CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. sgRNA Design for the Vacuolar Invertase and Polyphenol Oxidase 2 Genes of Solanum tuberosum cv. Atlantic and cv. Spunta
2.1.1. Vacuolar Invertase Gene (InvVac)
2.1.2. Polyphenol Oxidase 2 Gene (PPO2)
2.2. Protoplast Transfection and Plant Regeneration
2.3. Identification of Edited Lines and Sequencing Analysis
2.3.1. High-Resolution Fragment Analysis (HRFA)
2.3.2. Detection of CRISPR-Induced Mutations
2.4. Plant Growth Conditions and Tuber Harvesting
2.5. Fried Product Characterization
2.6. HPLC-Based Determination of Sucrose and Reducing Sugars
2.7. Enzymatic Browning and PPO Activity
2.8. Field Trial of Line 6A
2.9. Statistical Analyses
3. Results
3.1. Identification of Edited Lines and Sequencing Analysis
3.1.1. Single Editing for the Vacuolar Invertase Gene
3.1.2. Multi-Target Editing for the Vacuolar Invertase and Polyphenol Oxidase 2 Genes
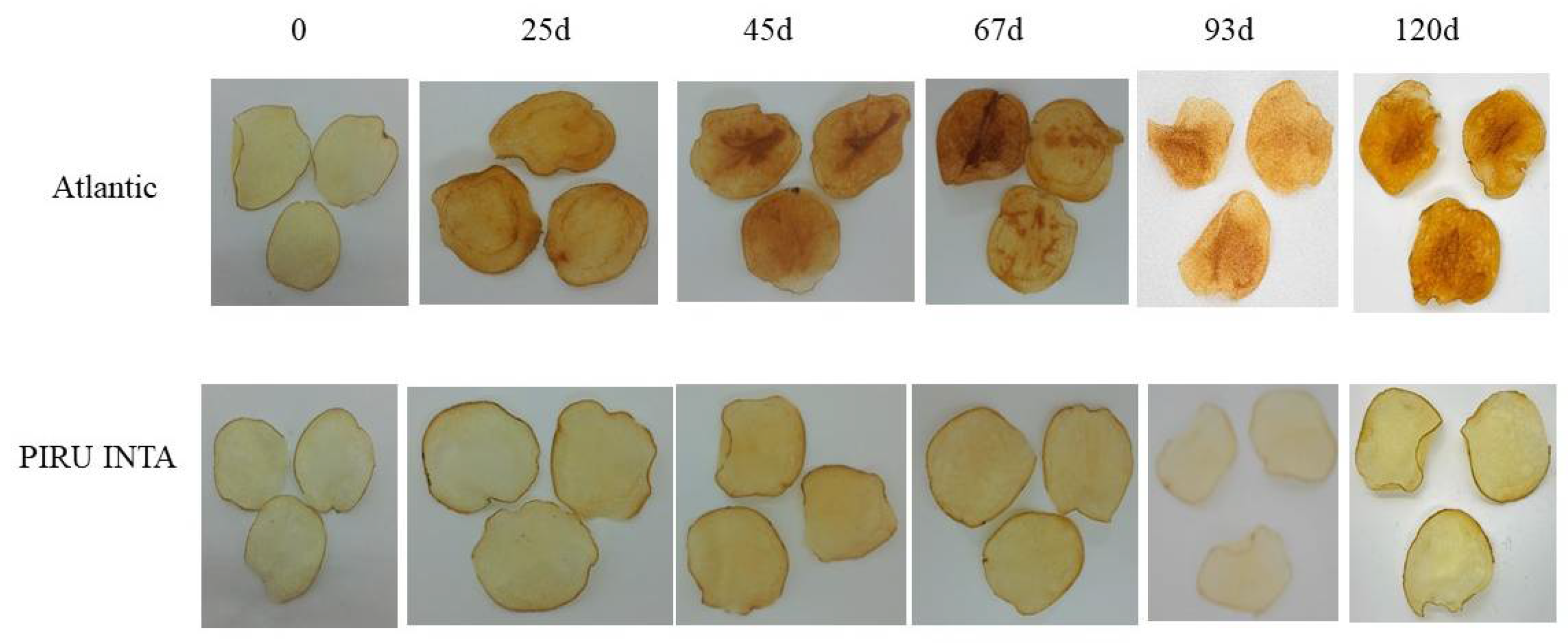
3.2. Tuber Production and Fried Product Characterization
3.3. HPLC-Based Determination of Sucrose and Reducing Sugars
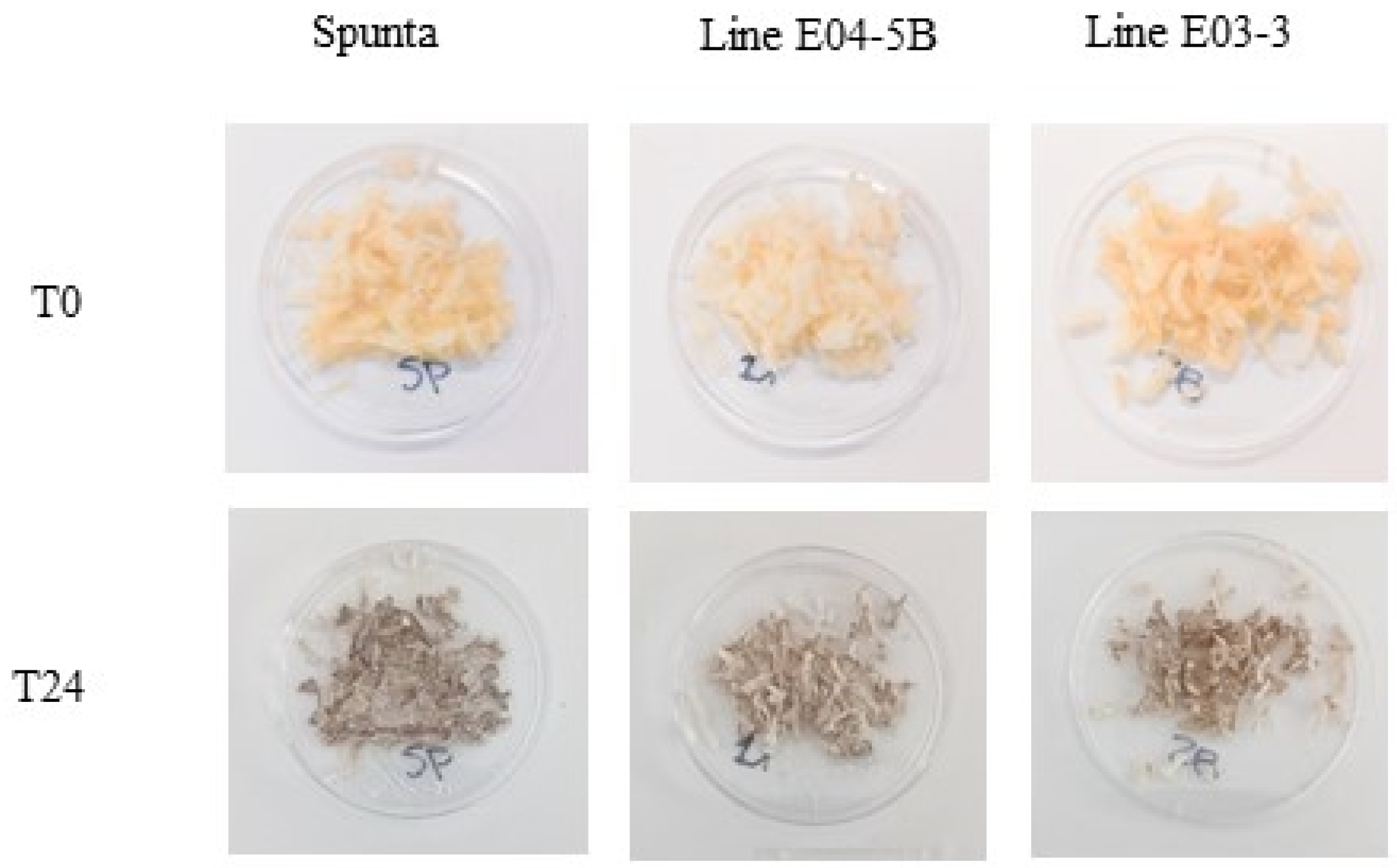
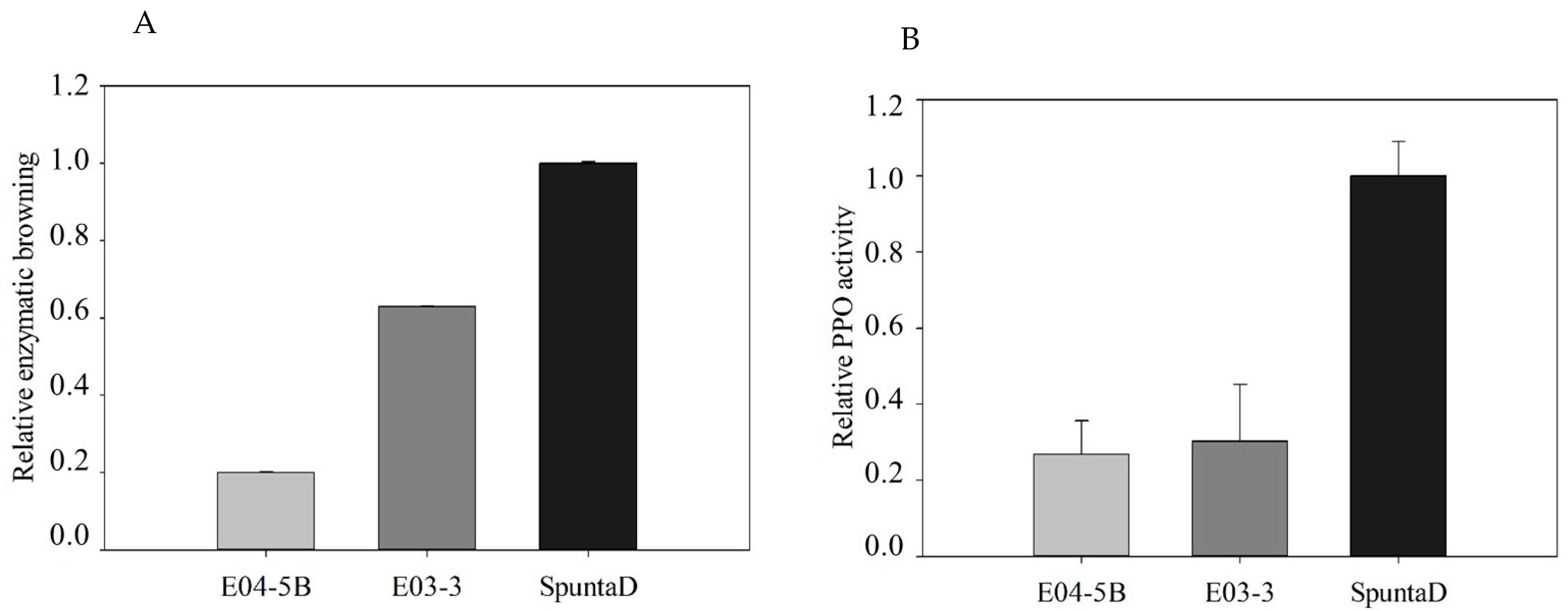
3.4. Enzymatic Browning and PPO Activity
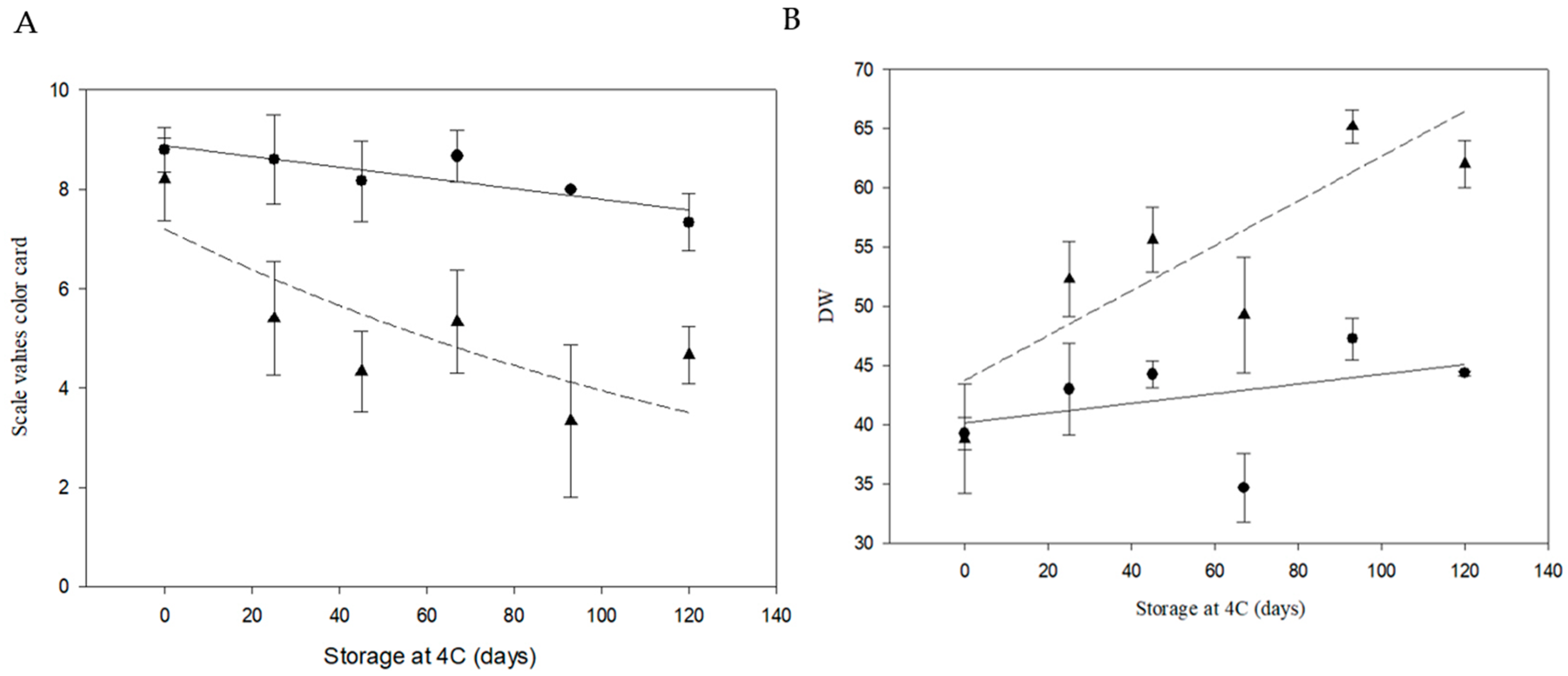
3.5. Field Trial of Line 6A Edited in the InvVac Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated nuclease 9 |
| GMO | Genetically modified organisms |
| RS | Reducing sugar |
| cv. | Cultivar |
| CIS | Cold-induced sweetening |
| PPO2 | Polyphenol oxidase 2 |
| InvVac | Vacuolar invertase |
| sgRNA | Single guide RNA |
| HPLC | High-performance liquid chromatography |
| HRFA | High-resolution fragment analysis |
| NGS | Next-generation sequencing |
| RNP | Ribonucleoprotein complex |
References
