Trimethylornithine Membrane Lipids: Discovered in Planctomycetes and Identified in Diverse Environments
Abstract
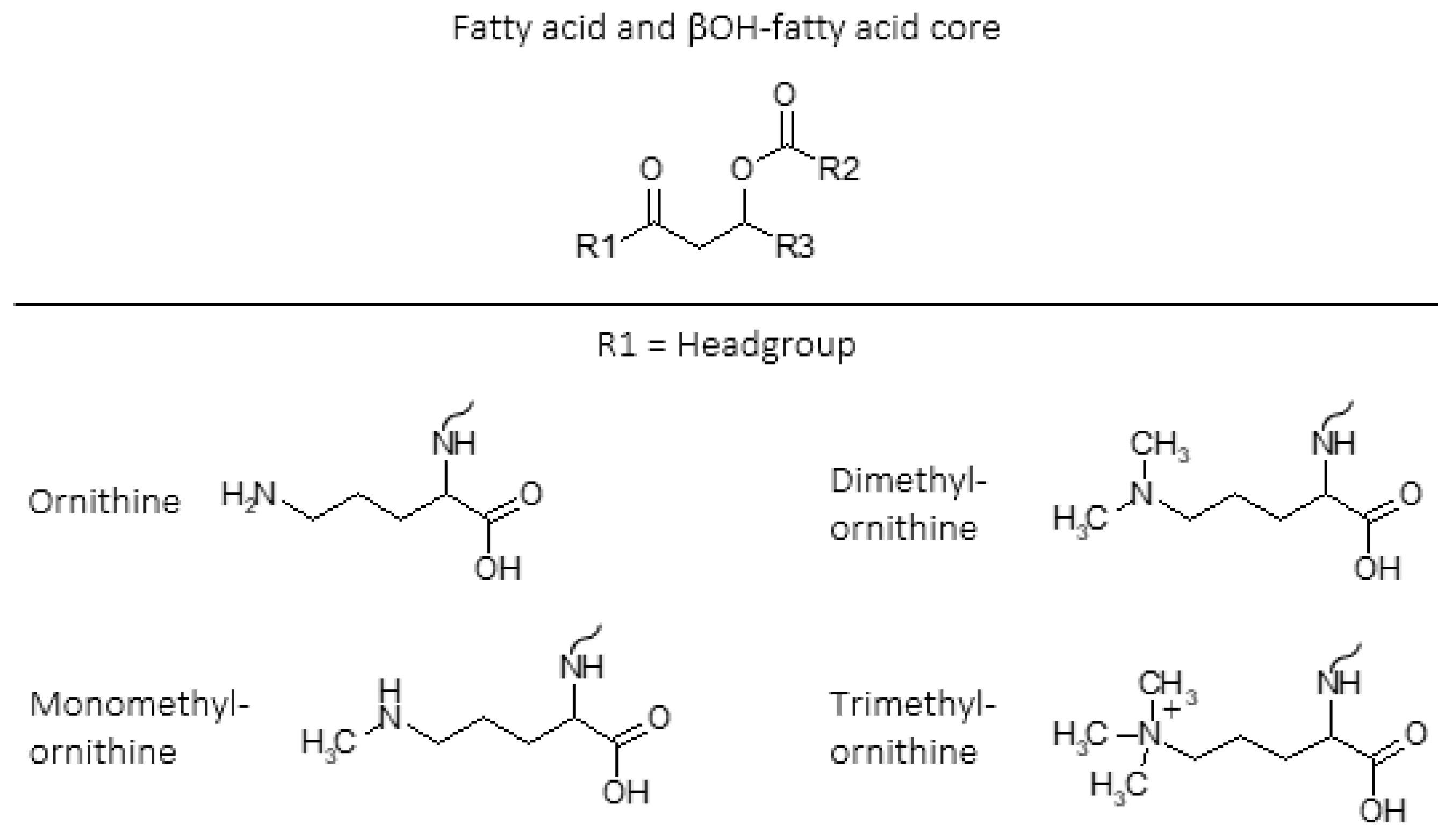
1. Introduction—Amino Acid Containing Membrane Lipids
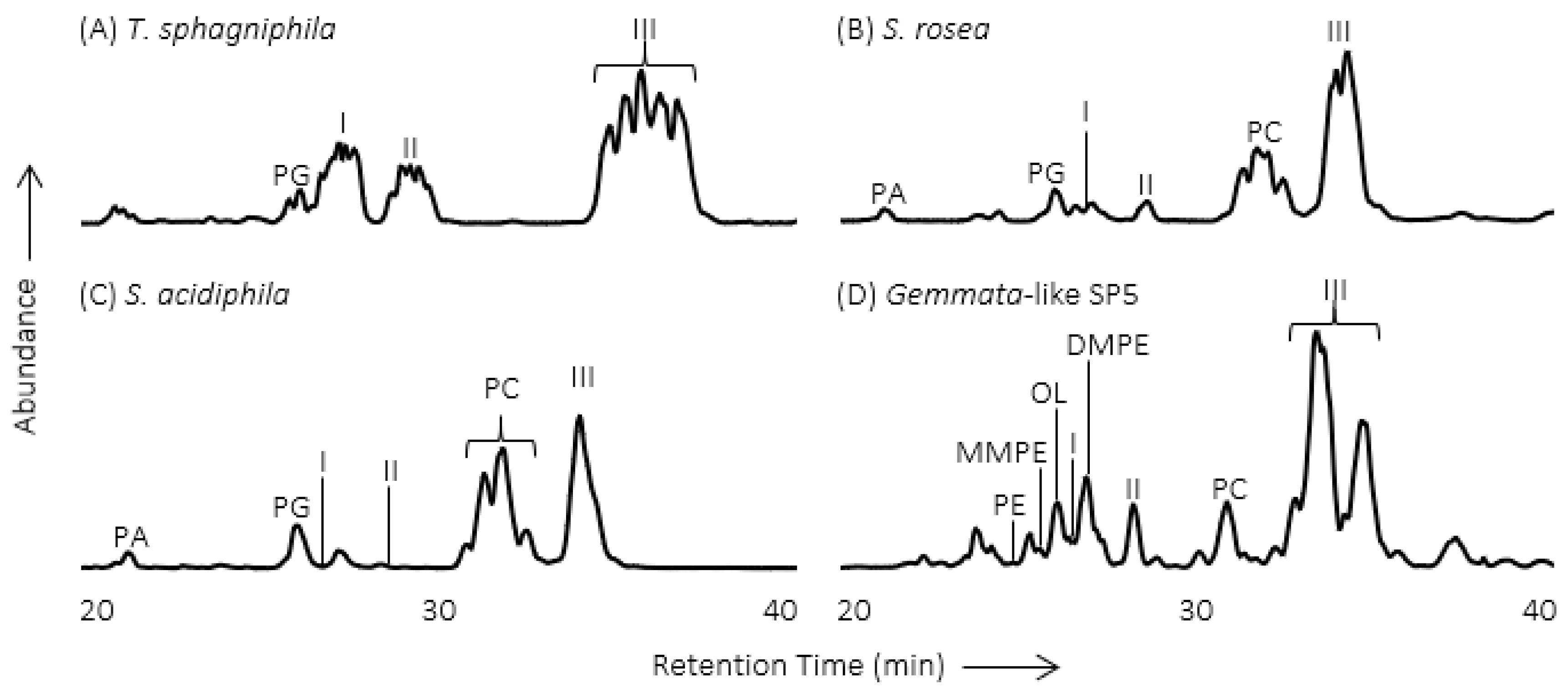
2. Methylated Ornithine Lipids Discovered in Northern Wetland Planctomycetes
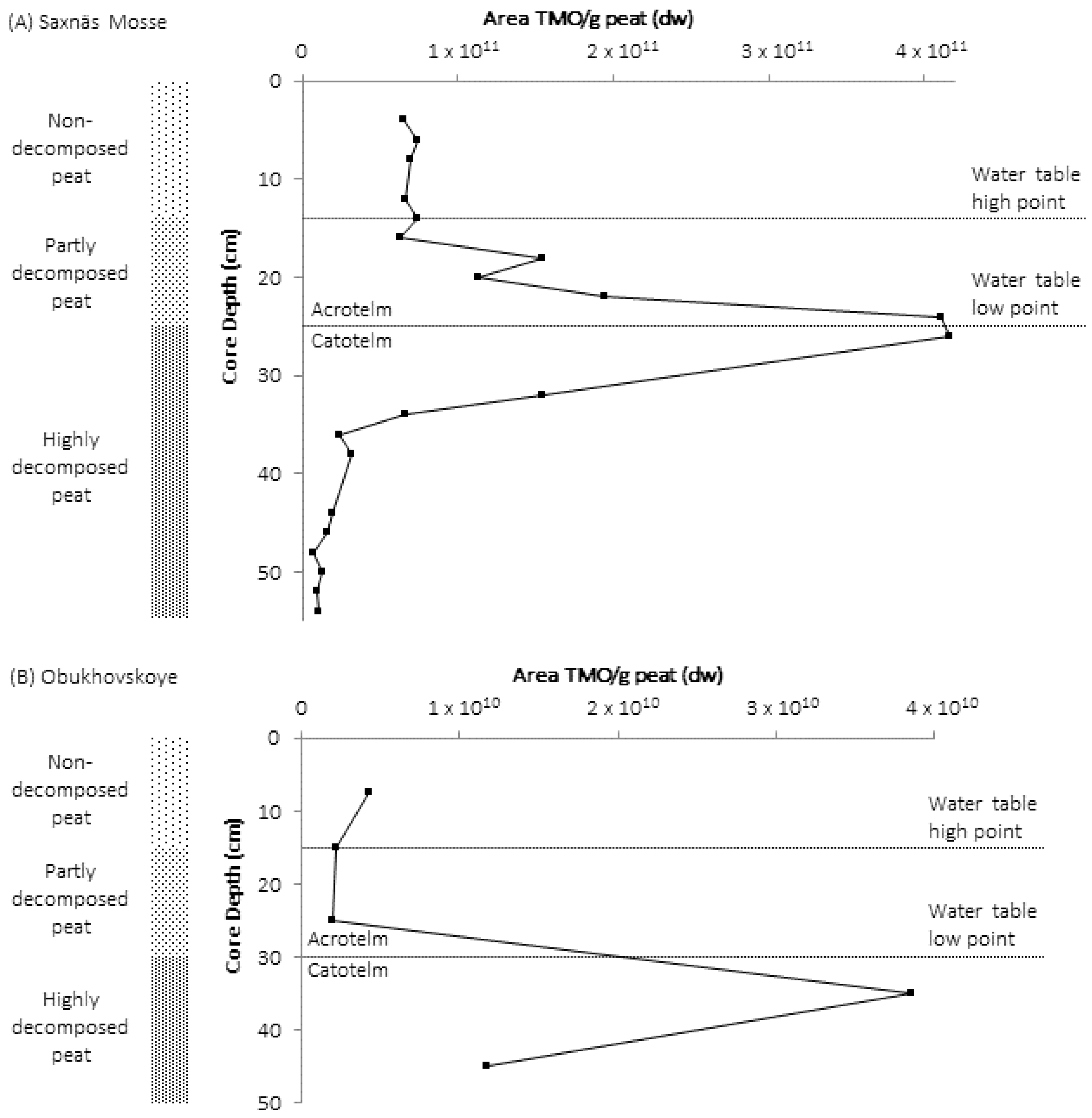
3. High Abundance of TMOs at the Oxic/Anoxic Interface of Northern Wetlands
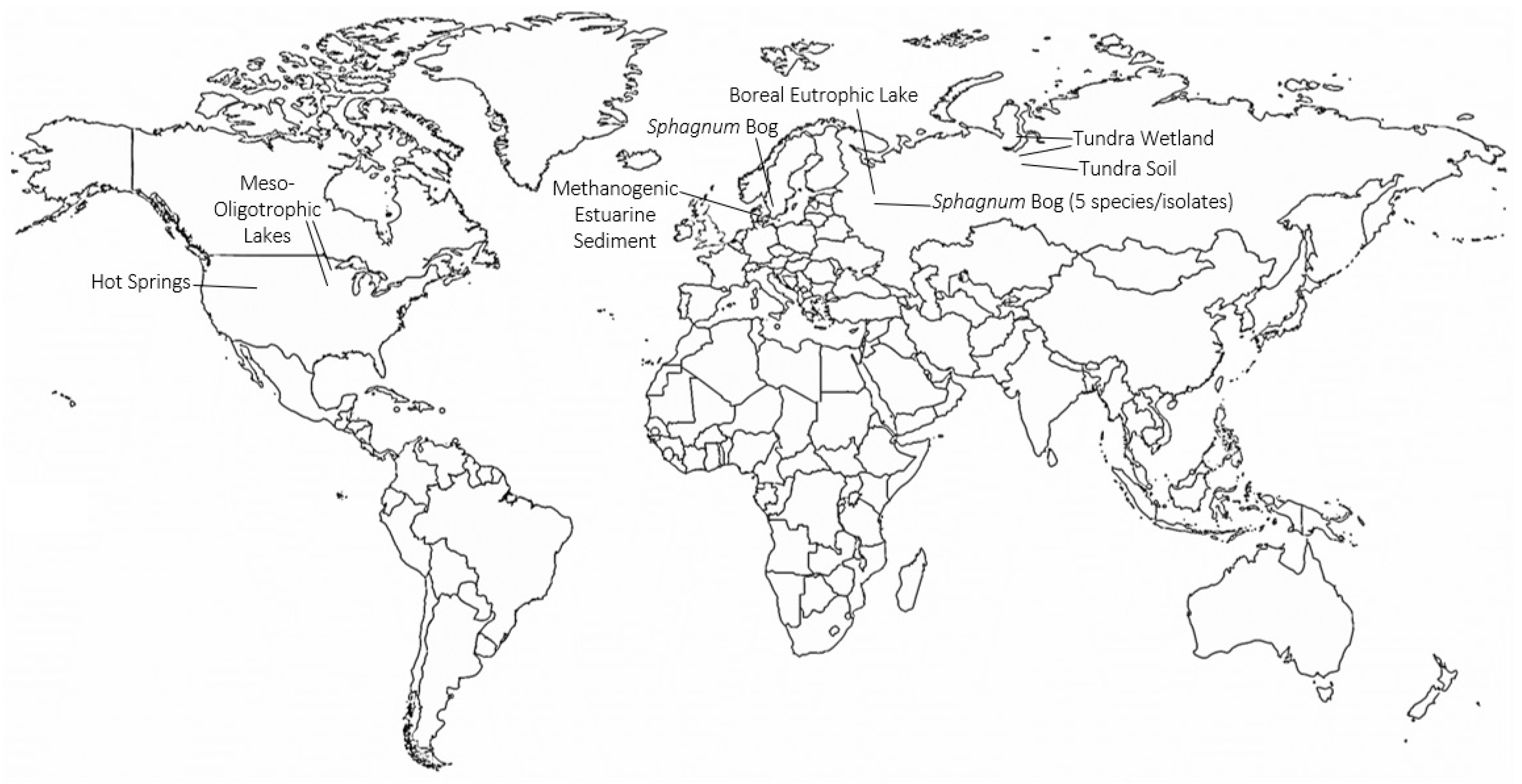
4. Identification of TMOs in Diverse Environments
5. TMOs: Specialized Lipids with Potential Broad Distribution
Funding
Acknowledgments
Conflicts of Interest
References
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000; ISBN 978-0-87893-106-4. [Google Scholar]
- Sturt, H.F.; Summons, R.E.; Smith, K.; Elvert, M.; Hinrichs, K.-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—New biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 2004, 18, 617–628. [Google Scholar] [CrossRef]
- Schubotz, F.; Wakeham, S.G.; Lipp, J.S.; Fredricks, H.F.; Hinrichs, K.-U. Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ. Microbiol. 2009, 11, 2720–2734. [Google Scholar] [CrossRef] [PubMed]
- White, D.C.; Davis, W.M.; Nickels, J.S.; King, J.D.; Bobbie, R.J. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 1979, 40, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Harvey, H.R.; Fallon, R.D.; Patton, J.S. The effect of organic matter and oxygen on the degradation of bacterial membrane lipids in marine sediments. Geochim. Cosmochim. Acta 1986, 50, 795–804. [Google Scholar] [CrossRef]
- Cevc, G. (Ed.) Phospholipids Handbook; CRC Press: Boca Raton, FL, USA, 1993. [Google Scholar]
