Comparative Transcriptome Analysis of the Response to Vibrio parahaemolyticus and Low-Salinity Stress in the Swimming Crab Portunus trituberculatus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Library Preparation and RNA-Seq
2.3. Data Analysis
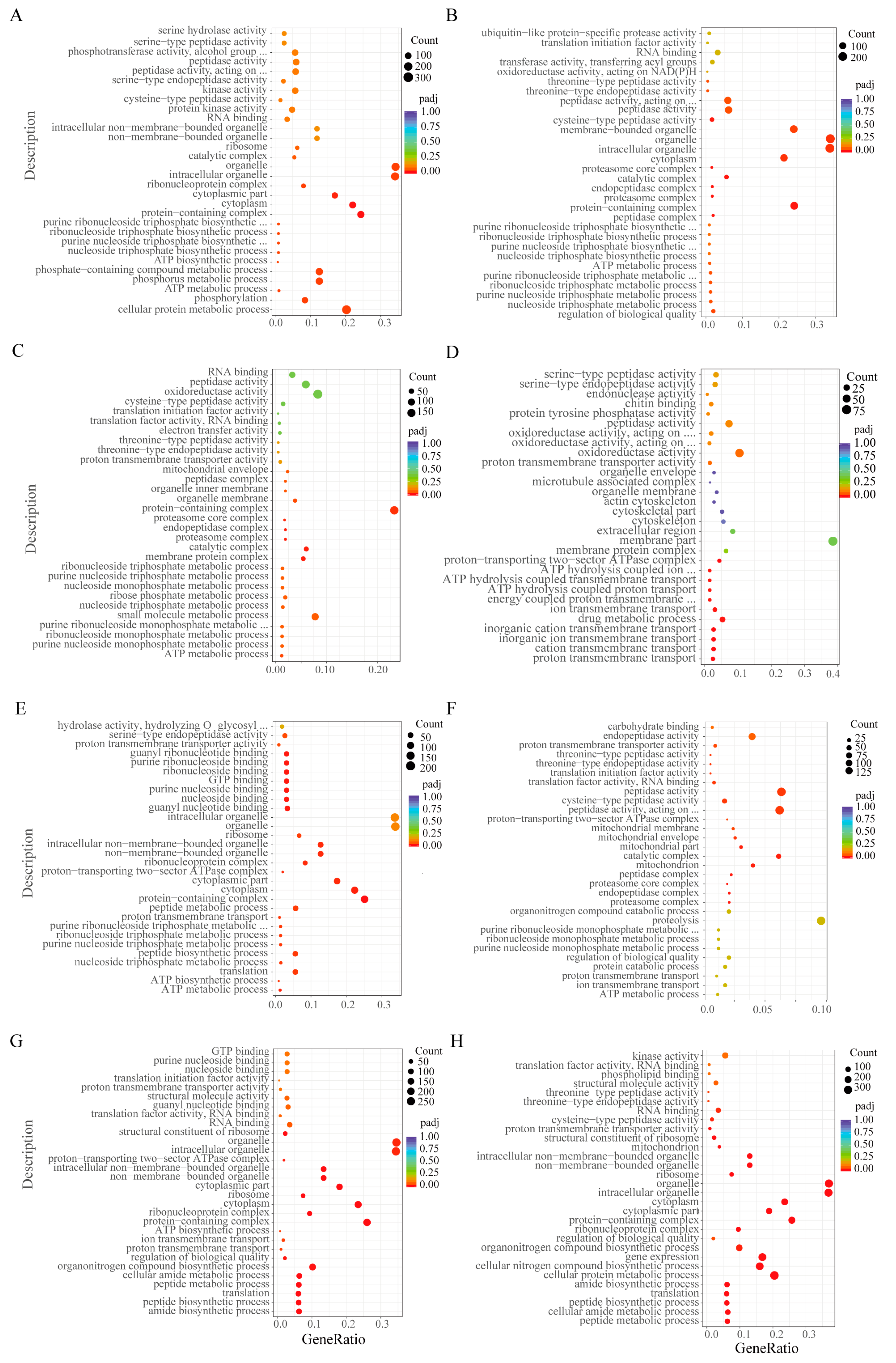
2.4. GO and KEGG Enrichment Analysis of DEGs
2.5. Quantitative Real-Time PCR Validation
3. Result
3.1. Information Obtained from the Sequencing Data
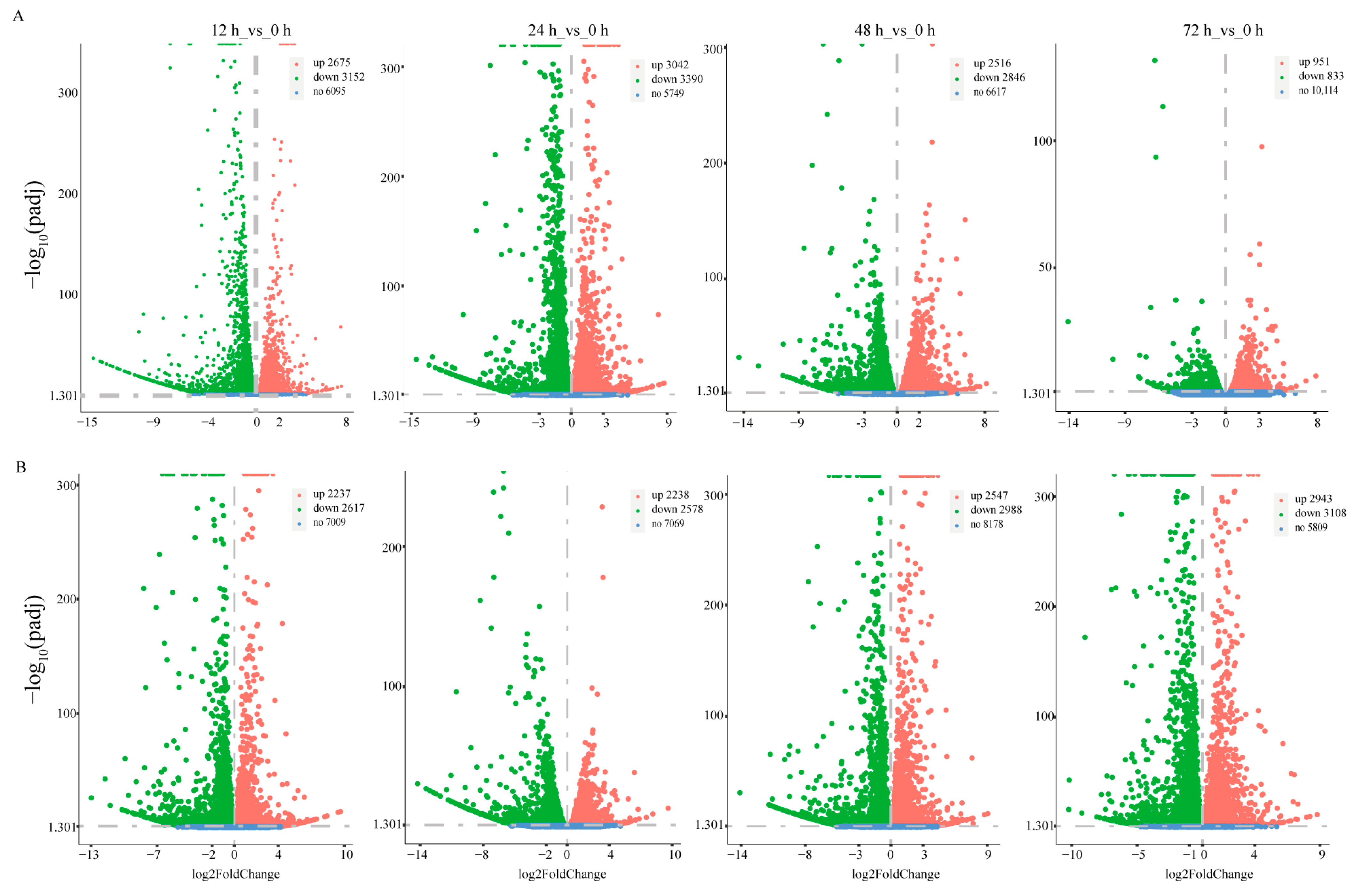
3.2. Dynamic Expression of the Transcriptome under Low Salinity and Pathogenic Challenge
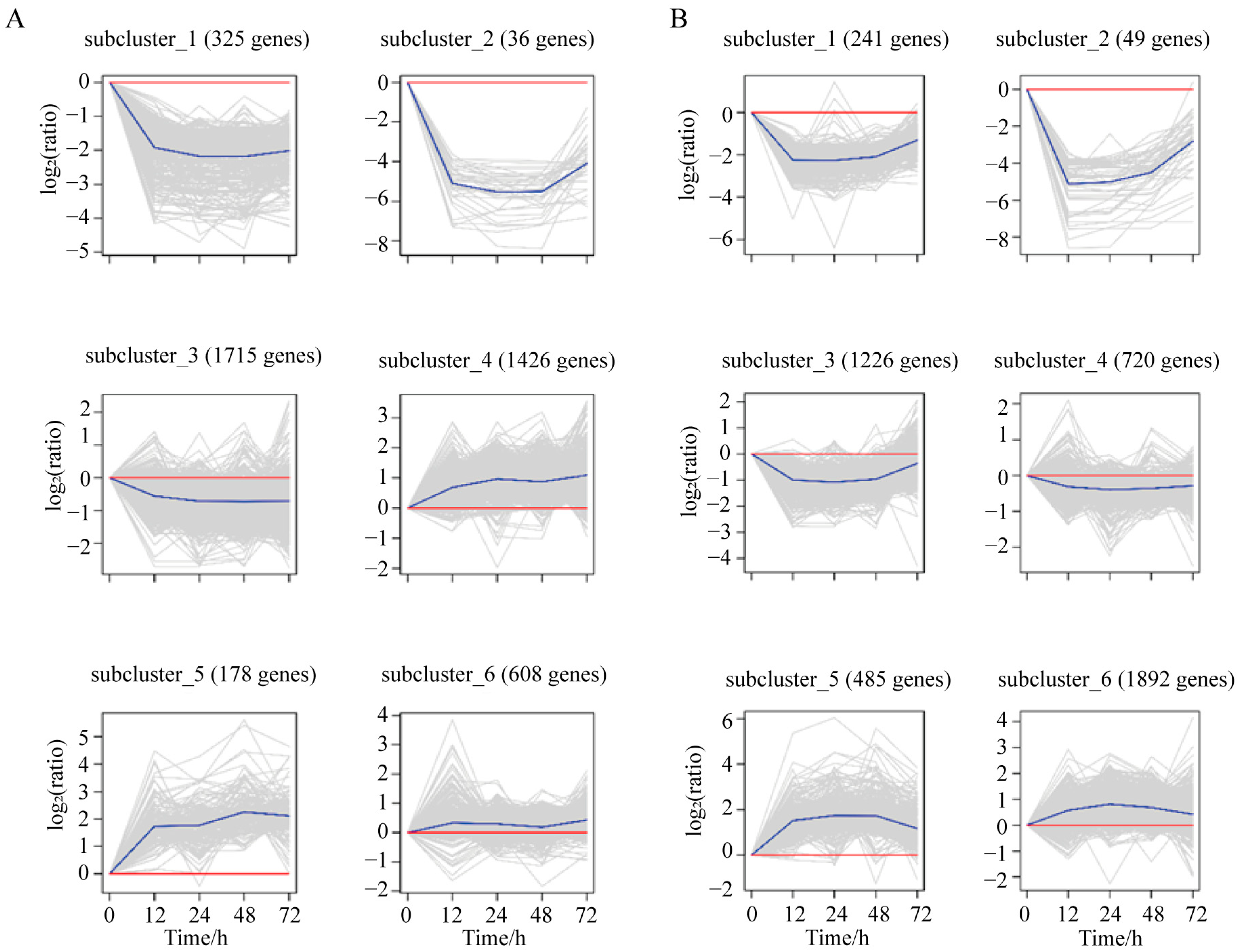
3.3. Expression Pattern Clustering of DEGs
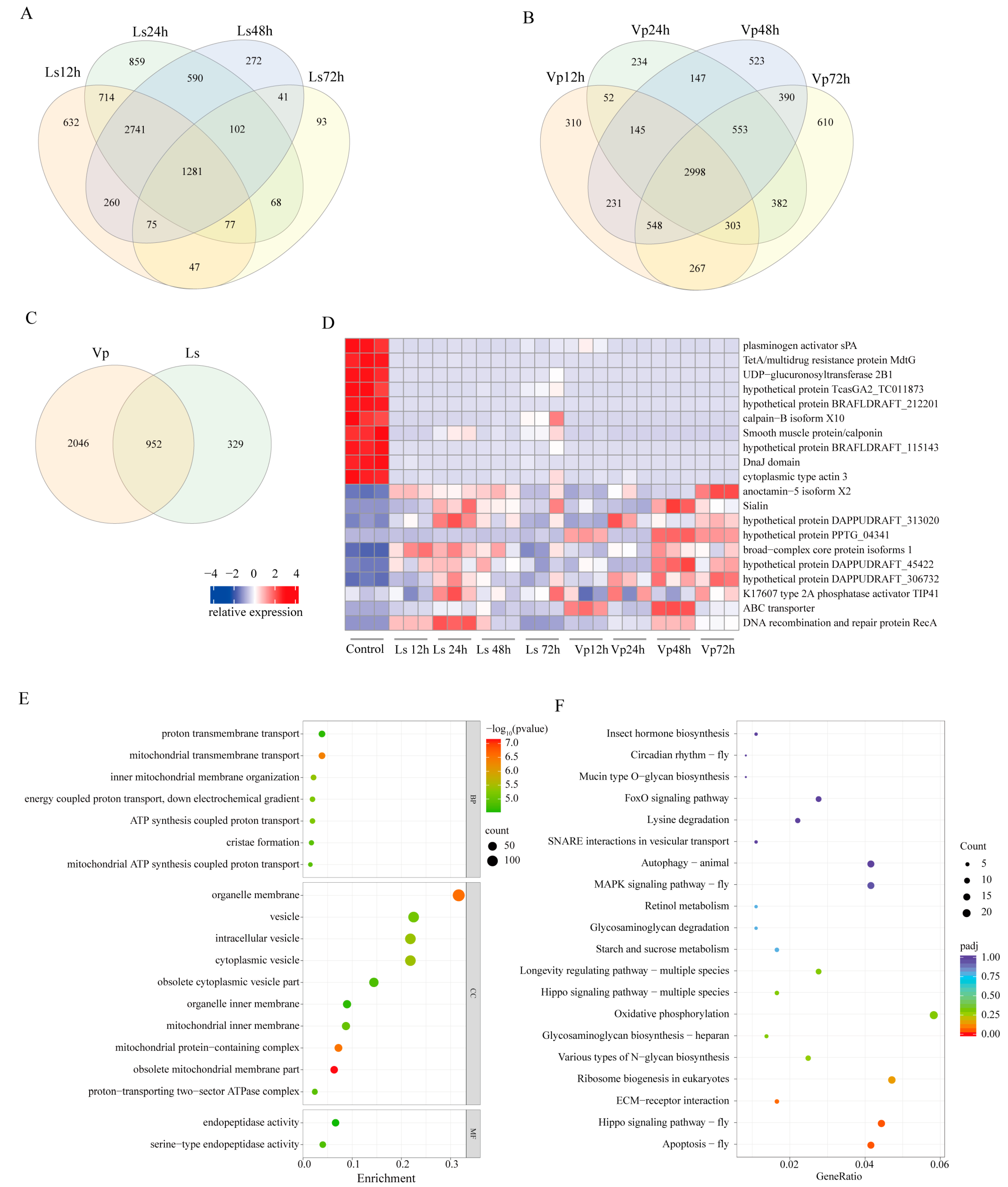
3.4. Screening of Reverse DEGs after Exposure to the Two Kinds of Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Senapin, S.; Phiwsaiya, K.; Gangnonngiw, W.; Flegel, T.W. False rumours of disease outbreaks caused by infectious myonecrosis virus (IMNV) in the whiteleg shrimp in Asia. J. Negat. Results Biomed. 2011, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T.; Chen, I.T.; Yang, Y.T.; Ko, T.P.; Huang, Y.T.; Huang, J.Y.; Huang, M.F.; Lin, S.J.; Chen, C.Y.; Lin, S.S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [PubMed]
- Theethakaew, C.; Nakamura, S.; Motooka, D.; Matsuda, S.; Kodama, T.; Chonsin, K.; Suthienkul, O.; Iida, T. Plasmid dynamics in Vibrio parahaemolyticus strains related to shrimp Acute Hepatopancreatic Necrosis Syndrome (AHPNS). Infect. Genet. Evol. 2017, 51, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.; Yao, D.; Zhao, Y.; Tran, N.T.; Li, S.; Ma, H.; Aweya, J.J. Metabolic reprogramming in crustaceans: A vital immune and environmental response strategy. Rev. Aquac. 2021, 14, 1094–1119. [Google Scholar] [CrossRef]
