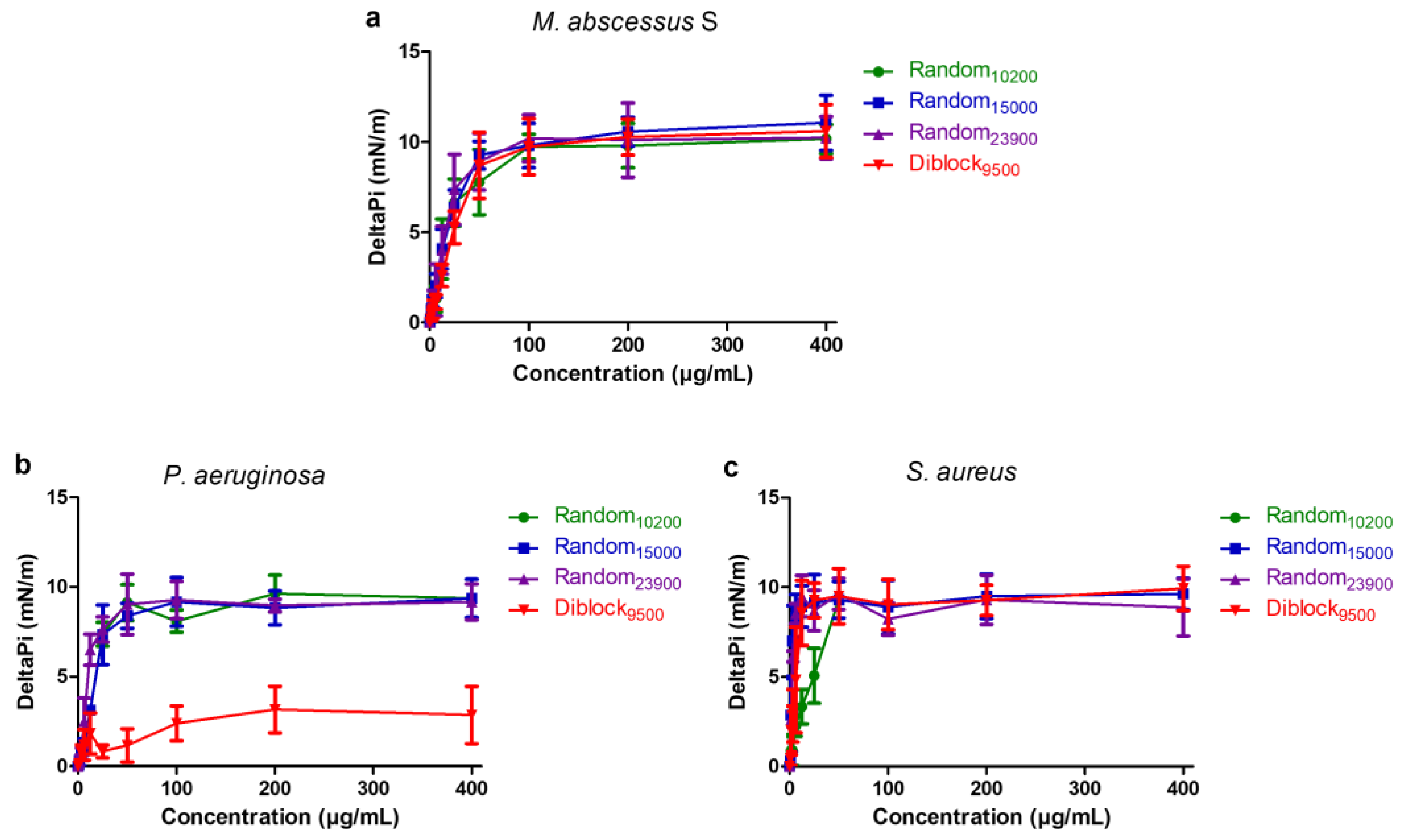
2.5. Insertion in Lipid Monolayers
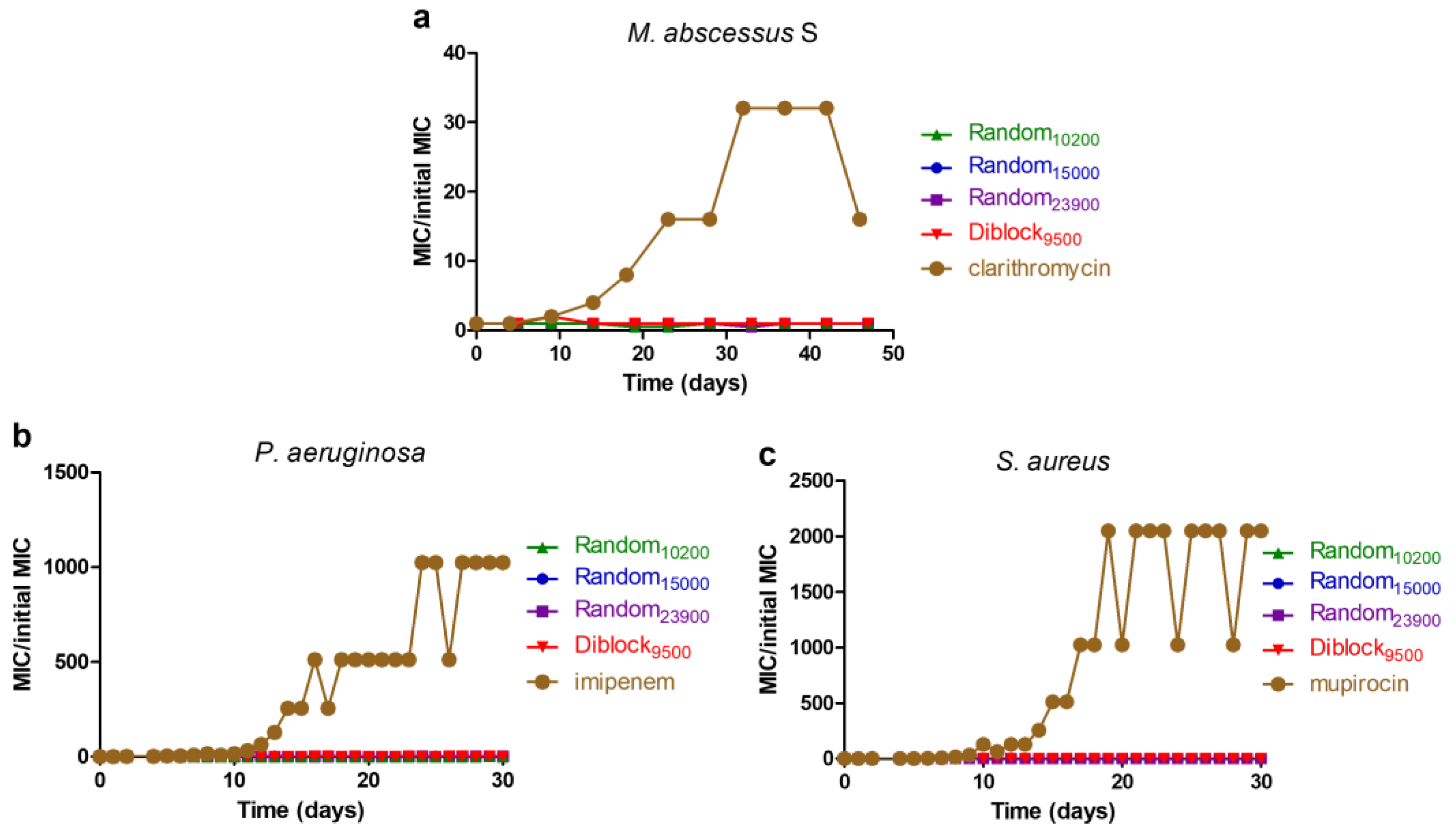
2.6. Resistance Induction Assay
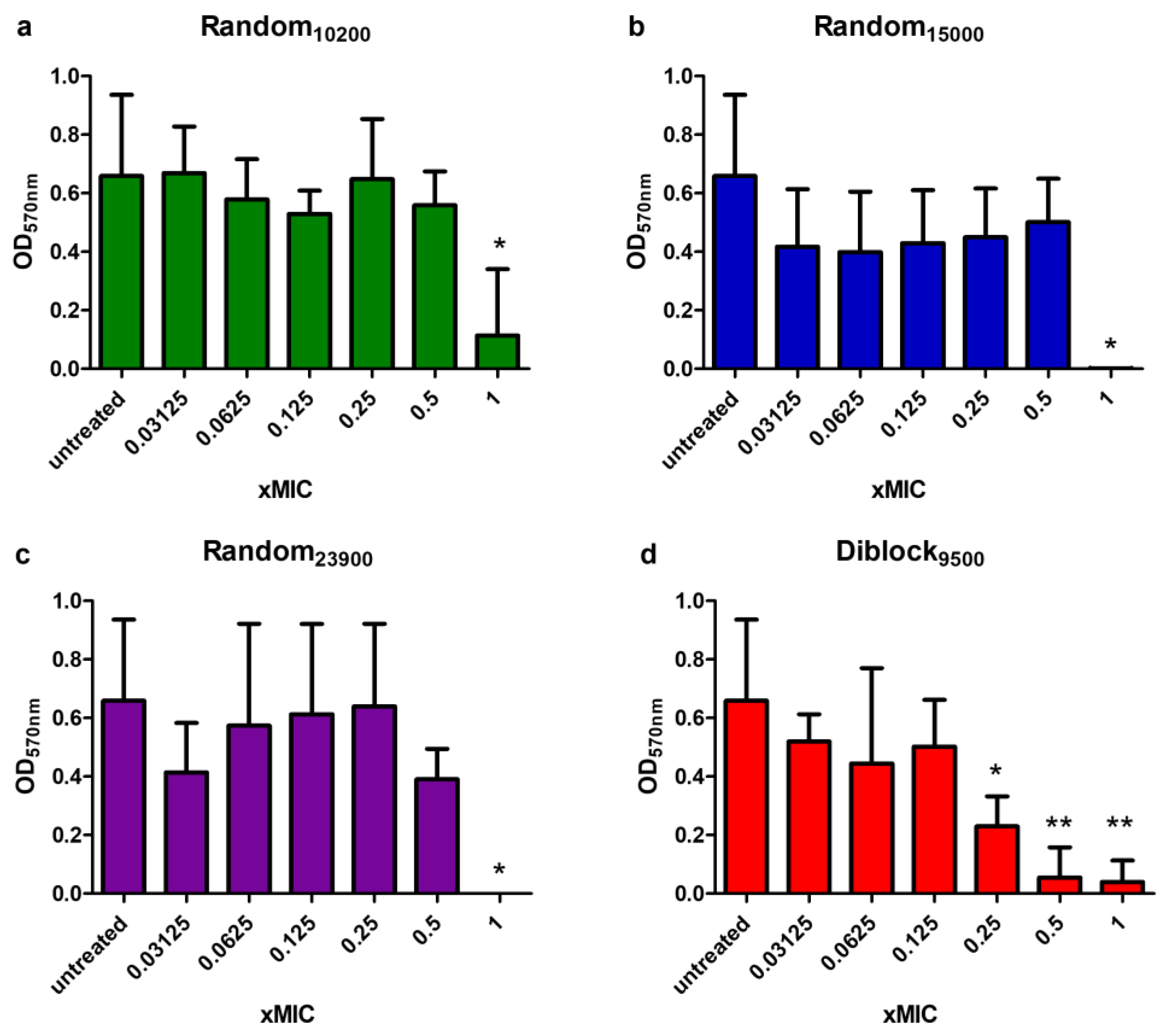
2.7. Anti-Biofilm Activity
3. Discussion
4. Materials and Methods
4.1. Synthesis and Characterization of the Cationic Amphiphilic Polymers
4.2. Bacterial Strains and Growth Conditions
4.3. Minimal Inhibitory Concentration (MIC) Determination
4.4. Evaluation of the Cytotoxic Effect of Polymers Using Human Cells
4.5. Time–Kill Assays
4.6. Bacterial Membrane Permeabilization Assay
4.7. Induction of Resistance Assay
4.8. Lipid Insertion Assay
4.9. Antibiofilm Formation Assay
4.10. Antibiofilm Disruption Assay
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shteinberg, M.; Haq, I.J.; Polineni, D.; Davies, J.C. Cystic Fibrosis. Lancet 2021, 397, 2195–2211. [Google Scholar] [CrossRef]
- Southern, K.W.; Munck, A.; Pollitt, R.; Travert, G.; Zanolla, L.; Dankert-Roelse, J.; Castellani, C.; ECFS CF Neonatal Screening Working Group. A Survey of Newborn Screening for Cystic Fibrosis in Europe. J. Cyst. Fibros 2007, 6, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Scotet, V.; Gutierrez, H.; Farrell, P.M. Newborn Screening for CF across the Globe-Where Is It Worthwhile? Int. J. Neonatal Screen 2020, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetz, D.; Ren, C.L. Review of Cystic Fibrosis. Pediatr. Ann. 2019, 48, e154–e161. [Google Scholar] [CrossRef] [PubMed]
- McElvaney, O.J.; Wade, P.; Murphy, M.; Reeves, E.P.; McElvaney, N.G. Targeting Airway Inflammation in Cystic Fibrosis. Expert Rev. Respir. Med. 2019, 13, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Lobo, L.J.; Noone, P.G. Respiratory Infections in Patients with Cystic Fibrosis Undergoing Lung Transplantation. Lancet Respir. Med. 2014, 2, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M. Which Pathogens Should We Worry About? Paediatr. Respir. Rev. 2019, 31, 15–17. [Google Scholar] [CrossRef]
- Lipuma, J.J. The Changing Microbial Epidemiology in Cystic Fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirgwin, M.E.; Dedloff, M.R.; Holban, A.M.; Gestal, M.C. Novel Therapeutic Strategies Applied to Pseudomonas Aeruginosa Infections in Cystic Fibrosis. Materials 2019, 12, 4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.L.; Morgan, W.J.; Konstan, M.W.; Schechter, M.S.; Wagener, J.S.; Fisher, K.A.; Regelmann, W.E. Presence of Methicillin Resistant Staphylococcus Aureus in Respiratory Cultures from Cystic Fibrosis Patients Is Associated with Lower Lung Function. Pediatr. Pulmonol. 