Isolation of OsMetAP10, a Peptidase_M24 Superfamily Gene, Regulating Heading Date in Rice
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Population Construction and Genome Sequencing
2.3. MutMap+ Analysis and Phenotype Association Verification
2.4. Plasmid Construction and Transformation
2.5. Phylogenetic Analysis
2.6. Histochemical GUS (β-Glucuronidase) Staining
2.7. Diurnal Expression Analysis
2.8. Total RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
2.9. Transient Expression Assay in Rice Protoplasts
2.10. RNA-Seq Analysis
3. Results
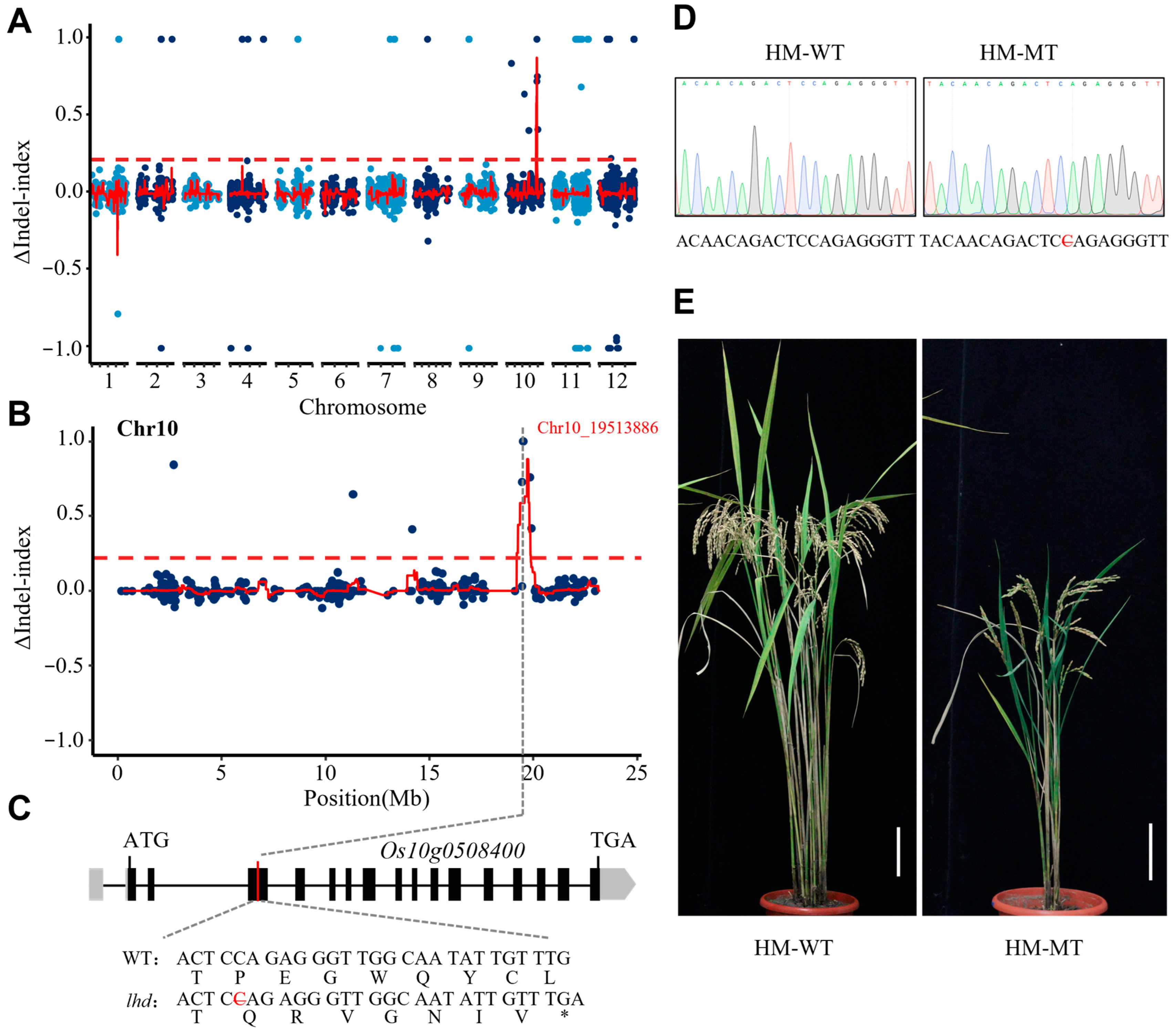
3.1. Identification of a Rice Late-Heading-Date Mutant
3.2. Gene Mapping and Candidate Gene Analysis for Late Flowering of lhd Mutant
3.3. Phylogenetic Analysis for Os10g0508400 Homologous Genes
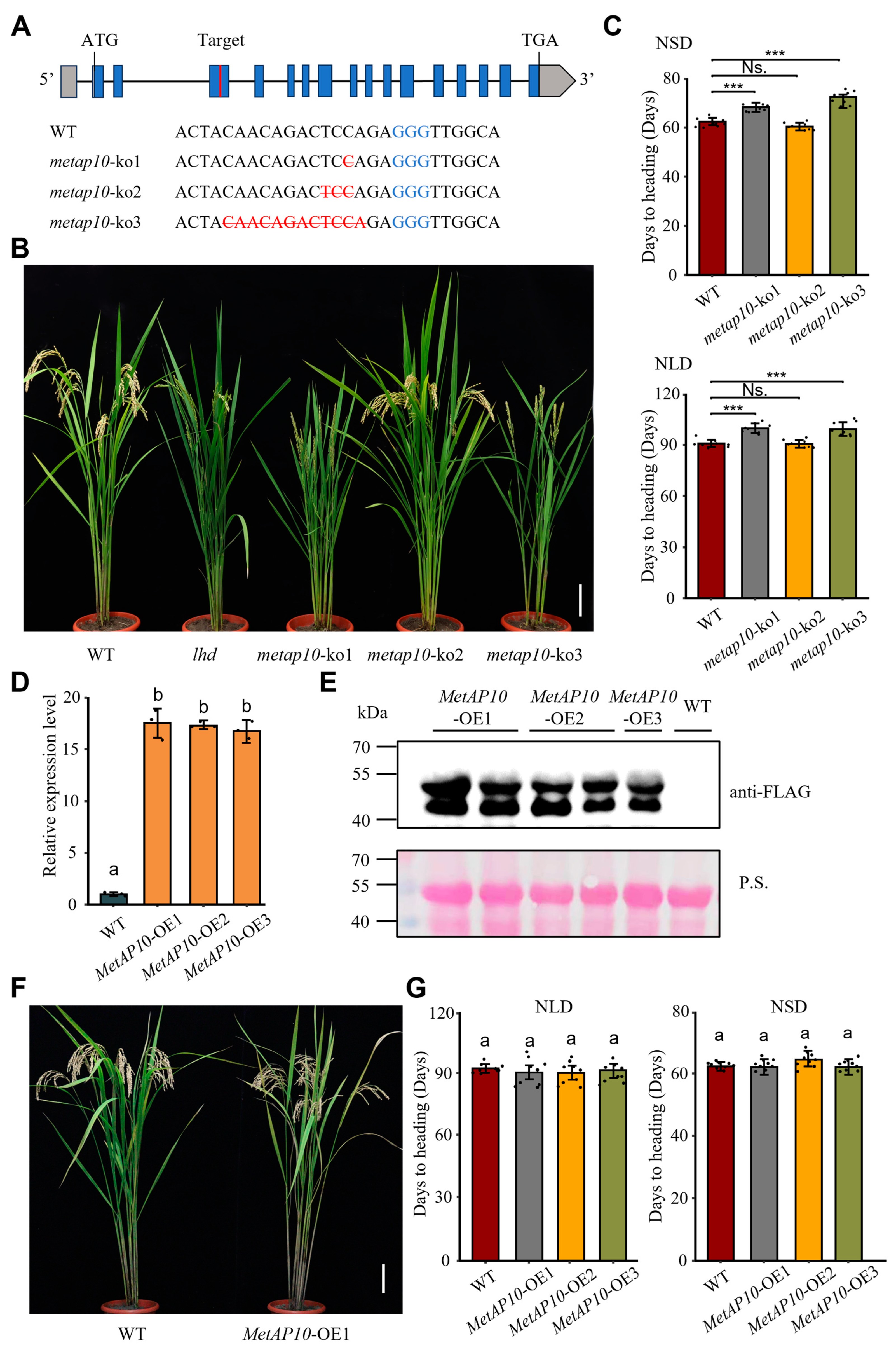
3.4. OsMetAP10 Mutation Accounts for the Late Heading and Inferior Performance of lhd Mutant
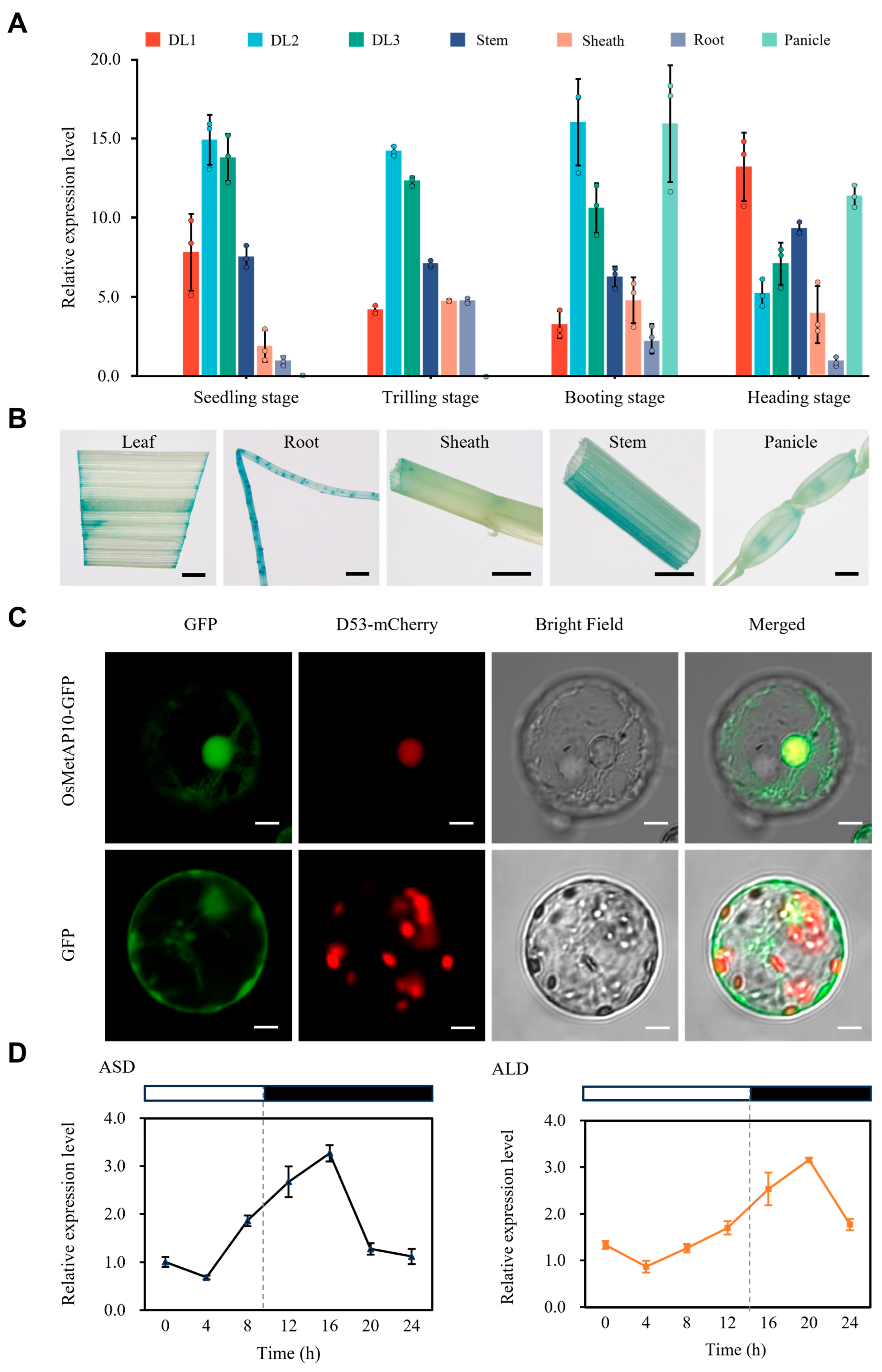
3.5. Spatiotemporal Expression and Subcellular Localization of OsMetAP10 Protein
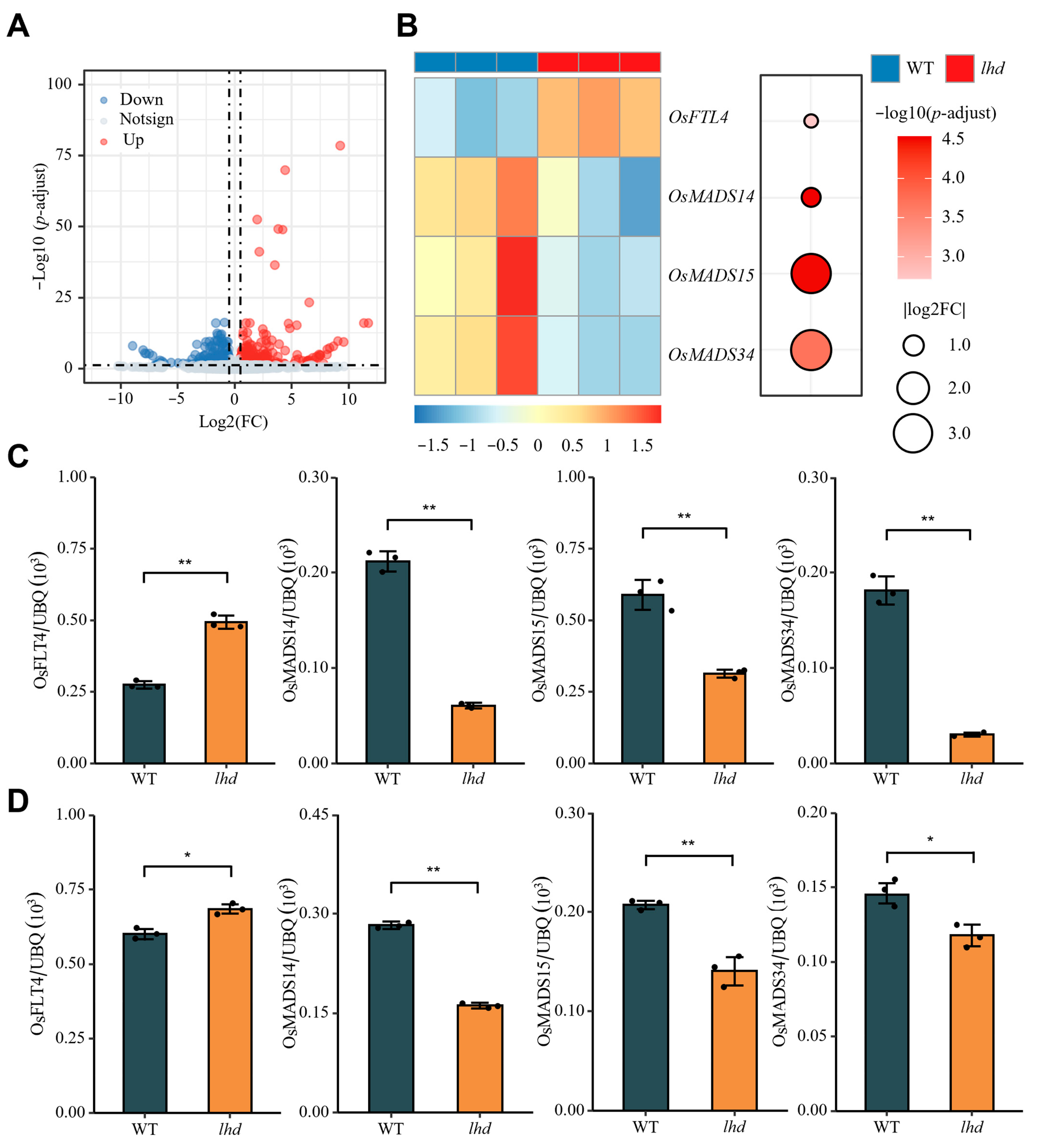
3.6. Analysis of Downstream Signals of OsMetAP10 Mutation Leading to Late Heading
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ND | Natural day |
| LD | Long day |
| SD | Short day |
| FT | FLOWERING LOCUS T |
| BVP | Basic vegetative growth phase |
| Hd3a | Heading date 3a |
| RFT1 | RICE FLOWERING LOCUS T1 |