- Wang, F.I.; Chen, J.C. Effect of salinity on the immune response of tiger shrimp Penaeus monodon and its susceptibility to Photobacterium damselae subsp. damselae. Fish Shellfish Immunol. 2006, 20, 671–681. [Google Scholar] [CrossRef]
- Zhou, Z.K.; Gu, W.B.; Cong, W.; Zhou, Y.L.; Tu, D.D.; Liu, Z.P.; Zhu, Q.H.; Shu, M.A. Seven transcripts from the chitinase gene family of the mud crab Scylla paramamosain: Their expression profiles during development and moulting and under environmental stresses. Aquac. Res. 2018, 49, 3296–3308. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Y.; Li, F.; Huang, B.; Xiang, J. Molecular characterization and expression analysis of chitinase (Fcchi-3) from Chinese shrimp, Fenneropenaeus chinensis. Mol. Biol. Rep. 2010, 37, 1913–1921. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, D.; Zhang, X.; Li, G.; Zhang, Y.; Huang, C.; Zhang, Z.; Tian, C. A first insight into the gonad transcriptome of Hong Kong Catfish (Clarias fuscus). Animals 2021, 11, 1131. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Peng, L.; Ren, L.; Zhang, H.; Zou, L.; Liu, W.; Xiao, Y. Comparative transcriptome analysis of molecular mechanism underlying gray-to-red body color formation in red crucian carp (Carassius auratus, red var.). Fish Physiol. Biochem. 2017, 43, 1387–1398. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Shen, Y.; Bi, Y.; Hou, W.; Pan, G.; Wu, X. Transcriptional responses to low-salinity stress in the gills of adult female Portunus trituberculatus. Comp. Biochem. Physiol. Part D 2019, 29, 86–94. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, X.; Yu, Y.; Huang, H.; Li, F.; Xiang, J. Comparative transcriptomic characterization of the early development in Pacific white shrimp Litopenaeus vannamei. PLoS ONE 2014, 9, e106201. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Jiang, K.; Li, D.; Yan, X.; Nie, H. Transcriptomic analysis of Manila clam Ruditapes philippinarum under lipopolysaccharide challenge provides molecular insights into immune response. Fish Shellfish. Immunol. 2020, 106, 110–119. [Google Scholar] [CrossRef]