2007, 42, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.E.; Aksamit, T.R. Understanding Nontuberculous Mycobacterial Lung Disease: It’s Been a Long Time Coming. F1000Res 2016, 5, 2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, S.T.; Rhoades, E.; Recht, J.; Pang, X.; Alsup, A.; Kolter, R.; Lyons, C.R.; Byrd, T.F.Y. Spontaneous Reversion of Mycobacterium Abscessus from a Smooth to a Rough Morphotype Is Associated with Reduced Expression of Glycopeptidolipid and Reacquisition of an Invasive Phenotype. Microbiology 2006, 152, 1581–1590. [Google Scholar] [CrossRef] [Green Version]
- Roux, A.-L.; Viljoen, A.; Bah, A.; Simeone, R.; Bernut, A.; Laencina, L.; Deramaudt, T.; Rottman, M.; Gaillard, J.-L.; Majlessi, L.; et al. The Distinct Fate of Smooth and Rough Mycobacterium Abscessus Variants inside Macrophages. Open Biol. 2016, 6, 160185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catherinot, E.; Clarissou, J.; Etienne, G.; Ripoll, F.; Emile, J.-F.; Daffé, M.; Perronne, C.; Soudais, C.; Gaillard, J.-L.; Rottman, M. Hypervirulence of a Rough Variant of the Mycobacterium Abscessus Type Strain. Infect. Immun. 2007, 75, 1055–1058. [Google Scholar] [CrossRef] [Green Version]
- Catherinot, E.; Roux, A.-L.; Macheras, E.; Hubert, D.; Matmar, M.; Dannhoffer, L.; Chinet, T.; Morand, P.; Poyart, C.; Heym, B.; et al. Acute Respiratory Failure Involving an R Variant of Mycobacterium Abscessus. J. Clin. Microbiol. 2009, 47, 271–274. [Google Scholar] [CrossRef] [Green Version]
- Nessar, R.; Cambau, E.; Reyrat, J.M.; Murray, A.; Gicquel, B. Mycobacterium Abscessus: A New Antibiotic Nightmare. J. Antimicrob. Chemother. 2012, 67, 810–818. [Google Scholar] [CrossRef] [Green Version]
- Sanguinetti, M.; Ardito, F.; Fiscarelli, E.; La Sorda, M.; D’Argenio, P.; Ricciotti, G.; Fadda, G. Fatal Pulmonary Infection Due to Multidrug-Resistant Mycobacterium Abscessus in a Patient with Cystic Fibrosis. J. Clin. Microbiol. 2001, 39, 816–819. [Google Scholar] [CrossRef] [Green Version]
- Jeon, K.; Kwon, O.J.; Lee, N.Y.; Kim, B.-J.; Kook, Y.-H.; Lee, S.-H.; Park, Y.K.; Kim, C.K.; Koh, W.-J. Antibiotic Treatment of Mycobacterium Abscessus Lung Disease. Am. J. Respir. Crit. Care Med. 2009, 180, 896–902. [Google Scholar] [CrossRef]