| SAM | Shoot apical meristem |
| FAC | Flowering activation complex |
| OsGI | OsGIGANTEA |
| Hd1 | Heading date 1 |
| GI | GIGANTEA |
| CO | CONSTANS |
| DTH8 | Days to heading on chromosome 8 |
| Ghd7 | Grain number, plant height, and heading date 7 |
| Ehd1 | Early heading date 1 |
| OsCOL4 | OsCONSTANS-LIKE 4 |
| MAP | Methionine aminopeptidase |
| NME | N-terminal methionine (Met) excision |
| CLM38 | Calmodulin-like protein 38 |
| GUS | β-glucuronidase |
| lhd | Late-heading-date mutant |
| WT | Wild-type |
| NLD | Natural long day |
| NSD | Natural short day |
| ALD | Artificial long day |
| ASD | Artificial short day |
| HMM | Hidden Markov model |
References
- Zhang, X.; Feng, Q.; Miao, J.; Zhu, J.; Zhou, C.; Fan, D.; Lu, Y.; Tian, Q.; Wang, Y.; Zhan, Q.; et al. The WD40 Domain-Containing Protein Ehd5 Positively Regulates Flowering in Rice (Oryza sativa). Plant Cell 2023, 35, 4002–4019. [Google Scholar] [CrossRef]
- Chang, T.-T.; Li, C.-C.; Vergara, B.S. Component Analysis of Duration from Seeding to Heading in Rice by the Basic Vegetative Phase and the Photoperiod-Sensitive Phase. Euphytica 1969, 18, 79–91. [Google Scholar] [CrossRef]
- Cai, M.; Zhu, S.; Wu, M.; Zheng, X.; Wang, J.; Zhou, L.; Zheng, T.; Cui, S.; Zhou, S.; Li, C.; et al. DHD4, a CONSTANS-like Family Transcription Factor, Delays Heading Date by Affecting the Formation of the FAC Complex in Rice. Mol. Plant 2021, 14, 330–343. [Google Scholar] [CrossRef]