- Cao, M.; Zhang, M.; Yang, N.; Fu, Q.; Su, B.; Zhang, X.; Li, Q.; Yan, X.; Thongda, W.; Li, C. Full length transcriptome profiling reveals novel immune-related genes in black rockfish (Sebastes schlegelii). Fish Shellfish Immunol. 2020, 106, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Interaminense, J.A.; Vogeley, J.L.; Gouveia, C.K.; Portela, R.S.; Oliveira, J.P.; Silva, S.; Coimbra, M.R.M.; Peixoto, S.M.; Soares, R.B.; Buarque, D.S.; et al. Effects of dietary Bacillus subtilis and Shewanella algae in expression profile of immune-related genes from hemolymph of Litopenaeus vannamei challenged with Vibrio parahaemolyticus. Fish Shellfish Immunol. 2019, 86, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, A.; Sundaram, M.; Saranya, C.; Swain, S.; Dash, R.R.; Dayal, J.S. Carbohydrate sources deferentially influence growth performances, microbial dynamics and immunomodulation in Pacific white shrimp (Litopenaeus vannamei) under biofloc system. Fish Shellfish Immunol. 2019, 86, 1207–1216. [Google Scholar] [CrossRef]
- Pan, L.; Zhang, X.; Yang, L.; Pan, S. Effects of Vibro harveyi and Staphyloccocus aureus infection on hemocyanin synthesis and innate immune responses in white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 93, 659–668. [Google Scholar] [CrossRef]