- Ballarino, G.J.; Olivier, K.N.; Claypool, R.J.; Holland, S.M.; Prevots, D.R. Pulmonary Nontuberculous Mycobacterial Infections: Antibiotic Treatment and Associated Costs. Respir. Med. 2009, 103, 1448–1455. [Google Scholar] [CrossRef]
- Osmani, M.; Sotello, D.; Alvarez, S.; Odell, J.A.; Thomas, M. Mycobacterium Abscessus Infections in Lung Transplant Recipients: 15-Year Experience from a Single Institution. Transpl. Infect. Dis. 2018, 20, e12835. [Google Scholar] [CrossRef] [PubMed]
- Yung, D.B.Y.; Sircombe, K.J.; Pletzer, D. Friends or Enemies? The Complicated Relationship between Pseudomonas Aeruginosa and Staphylococcus Aureus. Mol. Microbiol. 2021, 116, 1–15. [Google Scholar] [CrossRef]
- Camus, L.; Briaud, P.; Bastien, S.; Elsen, S.; Doléans-Jordheim, A.; Vandenesch, F.; Moreau, K. Trophic Cooperation Promotes Bacterial Survival of Staphylococcus Aureus and Pseudomonas Aeruginosa. ISME J. 2020, 14, 3093–3105. [Google Scholar] [CrossRef]
- Brooks, B.D.; Brooks, A.E. Therapeutic Strategies to Combat Antibiotic Resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, J.L. Antimicrobial Peptides Stage a Comeback. Nat. Biotechnol. 2013, 31, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Duplantier, A.J.; van Hoek, M.L. The Human Cathelicidin Antimicrobial Peptide LL-37 as a Potential Treatment for Polymicrobial Infected Wounds. Front. Immunol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef]
- Shen, W.; He, P.; Xiao, C.; Chen, X. From Antimicrobial Peptides to Antimicrobial Poly(α-Amino Acid)s. Adv. Healthc. Mater. 2018, 7, 1800354. [Google Scholar] [CrossRef]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef]
- Azmi, F.; Skwarczynski, M.; Toth, I. Towards the Development of Synthetic Antibiotics: Designs Inspired by Natural Antimicrobial Peptides. Curr. Med. Chem. 2016, 23, 4610–4624. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Laird, D.; Riga, E.K.; Deng, Z.; Dorner, F.; Perez-Hernandez, H.