- Geiger, O.; González-Silva, N.; López-Lara, I.M.; Sohlenkamp, C. Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 2010, 49, 46–60. [Google Scholar] [CrossRef]
- López-Lara, I.M.; Sohlenkamp, C.; Geiger, O. Membrane Lipids in Plant-Associated Bacteria: Their Biosyntheses and Possible Functions. Mol. Plant-Microbe Interact. 2003, 16, 567–579. [Google Scholar] [CrossRef]
- Vences-Guzmán, M.Á.; Geiger, O.; Sohlenkamp, C. Ornithine lipids and their structural modifications: From A to E and beyond. FEMS Microbiol. Lett. 2012, 335, 1–10. [Google Scholar] [CrossRef]
- Vences-Guzmán, M.Á.; Guan, Z.; Escobedo-Hinojosa, W.I.; Bermúdez-Barrientos, J.R.; Geiger, O.; Sohlenkamp, C. Discovery of a bifunctional acyltransferase responsible for ornithine lipid synthesis in Serratia proteamaculans. Environ. Microbiol. 2015, 17, 1487–1496. [Google Scholar] [CrossRef]
- Tahara, Y.; Yamada, Y.; Kondo, K. A New Lysine-containing Lipid Isolated from Agrobacterium tumefaciens. Agric. Biol. Chem. 1976, 40, 1449–1450. [Google Scholar] [CrossRef]
- Moore, E.K.; Hopmans, E.C.; Rijpstra, W.I.C.; Sanchez Andrea, I.; Villanueva, L.; Wienk, H.; Schoutsen, F.; Stams, A.; Sinninghe Damste, J. Lysine and novel hydroxylysine lipids in soil bacteria: Amino acid membrane lipid response to temperature and pH in Pseudopedobacter saltans. Front. Microbiol. 2015, 6, 637. [Google Scholar] [CrossRef]
- Hilker, D.R.; Gross, M.L.; Knocke, H.W.; Shively, J.M. The interpretation of the mass spectrum of an ornithine-containing lipid from Thiobacillus thiooxidans. Biomed. Mass Spectrom. 1978, 5, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Cerny, R.L.; Tomer, K.B.; Gross, M.L. Desorption ionization combined with tandem mass spectrometry: Advantages for investigating complex lipids, disaccharides and organometallic complexes. Org. Mass Spectrom. 1986, 21, 655–660. [Google Scholar] [CrossRef]