- Komiya, R.; Ikegami, A.; Tamaki, S.; Yokoi, S.; Shimamoto, K. Hd3a and RFT1 Are Essential for Flowering in Rice. Development 2008, 135, 767–774. [Google Scholar] [CrossRef]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a Protein Is a Mobile Flowering Signal in Rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Tsuji, H.; Taoka, K.; Shimamoto, K. Regulation of Flowering in Rice: Two Florigen Genes, a Complex Gene Network, and Natural Variation. Curr. Opin. Plant Biol. 2011, 14, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 Proteins Act as Intracellular Receptors for Rice Hd3a Florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Taoka, K.; Shimamoto, K. Florigen in Rice: Complex Gene Network for Florigen Transcription, Florigen Activation Complex, and Multiple Functions. Curr. Opin. Plant Biol. 2013, 16, 228–235. [Google Scholar] [CrossRef]
- Pasriga, R.; Yoon, J.; Cho, L.-H.; An, G. Overexpression of RICE FLOWERING LOCUS T 1 (RFT1) Induces Extremely Early Flowering in Rice. Mol. Cells 2019, 42, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Zhu, C.; Liu, T.; Zhang, S.; Feng, S.; Wu, C. Phosphorylation of OsFD1 by OsCIPK3 Promotes the Formation of RFT1-Containing Florigen Activation Complex for Long-Day Flowering in Rice. Mol. Plant 2021, 14, 1135–1148. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Yi, J.; An, G. OsPhyA Modulates Rice Flowering Time Mainly through OsGI under Short Days and Ghd7 under Long Days in the Absence of Phytochrome B. Plant Mol. Biol. 2016, 91, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of Photoperiodic Control Pathways Produces Short-Day Flowering in Rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 Module Mediates Day-Length-Dependent Regulation of Rice Flowering. Mol. Plant 2017, 10, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H.; et al. Strong Photoperiod Sensitivity Is Controlled by Cooperation and Competition among Hd1, Ghd7 and DTH8 in Rice Heading. New Phytol. 2021, 229, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural Variation in Ghd7 Is an Important Regulator of Heading Date and Yield Potential in Rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-Type Response Regulator in Rice, Confers Short-Day Promotion of Flowering and Controls FT-like Gene Expression Independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Yamanouchi, U.; Wang, Z.-X.; Minobe, Y.; Izawa, T.; Yano, M. Ehd2, a Rice Ortholog of the Maize INDETERMINATE1 Gene, Promotes Flowering by Up-Regulating Ehd1. Plant Physiol. 2008, 148, 1425–1435. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, S.; Cui, S.; Hou, H.; Wu, H.; Hao, B.; Cai, L.; Xu, Z.; Liu, L.; Jiang, L.; et al. Transcriptional and Post-Transcriptional Regulation of Heading Date in Rice. New Phytol. 