-R.; Guevara-Solarte, D.L.; Steinberg, T.; Al-Ahmad, A.; Lienkamp, K. Antimicrobial and Cell-Compatible Surface-Attached Polymer Networks–How the Correlation of Chemical Structure to Physical and Biological Data Leads to a Modified Mechanism of Action. J. Mater. Chem. B 2015, 3, 6224–6238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tew, G.N.; Scott, R.W.; Klein, M.L.; DeGrado, W.F. De Novo Design of Antimicrobial Polymers, Foldamers, and Small Molecules: From Discovery to Practical Applications. Acc. Chem. Res. 2010, 43, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ahmad, A.; Laird, D.; Zou, P.; Tomakidi, P.; Steinberg, T.; Lienkamp, K. Nature-Inspired Antimicrobial Polymers--Assessment of Their Potential for Biomedical Applications. PLoS ONE 2013, 8, e73812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroda, K.; Caputo, G.A. Antimicrobial Polymers as Synthetic Mimics of Host-Defense Peptides. Wiley Interdiscip Rev. Nanomed Nanobiotechnol. 2013, 5, 49–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmona-Ribeiro, A.M.; de Melo Carrasco, L.D. Cationic Antimicrobial Polymers and Their Assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef] [Green Version]
- Deka, S.R.; Sharma, A.K.; Kumar, P. Cationic Polymers and Their Self-Assembly for Antibacterial Applications. Curr. Top. Med. Chem. 2015, 15, 1179–1195. [Google Scholar] [CrossRef]
- Ergene, C.; Palermo, E.F. Antimicrobial Synthetic Polymers: An Update on Structure-Activity Relationships. Curr. Pharm. Des. 2018, 24, 855–865. [Google Scholar] [CrossRef]
- Konai, M.M.; Bhattacharjee, B.; Ghosh, S.; Haldar, J. Recent Progress in Polymer Research to Tackle Infections and Antimicrobial Resistance. Biomacromolecules 2018, 19, 1888–1917. [Google Scholar] [CrossRef]
- Makobongo, M.O.; Gancz, H.; Carpenter, B.M.; McDaniel, D.P.; Merrell, D.S. The Oligo-Acyl Lysyl Antimicrobial Peptide C12K-2β12 Exhibits a Dual Mechanism of Action and Demonstrates Strong In Vivo Efficacy against Helicobacter Pylori. Antimicrob. Agents Chemother. 2012, 56, 378–390. [Google Scholar] [CrossRef]
- Chakrabarty, S.; King, A.; Kurt, P.; Zhang, W.; Ohman, D.E.; Wood, L.F.; Lovelace, C.; Rao, R.; Wynne, K.J. Highly Effective, Water-Soluble, Hemocompatible 1,3-Propylene Oxide-Based Antimicrobials: Poly[(3,3-Quaternary/PEG)-Copolyoxetanes]. Biomacromolecules 2011, 12, 757–769. [Google Scholar] [CrossRef] [PubMed]