- Zhang, X.; Ferguson-Miller, S.M.; Reid, G.E. Characterization of ornithine and glutamine lipids extracted from cell membranes of Rhodobacter sphaeroides. J. Am. Soc. Mass Spectrom. 2009, 20, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.K.; Hopmans, E.C.; Rijpstra, W.I.C.; Villanueva, L.; Dedysh, S.N.; Kulichevskaya, I.S.; Wienk, H.; Schoutsen, F.; Damsté, J.S.S. Novel Mono-, Di-, and Trimethylornithine Membrane Lipids in Northern Wetland Planctomycetes. Appl. Environ. Microbiol. 2013, 79, 6874–6884. [Google Scholar] [CrossRef]
- Moore, E.K.; Hopmans, E.C.; Rijpstra, W.I.C.; Villanueva, L.; Damsté, J.S.S. Elucidation and identification of amino acid containing membrane lipids using liquid chromatography/high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 739–750. [Google Scholar] [CrossRef]
- Weissenmayer, B.; Gao, J.-L.; López-Lara, I.M.; Geiger, O. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 2002, 45, 721–733. [Google Scholar] [CrossRef]
- Gao, J.-L.; Weissenmayer, B.; Taylor, A.M.; Thomas-Oates, J.; López-Lara, I.M.; Geiger, O. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol. Microbiol. 2004, 53, 1757–1770. [Google Scholar] [CrossRef]
- Vences-Guzmán, M.Á.; Guan, Z.; Ormeño-Orrillo, E.; González-Silva, N.; López-Lara, I.M.; Martínez-Romero, E.; Geiger, O.; Sohlenkamp, C. Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 2011, 79, 1496–1514. [Google Scholar] [CrossRef]
- González-Silva, N.; López-Lara, I.M.; Reyes-Lamothe, R.; Taylor, A.M.; Sumpton, D.; Thomas-Oates, J.; Geiger, O. The Dioxygenase-Encoding olsD Gene from Burkholderia cenocepacia Causes the Hydroxylation of the Amide-Linked Fatty Acyl Moiety of Ornithine-Containing Membrane Lipids. Biochemistry 2011, 50, 6396–6408. [Google Scholar] [CrossRef]