2021, 230, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1,a CONSTANS Ortholog in Rice, Functions as an Ehd1 Repressor through Interaction with Monocot-Specific CCT-Domain Protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef]
- Bazan, J.F.; Weaver, L.H.; Roderick, S.L.; Huber, R.; Matthews, B.W. Sequence and Structure Comparison Suggest That Methionine Aminopeptidase, Prolidase, Aminopeptidase P, and Creatinase Share a Common Fold. Proc. Natl. Acad. Sci. USA 1994, 91, 2473–2477. [Google Scholar] [CrossRef]
- Yan, L.-Y.; Guo, J.-G.; Zhang, X.; Liu, Y.; Xiong, X.-X.; Han, Y.-X.; Zhang, L.-L.; Zhang, X.-H.; Min, D.-H. Genome-Wide Analysis of the Peptidase M24 Superfamily in Triticum Aestivum Demonstrates That TaM24-9 Is Involved in Abiotic Stress Response. Int. J. Mol. Sci. 2022, 23, 6904. [Google Scholar] [CrossRef]
- Ross, S.; Giglione, C.; Pierre, M.; Espagne, C.; Meinnel, T. Functional and Developmental Impact of Cytosolic Protein N-Terminal Methionine Excision in Arabidopsis. Plant Physiol. 2005, 137, 623–637. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.; Qiang, X.; Li, X.; Li, X.; Zhu, S.; Wang, L.; Wang, Y.; Liao, H.; Luan, S.; et al. EBP1 Nuclear Accumulation Negatively Feeds Back on FERONIA-Mediated RALF1 Signaling. PLoS Biol. 2018, 16, e2006340. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.