- Pang, H.; Wang, G.; Zhou, S.; Wang, J.; Zhao, J.; Hoare, R.; Monaghan, S.J.; Wang, Z.; Sun, C. Survival and immune response of white shrimp Litopenaeus vannamei following single and concurrent infections with WSSV and Vibrio parahaemolyticus. Fish Shellfish Immunol. 2019, 92, 712–718. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, L.; Jiang, W.; Abasubong, K.P.; Zhang, C.; Zhang, D.; Li, X.; Jiang, G.; Chi, C.; Liu, W. Effects of dietary icariin supplementation on the ovary development-related transcriptome of Chinese mitten crab (Eriocheir sinensis). Comp. Biochem. Physiol. Part D 2021, 37, 100756. [Google Scholar] [CrossRef]
- Yang, Z.G.; Zhou, J.Y.; Wei, B.H.; Cheng, Y.X.; Zhang, L.; Zhen, X.M. Comparative transcriptome analysis reveals osmotic-regulated genes in the gill of Chinese mitten crab (Eriocheir sinensis). PLoS ONE 2019, 14, e0210469. [Google Scholar] [CrossRef]
- Kong, T.; Lin, S.; Ren, X.; Li, S.; Gong, Y. Transcriptome and metabolome integration analysis of mud crab Scylla paramamosain challenged to Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 2020, 103, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lv, J.J.; Wang, L.; Sun, D.F.; Gao, B.Q.; Liu, P. Characterization of a chitinase-1 gene (PtCht-1) from a marine crab Portunus trituberculatus and its response to immune stress. Gene 2020, 741, 144523. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Cerenius, L.; Jiravanichpaisal, P.; Liu, H.P.; Söderhill, I. Crustacean immunity. Adv. Exp. Med. Biol. 2010, 708, 239–259. [Google Scholar]
- Hao, J.W.; Gao, B.Q.; Wang, C.; Meng, X.L.; Wan, X.Y.; Li, X.P.; Liu, P.; Zhang, Q.L. Natural infection of Portunus trituberculatus with acute hepatopancreas necrosis disease causing by Vibrio parahaemolyticus (VpAHPND). J. Fish. China 2019, 43, 1647–1660. [Google Scholar]
- Ma, J.W.; Lv, J.J.; Liu, P.; Gao, B.Q.; Li, J. Effects of abrupt salinity stress on serum osmolarity and ion concentration of “Huangxuan No.1” Portunus trituberculatus. Prog. Fish. Sci. 2016, 37, 58–62. [Google Scholar]