- Gibbons, H.S.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. Oxygen Requirement for the Biosynthesis of theS-2-Hydroxymyristate Moiety in Salmonella typhimurium Lipid a function of LpxO, a new Fe2+/α-Ketoglutarate-Dependent Dioxygenase Homologue. J. Biol. Chem. 2000, 275, 32940–32949. [Google Scholar] [CrossRef]
- Kirschbaum, M.U.F. The temperature dependence of soil organic matter decomposition, and the effect of global warming on soil organic C storage. Soil Biol. Biochem. 1995, 27, 753–760. [Google Scholar] [CrossRef]
- Biasi, C.; Rusalimova, O.; Meyer, H.; Kaiser, C.; Wanek, W.; Barsukov, P.; Junger, H.; Richter, A. Temperature-dependent shift from labile to recalcitrant carbon sources of arctic heterotrophs. Rapid Commun. Mass Spectrom. 2005, 19, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Dorrepaal, E.; Toet, S.; van Logtestijn, R.S.P.; Swart, E.; van de Weg, M.J.; Callaghan, T.V.; Aerts, R. Carbon respiration from subsurface peat accelerated by climate warming in the subarctic. Nature 2009, 460, 616–619. [Google Scholar] [CrossRef]
- Lai, D.Y.F. Methane Dynamics in Northern Peatlands: A Review. Pedosphere 2009, 19, 409–421. [Google Scholar] [CrossRef]
- Gorham, E. Northern Peatlands: Role in the Carbon Cycle and Probable Responses to Climatic Warming. Ecol. Appl. 1991, 1, 182–195. [Google Scholar] [CrossRef]
- Bain, C.G.; Bonn, A.; Stoneman, R.; Chapman, S.; Coupar, A.; Evans, M.; Gearey, B.; Howat, M.; Joosten, H.; Keenleyside, C.; et al. IUCN UK Commission of Inquiry on Peatlands; IUCN UK Peatland Programme: Edinburgh, UK, 2011. [Google Scholar]