- Food and Agriculture Organization (FAO). The State of Food and Agriculture: Food Loss and Waste Reduction; FAO Report 2021; Food and Agriculture Organization (FAO): Rome, Italy, 2021. [Google Scholar]
- Birch, P.R.J.; Bryan, G.; Fenton, B.; Gilroy, E.M.; Hein, I.; Jones, J.T.; Prashar, A.; Taylor, M.A.; Torrance, L.; Toth, I.K. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 2012, 4, 477–508. [Google Scholar] [CrossRef]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A Review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef]
- Bhaskar, P.B.; Wu, L.; Busse, J.S.; Whitty, B.R.; Hamernik, A.J.; Jansky, S.H.; Buell, C.R.; Bethke, P.C.; Jiang, J. Suppression of the vacuolar invertase gene prevents cold induced sweetening in potato. Plant Physiol. 2010, 154, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Chefter, J.C.; Chefter, H. Introducción a la Bioquímica y Tecnología de Los Alimentos; Zaragoza: Acribia, Spain, 1976. [Google Scholar]
- Ohara-Takada, A.; Matsuura-Endo, C.; Chuda, Y.; Ono, H.; Yada, H.; Yoshida, M.; Kobayashi, A.; Tsuda, S.; Takigawa, S.; Noda, T.; et al. Change in Content of Sugars and Free Amino Acid in Potato Tubers under Short-Term Storage at Low Temperature and the Effect on Acrylamide Level after Frying. Biosci. Biotechnol. Biochem 2005, 69, 1232–1238. [Google Scholar] [CrossRef]
- Feingold, S.; Bonnecarrère, V.; Nepomuceno, A.; Hinrichsen, P.; Cardozo Tellez, L.; Molinari, H.; Barba, P.; Eyherabide, G.; Ceretta, S.; Dujack, C. Gene Editing: An Opportunity for the Region. Rev. Inv. Agrop. 2018, 44, 424–427. [Google Scholar]