- Chen, T.; Li, Z.; Liu, J.; Liang, C.; Yuan, L. Transcriptome of hepatopancreas in kuruma shrimp Marsupenaeus japonicus under low-salinity stress. J. Oceanol. Limnol. 2021, 40, 745–765. [Google Scholar] [CrossRef]
- Tang, D.; Wu, Y.; Huang, S.; Wu, L.; Luo, Y.; Wang, Z. Transcriptome reveals the mechanism of immunity in the low salinity stress of the Chinese shrimp (Fenneropenaeus chinensis). Thalass. Int. J. Mar. Sci. 2022, 38, 977–987. [Google Scholar] [CrossRef]
- Chen, D.; Guo, L.; Yi, C.; Wang, S.; Ru, Y.; Wang, H. Hepatopancreatic transcriptome analysis and humoral immune factor assays in red claw crayfish (Cherax quadricarinatus) provide insight into innate immunomodulation under Vibrio parahaemolyticus infection. Ecotoxicol. Environ. Saf. 2021, 217, 112266. [Google Scholar] [CrossRef]
- Ren, X.; Liu, P.; Li, J. Comparative transcriptomic analysis of Marsupenaeus japonicus hepatopancreas in response to Vibrio parahaemolyticus and white spot syndrome virus. Fish Shellfish Immunol. 2019, 87, 755–764. [Google Scholar] [CrossRef]
- Cheng, A.C.; Shiu, Y.L.; Chiu, S.T.; Ballantyne, R.C.; Liu, H. Effects of chitin from Daphnia similis and its derivative, chitosan on the immune response and disease resistance of white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2021, 119, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ni, M.; Zhang, P.; Bai, Y.; Zhou, B.; Zheng, J.; Cui, Z. Identification and functional characterization of C-type lectins and crustins provide new insights into the immune response of Portunus trituberculatus. Fish Shellfish Immunol. 2022, 129, 170–181. [Google Scholar] [CrossRef]