- Sundh, I.; Nilsson, M.; Granberg, G.; Svensson, B.H. Depth distribution of microbial production and oxidation of methane in northern boreal peatlands. Microb. Ecol. 1994, 27, 253–265. [Google Scholar] [CrossRef]
- Basiliko, N.; Henry, K.; Gupta, V.; Moore, T.; Driscoll, B.; Dunfield, P. Controls on bacterial and archaeal community structure and greenhouse gas production in natural, mined, and restored Canadian peatlands. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef]
- McCalley, C.K.; Woodcroft, B.J.; Hodgkins, S.B.; Wehr, R.A.; Kim, E.-H.; Mondav, R.; Crill, P.M.; Chanton, J.P.; Rich, V.I.; Tyson, G.W.; et al. Methane dynamics regulated by microbial community response to permafrost thaw. Nature 2014, 514, 478–481. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Pankratov, T.A.; Dedysh, S.N. Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Microbiology 2006, 75, 329–335. [Google Scholar] [CrossRef]
- Dedysh, S.N. Cultivating Uncultured Bacteria from Northern Wetlands: Knowledge Gained and Remaining Gaps. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- Serkebaeva, Y.M.; Kim, Y.; Liesack, W.; Dedysh, S.N. Pyrosequencing-Based Assessment of the Bacteria Diversity in Surface and Subsurface Peat Layers of a Northern Wetland, with Focus on Poorly Studied Phyla and Candidate Divisions. PLoS ONE 2013, 8, e63994. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Damsté, J.S.S. Acidobacteria. In eLS; American Cancer Society: Atlanta, GA, USA, 2018; pp. 1–10. ISBN 978-0-470-01590-2. [Google Scholar]