-J.; Shin, J.S.; Ok, S.H. Barley DNA-Binding Methionine Aminopeptidase, Which Changes the Localization from the Nucleus to the Cytoplasm by Low Temperature, Is Involved in Freezing Tolerance. Plant Sci. 2011, 180, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap+: Genetic Mapping and Mutant Identification without Crossing in Rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef]
- Xu, X.; Lu, L.; Zhu, B.; Xu, Q.; Qi, X.; Chen, X. QTL Mapping of Cucumber Fruit Flesh Thickness by SLAF-Seq. Sci. Rep. 2015, 5, 15829. [Google Scholar] [CrossRef]
- Jiang, M.; Hu, H.; Kai, J.; Traw, M.B.; Yang, S.; Zhang, X. Different Knockout Genotypes of OsIAA23 in Rice Using CRISPR/Cas9 Generating Different Phenotypes. Plant Mol. Biol. 2019, 100, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Walker, K.W.; Bradshaw, R.A. Chapter 335—Methionyl Aminopeptidase Type 1. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 1495–1500. ISBN 978-0-12-382219-2. [Google Scholar]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Zhu, S.; Li, C.; Wang, C.; Cai, M.; Zheng, X.; Zhou, L.; Zhang, H.; Sheng, P.; Wu, M.; et al. OsRE1 Interacts with OsRIP1 to Regulate Rice Heading Date by Finely Modulating Ehd1 Expression. Plant Biotechnol. J. 2021, 19, 300–310. [Google Scholar] [CrossRef]