- Lv, J.J.; Liu, P.; Wang, Y.; Gao, B.Q.; Chen, P.; Li, J. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS ONE 2013, 8, e82155. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Zhuang, X.; Liao, M.; Cui, Q.; Yan, C.; Huang, J.; Jiang, Z.; Huang, L.; Luo, W.; Liu, Y.; et al. Andrographis paniculata improves growth and non-specific immunity of shrimp Litopenaeus vannamei, and protects it from Vibrio alginolyticus by reducing oxidative stress and apoptosis. Dev. Comp. Immunol. 2022, 139, 104542. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhi, B.; Wu, W.; Zhang, X. Requirement for shrimp caspase in apoptosis against virus infection. Dev. Comp. Immunol. 2008, 32, 706–715. [Google Scholar] [CrossRef]
- Zuo, H.; Chang, C.; Yun, G.; Lin, J.; Jin, C.; Wei, W. Regulation of shrimp PjCaspase promoter activity by WSSV VP38 and VP41B. Fish Shellfish Immunol. 2011, 30, 1188–1191. [Google Scholar] [CrossRef]
- Nakamura, R.; Pedrosa-Gerasmio, I.R.; Alenton, R.R.R.; Nozaki, R.; Kondo, H.; Hirono, I. Anti-PirA-like toxin immunoglobulin (IgY) in feeds passively immunizes shrimp against acute hepatopancreatic necrosis disease. J. Fish Dis. 2019, 42, 1125–1132. [Google Scholar] [CrossRef]
- Rodríguez-Nunez, I.; Wcisel, D.J.; Litman, G.W.; Yoder, J.A. Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: Deciphering the differences. Dev. Comp. Immunol. 2014, 46, 24–34. [Google Scholar] [CrossRef]
- Piewbang, C.; Tattiyapong, P.; Techangamsuwan, S.; Surachetpong, W. Tilapia lake virus immunoglobulin G (TiLV IgG) antibody: Immunohistochemistry application reveals cellular tropism of TiLV infection. Fish Shellfish Immunol. 2021, 116, 115–123. [Google Scholar] [CrossRef]
- Wu, L.; Gao, A.; Li, L.; Chen, J.; Li, J.; Ye, J. A single-cell transcriptome profiling of anterior kidney leukocytes from Nile tilapia (Oreochromis niloticus). Front. Immunol. 2021, 12, 783196. [Google Scholar] [CrossRef]