- Nahirñak, V.; Almasia, N.I.; González, M.N.; Massa, G.A.; Décima Oneto, C.A.; Feingold, S.E.; Hopp, H.E.; Vazquez Rovere, C. State of the Art of Genetic Engineering in Potato: From the First Report to Its Future Potential. Front. Plant Sci. 2022, 12, 768233. [Google Scholar] [CrossRef] [PubMed]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Décima Oneto, C.A.; Turesson, H.; Storani, L.; Olsson, N.; Fält, A.-S.; Hofvander, P.; Feingold, S.E. Comparative potato genome editing: Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell Tiss. Organ Cult. 2021, 145, 291–305. [Google Scholar] [CrossRef]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Coffman, A. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotech. J. 2016, 14, 169–176. [Google Scholar] [CrossRef]
- Teper-Bamnolker, P.; Roitman, M.; Katar, O.; Peleg, N.; Aruchamy, K.; Suher, S.; Doron-Faigenboim, A.; Leibman, D.; Omid, A.; Belausov, E.; et al. Metabolic regulation of cold-induced sweetening in potato. Food Chem. 2023, 393, 133335. [Google Scholar]
- Ly, D.N.P.; Iqbal, S.; Fosu-Nyarko, J.; Milroy, S.; Jones MG, K. Multiplex genome editing of potato for improved post-harvest traits. Theo. Appl. Gen. 2023, 136, 1081–1094. [Google Scholar]
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Park, J.; Bae, S.; y Kim, J.-S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Starker, C.G.; Voytas, D.F.; Buell, C.R.; Douches, D.S. Genome editing in potato with CRISPR/Cas9. Plant genome editing with CRISPR systems. Methods Protoc. 2019, 664, 183–201. [Google Scholar]
- Haymes, K.M. Mini-prep method suitable for a plant breeding program. Plant Molecular. Biol. Rep. 1996, 14, 280–284. [Google Scholar]
- Inc, S.S. SigmaPlot for Windows (Development and Testing Procedures): Version 12.0; Triestram & Partner GmbH: Bochum, Germany, 2008. [Google Scholar]