- King, A.; Chakrabarty, S.; Zhang, W.; Zeng, X.; Ohman, D.E.; Wood, L.F.; Abraham, S.; Rao, R.; Wynne, K.J. High Antimicrobial Effectiveness with Low Hemolytic and Cytotoxic Activity for PEG/Quaternary Copolyoxetanes. Biomacromolecules 2014, 15, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Kozon, D.; Mierzejewska, J.; Kobiela, T.; Grochowska, A.; Dudnyk, K.; Głogowska, A.; Sobiepanek, A.; Kuźmińska, A.; Ciach, T.; Augustynowicz-Kopeć, E.; et al. Amphiphilic Polymethyloxazoline–Polyethyleneimine Copolymers: Interaction with Lipid Bilayer and Antibacterial Properties. Macromol. Biosci. 2019, 19, 1900254. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Chin, W.; Dong, H.; Xu, L.; Zhong, G.; Huang, Y.; Li, L.; Xu, K.; Wu, M.; Hedrick, J.L.; et al. Biodegradable Antimicrobial Polycarbonates with In Vivo Efficacy against Multidrug-Resistant MRSA Systemic Infection. Adv. Healthc. Mater. 2015, 4, 2128–2136. [Google Scholar] [CrossRef]
- Tan, J.P.K.; Coady, D.J.; Sardon, H.; Yuen, A.; Gao, S.; Lim, S.W.; Liang, Z.C.; Tan, E.W.; Venkataraman, S.; Engler, A.C.; et al. Broad Spectrum Macromolecular Antimicrobials with Biofilm Disruption Capability and In Vivo Efficacy. Adv. Healthc. Mater. 2017, 6, 1601420. [Google Scholar] [CrossRef]
- Yang, C.; Lou, W.; Zhong, G.; Lee, A.; Leong, J.; Chin, W.; Ding, B.; Bao, C.; Tan, J.P.K.; Pu, Q.; et al. Degradable Antimicrobial Polycarbonates with Unexpected Activity and Selectivity for Treating Multidrug-Resistant Klebsiella Pneumoniae Lung Infection in Mice. Acta Biomater. 2019, 94, 268–280. [Google Scholar] [CrossRef]
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of Antimicrobial Polymeric Materials by Dispersion of Well-Defined Amphiphilic Methacrylic SG1-Based Copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
- Zou, Y.-J.; He, S.-S.; Du, J.-Z. ε-Poly(L-Lysine)-Based Hydrogels with Fast-Acting and Prolonged Antibacterial Activities. Chin. J. Polym. Sci. 2018, 36, 1239–1250. [Google Scholar] [CrossRef]
- Ioannou, C.J.; Hanlon, G.W.; Denyer, S.P. Action of Disinfectant Quaternary Ammonium Compounds against Staphylococcus Aureus. Antimicrob. Agents Chemother. 2007, 51, 296–306. [Google Scholar] [CrossRef] [Green Version]
- Jagatia, H.; Cantillon, D. DNA Isolation from Mycobacteria. Mycobact. Protoc. 2021, 59–75. [Google Scholar] [CrossRef]
- Pietsch, F.; Heidrich, G.; Nordholt, N.; Schreiber, F. Prevalent Synergy and Antagonism Among Antibiotics and Biocides in Pseudomonas Aeruginosa. Front. Microbiol. 2021, 11, 615618. [Google Scholar] [CrossRef] [PubMed]