- Dedysh, S.N.; Ivanova, A.A. Planctomycetes in boreal and subarctic wetlands: Diversity patterns and potential ecological functions. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaia, I.S.; Belova, S.E.; Kevbrin, V.V.; Dedysh, S.N.; Zavarzin, G.A. Analysis of the bacterial community developing in the course of Sphagnum moss decomposition. Mikrobiologiia 2007, 76, 702–710. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Ivanova, A.O. Abundance, Diversity, and Depth Distribution of Planctomycetes in Acidic Northern Wetlands. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Bremer, J.; Greenberg, D.M. Methyl transfering enzyme system of microsomes in the biosynthesis of lecithin (phosphatidylcholine). Biochim. Biophys. Acta 1961, 46, 205–216. [Google Scholar] [CrossRef]
- Yamashita, S.; Oshima, A.; Nikawa, J.; Hosaka, K. Regulation of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae. Eur. J. Biochem. 1982, 128, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, P.M.; Gill, T.; Toutenhoofd, S.; Summers, E.F.; McGraw, P.; Homann, M.J.; Henry, S.A.; Carman, G.M. Regulation of phosphatidylethanolamine methyltransferase and phospholipid methyltransferase by phospholipid precursors in Saccharomyces cerevisiae. Biochim. Biophys. Acta Gene Struct. Expr. 1991, 1090, 326–332. [Google Scholar] [CrossRef]
- Escobedo-Hinojosa, W.I.; Vences-Guzmán, M.Á.; Schubotz, F.; Sandoval-Calderón, M.; Summons, R.E.; López-Lara, I.M.; Geiger, O.; Sohlenkamp, C. OlsG (Sinac_1600) Is an Ornithine LipidN-Methyltransferase from the PlanctomyceteSingulisphaera acidiphila. J. Biol. Chem. 2015, 290, 15102–15111. [Google Scholar] [CrossRef]
- Freer, E.; Moreno, E.; Moriyón, I.; Pizarro-Cerdá, J.; Weintraub, A.; Gorvel, J.P. Brucella-Salmonella lipopolysaccharide chimeras are less permeable to hydrophobic probes and more sensitive to cationic peptides and EDTA than are their native Brucella sp. counterparts. J. Bacteriol. 1996, 178, 5867–5876. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Ivanova, A.O.; Dedysh, S.N.; Liesack, W. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 2011, 13, 1800–1814. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Rütters, H.; Sass, H.; Cypionka, H.; Rullkötter, J. Phospholipid analysis as a tool to study complex microbial communities in marine sediments. J. Microbiol. Methods 2002, 48, 149–160. [Google Scholar] [CrossRef]
- Weijers, J.W.H.; Schouten, S.; van der Linden, M.; van Geel, B.; Sinninghe Damsté, J.S. Water table related variations in the abundance of intact archaeal membrane lipids in a Swedish peat bog. FEMS Microbiol. Lett. 2004, 239, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Peterse, F.; Hopmans, E.C.; Schouten, S.; Mets, A.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S. Identification and distribution of intact polar branched tetraether lipids in peat and soil. Org. Geochem. 2011, 42, 1007–1015. [Google Scholar] [CrossRef]
- Moore, E.K.; Villanueva, L.; Hopmans, E.C.; Rijpstra, W.I.C.; Mets, A.; Dedysh, S.N.; Damsté, J.S.S. Abundant Trimethylornithine Lipids and Specific Gene Sequences Are Indicative of Planctomycete Importance at the Oxic/Anoxic Interface in Sphagnum-Dominated Northern Wetlands. Appl. Environ. Microbiol. 2015, 81, 6333–6344. [Google Scholar] [CrossRef]
- Roslev, P.; King, G.M. Regulation of methane oxidation in a freshwater wetland by water table changes and anoxia. FEMS Microbiol. Ecol. 1996, 19, 105–115. [Google Scholar] [CrossRef]
- Angel, R.; Claus, P.; Conrad, R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012, 6, 847–862. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Suzina, N.E.