- Colman, L.S.; Massa, G.A.; Carboni, M.F.; Feingold, S.E. Cold sweetening diversity in Andean potato germplasm from Argentina. J. Sci. Food Agric. 2017, 97, 4744–4749. [Google Scholar] [CrossRef]
- Yasmeen, A.; Shakoor, S.; Azam, S.; Bakhsh, A.; Shahid, N.; Latif, A.; Shahid, A.A.; Husnain, A.; Rao, A.Q. CRISPR/Cas-mediated knockdown of vacuolar invertase gene expression lowers the cold-induced sweetening in potatoes. Planta 2022, 256, 107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Li, Z.; Liu, C.; Wan, H. Roles and Regulations of Acid Invertases in Plants: Current Knowledge and Future Perspectives. Plants 2025, 14, 320. [Google Scholar] [CrossRef]
- Zhao, X.; Jayarathna, S.; Turesson, H.; Fält, A.-S.; Nestor, G.; González, M.N.; Olsson, N.; Beganovic, M.; Hofvander, P.; Andersson, R.; et al. Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 2021, 11, 4311. [Google Scholar] [CrossRef]
- Wszelaczyńska, E.; Wichrowska, D.; Pińska, M.; Rogozińska, I. Evaluation of Enzymatic browning degree of edible potato tubers induced by herbicides, mechanical damages and storage by means of instrumental and sensory methods. Pol. J. Food Nutr. Sci. 2007, 57, 163–166. [Google Scholar]
- Whelan, A.I.; Lema, M.A. Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food. 2015, 6, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Lema, M.A. Regulatory aspects of gene editing in Latin America. GMO Crops Food 2019, 658, 52–60. [Google Scholar]
- European Food Safety Authority (EFSA). Acrylamide in food: Health risks and mitigation strategies. EFSA J. 2020, 18, e06049. [Google Scholar]