- Dengler, W.A.; Schulte, J.; Berger, D.P.; Mertelsmann, R.; Fiebig, H.H. Development of a Propidium Iodide Fluorescence Assay for Proliferation and Cytotoxicity Assays. Anti-Cancer Drugs 1995, 6, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Dennison, S.R.; Kim, Y.S.; Cha, H.J.; Phoenix, D.A. Investigations into the Ability of the Peptide, HAL18, to Interact with Bacterial Membranes. Eur. Biophys. J. 2008, 38, 37. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas Aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Oluyombo, O.; Penfold, C.N.; Diggle, S.P. Competition in Biofilms between Cystic Fibrosis Isolates of Pseudomonas Aeruginosa Is Shaped by R-Pyocins. mBio 2019, 10, e01828-18. [Google Scholar] [CrossRef] [Green Version]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chem. Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug Evolutionary Strategies to Reverse Antibiotic Resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Roblin, C.; Chiumento, S.; Jacqueline, C.; Pinloche, E.; Nicoletti, C.; Olleik, H.; Courvoisier-Dezord, E.; Amouric, A.; Basset, C.; Dru, L.; et al. The Multifunctional Sactipeptide Ruminococcin C1 Displays Potent Antibacterial Activity In Vivo as Well as Other Beneficial Properties for Human Health. Int. J. Mol. Sci. 2021, 22, 3253. [Google Scholar] [CrossRef]
- Mulkern, A.J.; Oyama, L.B.; Cookson, A.R.; Creevey, C.J.; Wilkinson, T.J.; Olleik, H.; Maresca, M.; da Silva, G.C.; Fontes, P.P.; Bazzolli, D.M.S.; et al. Microbiome-Derived Antimicrobial Peptides Offer Therapeutic Solutions for the Treatment of Pseudomonas Aeruginosa Infections. npj Biofilms Microbiomes 2022, 8, 70. [Google Scholar] [CrossRef]
- Olleik, H.; Yacoub, T.; Hoffer, L.; Gnansounou, S.M.; Benhaiem-Henry, K.; Nicoletti, C.; Mekhalfi, M.; Pique, V.; Perrier, J.; Hijazi, A.; et al. Synthesis and Evaluation of the Antibacterial Activities of 13-Substituted Berberine Derivatives. Antibiotics 2020, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Oyama, L.B.; Olleik, H.; Teixeira, A.C.N.; Guidini, M.M.; Pickup, J.A.; Hui, B.Y.P.; Vidal, N.; Cookson, A.R.; Vallin, H.; Wilkinson, T.; et al. In Silico Identification of Two Peptides with Antibacterial Activity against Multidrug-Resistant Staphylococcus Aureus. npj Biofilms Microbiomes 2022, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins 2019, 11, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyama, L.B.; Girdwood, S.E.; Cookson, A.R.; Fernandez-Fuentes, N.; Privé, F.; Vallin, H.E.; Wilkinson, T.J.; Golyshin, P.N.; Golyshina, O.V.; Mikut, R.; et