- Nakagawa, M.; Shimamoto, K.; Kyozuka, J. Overexpression of RCN1 and RCN2, Rice TERMINAL FLOWER 1/CENTRORADIALIS Homologs, Confers Delay of Phase Transition and Altered Panicle Morphology in Rice. Plant J. 2002, 29, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zhang, K.; Chen, J.; Gull, S.; Chen, C.; Hou, Y.; Li, X.; Miao, J.; Zhou, Y.; Liang, G. OsFTL4, an FT-like Gene, Regulates Flowering Time and Drought Tolerance in Rice (Oryza sativa L.). Rice 2022, 15, 47. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Zhao, J.; Wang, G.; Wang, Y.; Zhu, Y.; Yan, Y.; Chen, Z.; Chen, Z.; Feng, Z.; Zuo, S. Isolation of OsMetAP10, a Peptidase_M24 Superfamily Gene, Regulating Heading Date in Rice. Biology 2025, 14, 178. https://doi.org/10.3390/biology14020178
Sun Q, Zhao J, Wang G, Wang Y, Zhu Y, Yan Y, Chen Z, Chen Z, Feng Z, Zuo S. Isolation of OsMetAP10, a Peptidase_M24 Superfamily Gene, Regulating Heading Date in Rice. Biology. 2025; 14(2):178. https://doi.org/10.3390/biology14020178
Chicago/Turabian StyleSun, Quanyi, Jianhua Zhao, Guangda Wang, Yu Wang, Yuntao Zhu, Yu Yan, Zihang Chen, Zongxiang Chen, Zhiming Feng, and Shimin Zuo. 2025. "Isolation of OsMetAP10, a Peptidase_M24 Superfamily Gene, Regulating Heading Date in Rice" Biology 14, no. 2: 178. https://doi.org/10.3390/biology14020178
APA StyleSun, Q., Zhao, J., Wang, G., Wang, Y., Zhu, Y., Yan, Y., Chen, Z., Chen, Z., Feng, Z., & Zuo, S. (2025). Isolation of OsMetAP10, a Peptidase_M24 Superfamily Gene, Regulating Heading Date in Rice. Biology, 14(2), 178. https://doi.org/10.3390/biology14020178