- Zhu, X.; Chen, A.; Butler, N.M.; Zeng, Z.; Xin, H.; Wang, L.; Lv, Z.; Eshel, D.; Douches, D.S.; Jiang, J. Molecular dissection of an intronic enhancer governing cold-induced expression of the vacuolar invertase gene in potato. Plant Cell 2024, 36, 1985–1999. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Regulation (EU) 2020/1085 Concerning Maximum Residue Levels for Chlorpropham in or on Certain Products; Official Journal of the European Union: Luxembourg, 2020. [Google Scholar]
- Dushyant, D.; Roy, S.; Mahan, N.; Rani, D.V.; Kumar, R.; Gupta, D.K.; Sharma, K.K.; Kumar, M. Impact of Climate Change on Global Potato Production: A Review. J. Glob. Agric. Ecol. 2024, 16, 14–28. [Google Scholar] [CrossRef]
- Hoekstra, A.Y.; Mekonnen, M.M. The water footprint of humanity. Proc. Natl. Acad. Sci. USA 2021, 109, 3232–3237. [Google Scholar] [CrossRef]




| Primer Name | Sequence (5’–3’) | Purpose |
|---|---|---|
| InvVac-F1 | CAATTCAGTTGCCCCCTGTC | Sequence analysis of InvVac gene of Solanum tubersoum cv. Spunta and cv. Atlantic. |
| InvVac-R5 | CGCACGATTATTGTGTATGGTGCA | |
| sgRNAG0 | CCTCCCATTACACATTCCTC | sgRNA guide for InvVac. |
| sgRNAG1 | CTATTTGGGGAAATATCACA | sgRNA guide for InvVac. |
| sgRNAG4 | GAAGAAACAACGAAGAGTAC | sgRNA guide for InvVac. |
| sgRNAG10 | GGTCAAGTACAAAGGCAACC | sgRNA guide for InvVac. |
| sgRNA157 | TTTTCGATGTAACACGTGAC | sgRNA guide for PPO2 from González et al., 2020 [9]. |
| HRFAG0R-FAM | TCGGAAAGAAGGCTACAGAAAG | Amplification of InvVac gene fragment spanning the sgRNAG0 target site for HRFA and NGS. This primer was combined with InvVac-F1. |
| HRFAG4F-NED | TGGGTTGAAGCTGGATTATGG | Amplification of InvVac gene fragment spanning the sgRNAG4 target site for HRFA. This primer was combined with InvVac-R5. |
| HRFAG1R-VIC | ATCGTACCATTGATCAGGAACC | Amplification of InvVac gene fragment spanning the sgRNAG1 target site for HRFA. |
| InvVac-F4 | TTGGTCAACAGGTCCATTGT | |