al. The Rumen Microbiome: An Underexplored Resource for Novel Antimicrobial Discovery. npj Biofilms Microbiomes 2017, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahfoud, R.; Maresca, M.; Santelli, M.; Pfohl-Leszkowicz, A.; Puigserver, A.; Fantini, J. PH-Dependent Interaction of Fumonisin B1 with Cholesterol: Physicochemical and Molecular Modeling Studies at the Air-Water Interface. J. Agric. Food Chem. 2002, 50, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Santucci, P.; Point, V.; Poncin, I.; Guy, A.; Crauste, C.; Serveau-Avesque, C.; Galano, J.M.; Spilling, C.D.; Cavalier, J.-F.; Canaan, S. LipG a Bifunctional Phospholipase/Thioesterase Involved in Mycobacterial Envelope Remodeling. Biosci. Rep. 2018, 38, BSR20181953. [Google Scholar] [CrossRef] [Green Version]
- Meniche, X.; Labarre, C.; de Sousa-d’Auria, C.; Huc, E.; Laval, F.; Tropis, M.; Bayan, N.; Portevin, D.; Guilhot, C.; Daffé, M.; et al. Identification of a Stress-Induced Factor of Corynebacterineae That Is Involved in the Regulation of the Outer Membrane Lipid Composition. J. Bacteriol. 2009, 191, 7323–7332. [Google Scholar] [CrossRef] [PubMed]
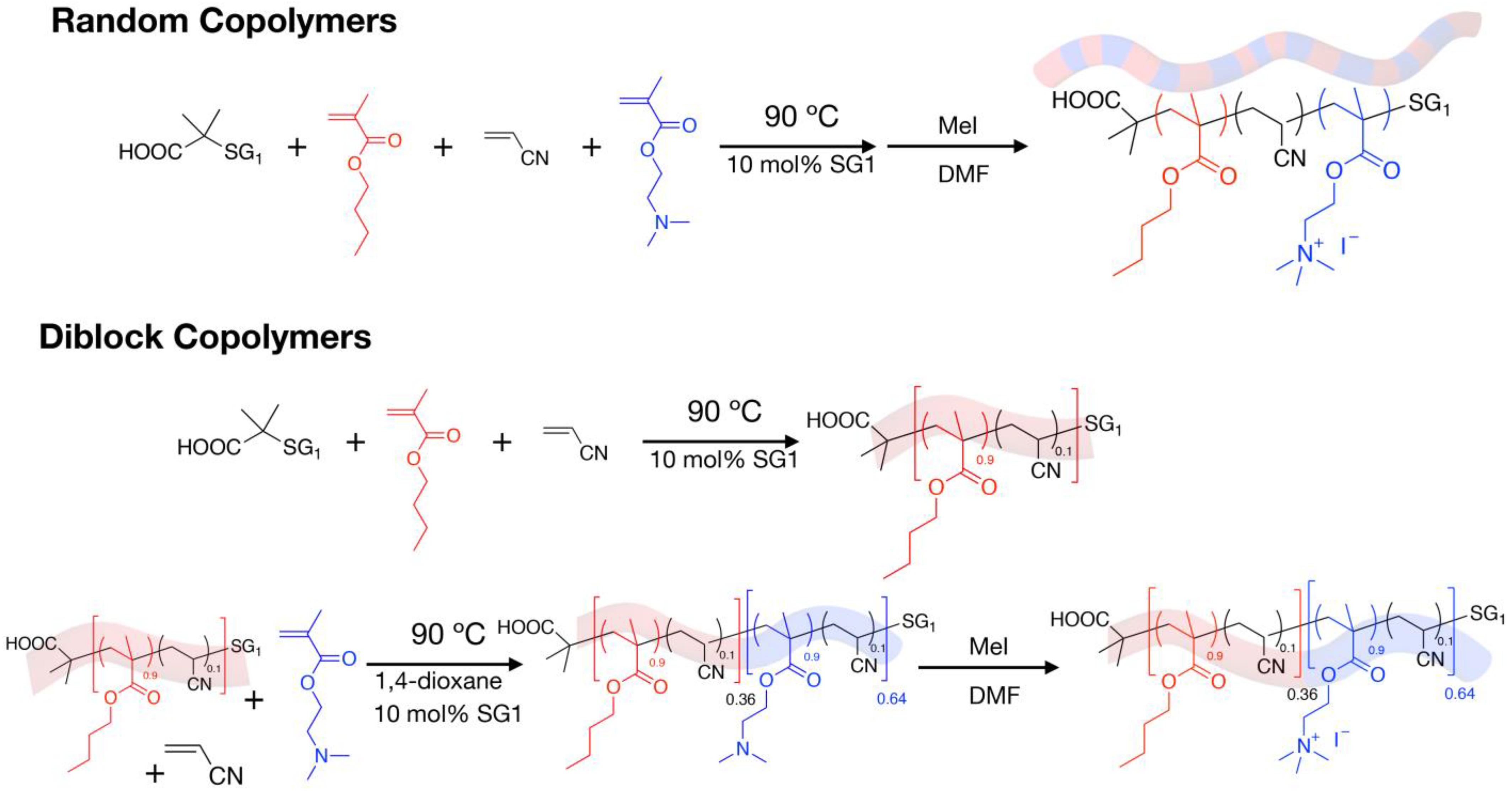
| Copolymer Name | Mn (g/mol) (SEC/DMF) | FDMAEMA |
|---|---|---|
| Random2800 | 2800 | 0.67 |
| Random3300 | 3300 | 0.63 |
| Random9300 | 9300 | 0.65 |
| Random10200 | 10,200 | 0.64 |
| Random14600 | 14,600 | 0.65 |
| Random15000 | 15,000 | 0.56 |
| Random18500 | 18,500 | 0.68 |
| Random23900 | 23,900 | 0.61 |
| Diblock6800 | 6800 | 0.65 |
| Diblock9500 | 9500 | 0.62 |
| Diblock9900 | 9900 | 0.72 |
| Diblock14700 | 14,700 | 0.78 |
| Diblock17900 | 17,900 | 0.62 |
| Diblock18000 | 18,000 | 0.8 |
| Diblock18400 | 18,400 | 0.64 |
| Diblock18800 | 18,800 | 0.57 |
| Diblock19200 | 19,200 | 0.74 |
| Diblock20500 | 20,500 | 0.64 |
| Diblock21900 | 21,900 | 0.58 |
| Copolymer Name | M. abscessus S | P. aeruginosa | S. aureus |
|---|---|---|---|
| Random2800 | 6.25 | 200 | 200 |
| Random3300 | 25 | 400 | 200 |
| Random9300 | 25 | 25 | 50 |
| Random10200 | 12.5 | 25 | 50 |
| Random14600 | 25 | 25 | 12.5 |
| Random15000 | 25 | 25 | 25 |
| Random18500 | 25 | 12.5 | 12.5 |
| Random23900 | 25 | 12.5 | 12.5 |
| Diblock6800 | 25 | 100 | 12.5 |
| Diblock9500 | 50 | 50 | 12.5 |
| Diblock9900 | 50 | 50 | 12.5 |
| Diblock14700 | 50 | 50 | 12.5 |