- Yoder, J.A.; Mueller, M.G.; Wei, S.; Corliss, B.C.; Prather, D.M.; Willis, T.R.; Litman, T.; Djeu, J.Y.; Litman, G.W. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc. Natl. Acad. Sci. USA 2001, 98, 6771–6776. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.C.; Yang, H.T.; Li, J.; Sun, J.J.; Bi, W.J.; Niu, G.J.; Zhang, Q.; Shi, X.Z.; Zhao, X.F.; Wang, J.X. Scavenger receptor C promotes bacterial clearance in kuruma shrimp Marsupenaeus japonicus by enhancing hemocyte phagocytosis and AMP expression. Fish Shellfish Immunol. 2017, 67, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Li, X.; Yang, L.; Wang, Q.; Li, W. A class B scavenger receptor mediates antimicrobial peptide secretion and phagocytosis in Chinese mitten crab (Eriocheir sinensis). Dev. Comp. Immunol. 2020, 103, 103496. [Google Scholar] [CrossRef] [PubMed]
- Chaikeeratisak, V.; Somboonwiwat, K.; Tassanakajon, A. Shrimp alpha-2-macroglobulin prevents the bacterial escape by inhibiting fibrinolysis of blood clots. PLoS ONE 2012, 7, e47384. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Lv, J.; Li, Y.; Wu, J.; Liu, P.; Gao, B. Comparative Transcriptome Analysis of the Response to Vibrio parahaemolyticus and Low-Salinity Stress in the Swimming Crab Portunus trituberculatus. Biology 2023, 12, 1518. https://doi.org/10.3390/biology12121518
Sun D, Lv J, Li Y, Wu J, Liu P, Gao B. Comparative Transcriptome Analysis of the Response to Vibrio parahaemolyticus and Low-Salinity Stress in the Swimming Crab Portunus trituberculatus. Biology. 2023; 12(12):1518. https://doi.org/10.3390/biology12121518
Chicago/Turabian StyleSun, Dongfang, Jianjian Lv, Yukun Li, Jie Wu, Ping Liu, and Baoquan Gao. 2023. "Comparative Transcriptome Analysis of the Response to Vibrio parahaemolyticus and Low-Salinity Stress in the Swimming Crab Portunus trituberculatus" Biology 12, no. 12: 1518. https://doi.org/10.3390/biology12121518