| PPO2_2Bf | GCTCCATTTCGGTGACTTT | Amplification of PPO2 gene fragment spanning the sgRNA157 target site for NGS from González et al., 2020 [9]. |
| PPO2_2Br | TGGTGGCAAAGAGTTACAAG | |
| G2-R | TGGTTCCTGATCAATGGTAC | Amplification of InvVac gene fragment spanning the sgRNAG10 target site for NGS. |
| G3-R | GTCCAAGCAGTGGTGGGGTC |
| Cultivar | Line | Allelic Variants in Target Site sgRNAG0 by HRFA | Allelic Variants in Target Site sgRNAG4 by HRFA | Allelic Variants in Target Site sgRNAG0 by NGS | Allelic Variants in Target Site sgRNAG4 by NGS |
|---|---|---|---|---|---|
| Spunta | 37S | −2; −5; −6; −12 | 0 | −2; −5; −6; −12 | 0 |
| 38S | −3 | −2; −3; −5 | ND | −2; −3; −5 | |
| 44S | −2; 0 | 0 | ND | ND | |
| 75S | −1; −3; −7 | 0 | −1; −3; −7 | 0 | |
| Atlantic | 6A | +2; −2; −4 | 0 | +1; −2 | 0 |
| 13A | −3; −4 | −6; −12; −28; 0 | ND | ND | |
| 16A | 0; −3; −6 | 0 | ND | ND | |
| 30A | 0; −3 | 0 | ND | ND | |
| 38A | +1; −1 | 0 | +1; −1; 0 | 0 |
| Line | Allelic Variants G0 | Allelic Variants G157 | Line | Allelic Variants G10 | Allelic Variants G157 |
|---|---|---|---|---|---|
| E03-2A | −1; 0 | −2; 0 | E04-2A | −1; −1; 0 | −2; 0 |
| E03-3 | +3; −5; −4; −2 | −2; –1; 0 | E04-2B | 0 | −1; −1; 0 |
| E03-4 | 0 | 2; 0 | E04-3 | −1; 0 | NR |
| E03-5A | 0 | −2; 0 | E04-4A | −1; −1; 0 | −2; 0 |
| E03-5B | 0 | −2; 0 | E04-4B | −1; −1; 0 | −2; 0 |
| E03-6A | −2; −1; 0 | −2; 0 | E04-4C | −1; 0 | NR |
| E03-7A | 0 | −2; 0 | E04-4D | 0 | −2; −1; 0 |
| E03-7B | −1; 0 | −2; 0 | E04-5A | +1; −1; 0 | +1; −1; −2; 0 |
| E03-8A | NR | −2; 0 | E04-5B | +1; −1; 0 | +1; −1; −2; 0 |
| E03-8D | 0 | −2; 0 | E04-5C | +1; −1; 0 | +1; −1; −2; 0 |
| E03-10B | NR | −2; 0 | E04-5D | +1; −1; 0 | NR |
| E03-11C | 0 | −2; −47; −49; 0 | E04-6A | 0 | −2; 0 |