| Diblock17900 | 100 | 50 | 12.5 |
| Diblock18000 | 50 | 50 | 12.5 |
| Diblock18400 | 100 | 50 | 25 |
| Diblock18800 | 100 | 50 | 12.5 |
| Diblock19200 | 50 | 50 | 12.5 |
| Diblock20500 | 50 | 100 | 12.5 |
| Diblock21900 | 100 | 50 | 12.5 |
| Copolymer Name | IC50 BEAS-2B | TI M. abscessus S | TI P. aeruginosa | TI S. aureus |
|---|---|---|---|---|
| Random2800 | >400 | >64 | >2 | >2 |
| Random3300 | >400 | >16 | >1 | >2 |
| Random9300 | 195.1 | 7.8 | 7.8 | 3.9 |
| Random10200 | 231.1 | 18.5 | 9.2 | 4.6 |
| Random14600 | 133.7 | 5.3 | 5.3 | 10.7 |
| Random15000 | 148.3 | 5.9 | 5.9 | 5.9 |
| Random18500 | 124.7 | 5.0 | 10.0 | 10.0 |
| Random23900 | 204.2 | 8.2 | 16.3 | 16.3 |
| Diblock6800 | 228.4 | 9.1 | 2.3 | 18.3 |
| Diblock9500 | 250.6 | 5.0 | 5.0 | 20.0 |
| Diblock9900 | 244.5 | 4.9 | 4.9 | 19.6 |
| Diblock14700 | 211.5 | 4.2 | 4.2 | 16.9 |
| Diblock17900 | 172.8 | 1.7 | 3.4 | 13.8 |
| Diblock18000 | 219.2 | 4.4 | 4.4 | 17.5 |
| Diblock18400 | 242.1 | 2.4 | 4.8 | 9.7 |
| Diblock18800 | 232.1 | 2.3 | 4.6 | 18.6 |
| Diblock19200 | 161.8 | 3.2 | 3.2 | 12.9 |
| Diblock20500 | 222.7 | 4.4 | 2.2 | 17.8 |
| Diblock21900 | 239 | 2.4 | 4.8 | 19.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casanova, M.; Olleik, H.; Hdiouech, S.; Roblin, C.; Cavalier, J.-F.; Point, V.; Jeannot, K.; Caron, B.; Perrier, J.; Charriau, S.; et al. Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics 2023, 12, 120. https://doi.org/10.3390/antibiotics12010120
Casanova M, Olleik H, Hdiouech S, Roblin C, Cavalier J-F, Point V, Jeannot K, Caron B, Perrier J, Charriau S, et al. Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics. 2023; 12(1):120. https://doi.org/10.3390/antibiotics12010120
Chicago/Turabian StyleCasanova, Magali, Hamza Olleik, Slim Hdiouech, Clarisse Roblin, Jean-François Cavalier, Vanessa Point, Katy Jeannot, Baptiste Caron, Josette Perrier, Siméon Charriau, and et al. 2023. "Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis" Antibiotics 12, no. 1: 120. https://doi.org/10.3390/antibiotics12010120
APA StyleCasanova, M., Olleik, H., Hdiouech, S., Roblin, C., Cavalier, J.-F., Point, V., Jeannot, K., Caron, B., Perrier, J., Charriau, S., Lafond, M., Guillaneuf, Y., Canaan, S., Lefay, C., & Maresca, M. (2023). Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics, 12(1), 120. https://doi.org/10.3390/antibiotics12010120