| E03-14B | 0 | −2; −49; −47; 0 | E04-6B | −2; 0 | −2; 0 |
| E03-15C | −1; 0 | −2; −35; −49; 0 | E04-6C | 0 | −2; 0 |
| E03-17B | 0 | −2; −14; 0 | E04-6D | −1; 0 | −2; −1; 0 |
| E03-17D | 0 | −2; 0 | E04-6E | 0 | −2; 0 |
| E03-20B | −1; −2; 0 | −2; 0 | E04-6F | −1; 0 | −2; 0 |
| E03-20C | 0 | −2; −1; 0 | E04-7A | 0 | −2; 0 |
| E03-21B | 0 | −2; −5; 0 | E04-7B | −1; 0 | −2; −1; 0 |
| E03-28 | 0 | −2; 0 | E04-8A | 0 | −2; −1; 0 |
| E03-29 | 0 | −2; 0 | E04-8B | 0 | −1; 0 |
| E03-33B | 0 | NR | E04-8C | −1; 0 | −2; −1; 0 |
| E03-34 | 0 | −2; 0 |
| Line | Storage at 4 °C (Days) | Average Scale Values of Color Card | Average DW | Average Reducing Sugar (mg/gr. FW) | Average Sucrose (mg/gr. FW) |
|---|---|---|---|---|---|
| Atlantic | 0 | 8.8 ± 0.45 A | 36.6 ± 3.4 A | 0.6 ± 0.02 A | 2.6 ± 0.75 A |
| 15 | 4 ± 0 B | 52.1 ± 3.5 B | 7.2 ± 0.69 B | 6.1 ± 0.46 B | |
| 6A | 0 | 8.3 ± 0.5 A | 44.6 ± 1.7 A | 0.4 ± 0.01 A | 7.7 ± 1.75 A |
| 15 | 8.3 ± 0.96 A | 41.9 ± 3.5 A | 0.7 ± 0.18 A | 3.5 ± 0.27 B | |
| 60 | 8 ± 0.71 A | 42.1 ± 4.9 A | 0.9 ± 0.03 A | 7.8 ± 1.39 A | |
| 13A | 0 | 8.3 ± 0.96 A | 41.7 ± 2.7 A | 0.2 ± 0.07 A | 1.8 ± 1.13 A |
| 15 | 6.3 ± 0.5 B | 47.3 ± 4.3 A | 1.8 ± 0.61 B | 4.7 ± 1.45 A | |
| 60 | 3 ± 0 C | 54.7 ± 4.7 B | 6.3 ± 1.33 C | 3.0 ± 0.59 A | |
| 38A | 0 | 8.8 ± 0.5 A | 36.2 ± 1.7 A | 0.1 ± 0.02 A | 1.1 ± 0.10 A |
| 15 | 8 ± 0.82 A | 45.2 ± 4.3 B | 0.8 ± 0.27 A | 5.5 ± 2.14 B | |
| 60 | 6.4 ± 0.55 B | 47.3 ± 1.3 B | 2.4 ± 0.52 B | 5.5 ± 2.20 B | |
| Spunta | 0 | 7.8 ± 1.5 A | 45.1 ± 4.0 A | 0.7 ± 0.04 A | 2.9 ± 0.87 A |
| 15 | 1.3 ± 0.5 B | 59.9 ± 2.2 B | 7.9 ± 0.58 B | 2.2 ± 0.31 A | |
| 37S | 0 | 7.3 ± 1.5 A | 47.9 ± 0.8 A | 2.7 ± 0.64 A | 2.7 ± 1.32 A |
| 15 | 2 ± 1.41 B | 63.5 ± 1.8 B | 6.8 ± 1.61 A | 7.7 ± 1.76 B | |
| 75S | 0 | 7.3 ± 1.5 A | 42.7 ± 6.4 A | 0.9 ± 0.05 A | 2.2 ± 0.42 A |
| 15 | 1.5 ± 1 B | 64.9 ± 0.2 B | 8.6 ± 0.55 A | 6.8 ± 0.20 B | |
| SpuntaD | 0 | 4.8 ± 0.4 A | 53.5 ± 1.84 A | 2.3 ± 0.26 A | 1.5 ± 0.55 A |
| 15 | 1.4 ± 0.5 B | 61.1 ± 0.96 B | 10 ± 2.61 A | 1.7 ± 0.15 A | |
| E04-5B | 0 | 6 ± 0 A | 45.9 ± 1.56 A | ND | ND |
| 15 | 4 ± 0.7 B | 57.6 ± 2.14 B | ND | ND | |
| E03-3 | 0 | 9 ± 0 A | 41.6 ± 3.97 A | 0.4 ± 0.03 A | 1.7 ± 0.36 A |
| 15 | 7 ± 0 B | 44.7 ± 2.32 A | 1.5 ± 0.89 A | 7.1 ± 0.03 B | |
| 60 | 5.3 ± 0.6 C | 58.1 ± 0.68 B | 2.9 ± 0.13 B | 9.1 ± 0.21 B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massa, G.A.; Décima Oneto, C.A.; González, M.N.; Poulsen Hornum, A.; Arizmendi, A.; Sucar, S.; Divito, S.B.; Feingold, S.E. CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance. Biology 2025, 14, 445. https://doi.org/10.3390/biology14040445
Massa GA, Décima Oneto CA, González MN, Poulsen Hornum A, Arizmendi A, Sucar S, Divito SB, Feingold SE. CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance. Biology. 2025; 14(4):445. https://doi.org/10.3390/biology14040445
Chicago/Turabian StyleMassa, Gabriela Alejandra, Cecilia Andrea Décima Oneto, Matías Nicolás González, Anabela Poulsen Hornum, Ailín Arizmendi, Sofía Sucar, Silvina Beatriz Divito, and Sergio Enrique Feingold. 2025. "CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance" Biology 14, no. 4: 445. https://doi.org/10.3390/biology14040445
APA StyleMassa, G. A., Décima Oneto, C. A., González, M. N., Poulsen Hornum, A., Arizmendi, A., Sucar, S., Divito, S. B., & Feingold, S. E. (2025). CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance. Biology, 14(4), 445. https://doi.org/10.3390/biology14040445