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dedysh, S.N. Paludisphaera borealis gen. nov., sp. nov., a hydrolytic planctomycete from northern wetlands, and proposal of Isosphaeraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 837–844. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Detkova, E.N.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dedysh, S.N. Tundrisphaera lichenicola gen. nov., sp. nov., a psychrotolerant representative of the family Isosphaeraceae from lichen-dominated tundra soils. Int. J. Syst. Evol. Microbiol. 2017, 67, 3583–3589. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Baulina, O.I.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dedysh, S.N. Fimbriiglobus ruber gen. nov., sp. nov., a Gemmata-like planctomycete from Sphagnum peat bog and the proposal of Gemmataceae fam. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 218–224. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Naumoff, D.G.; Miroshnikov, K.K.; Ivanova, A.A.; Philippov, D.A.; Hakobyan, A.; Rijpstra, W.I.C.; Damsté, J.S.S.; Liesack, W.; Dedysh, S.N. Limnoglobus roseus gen. nov., sp. nov., a novel freshwater planctomycete with a giant genome from the family Gemmataceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Ivanova, A.A.; Naumoff, D.G.; Beletsky, A.V.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Mardanov, A.V.; Ravin, N.V.; Dedysh, S.N. Frigoriglobus tundricola gen. nov., sp. nov., a psychrotolerant cellulolytic planctomycete of the family Gemmataceae from a littoral tundra wetland. Syst. Appl. Microbiol. 2020, 43, 126129. [Google Scholar] [CrossRef] [PubMed]
- Bale, N.J.; Hopmans, E.C.; Schoon, P.L.; Kluijver, A.d.; Downing, J.A.; Middelburg, J.J.; Damsté, J.S.S.; Schouten, S. Impact of trophic state on the distribution of intact polar lipids in surface waters of lakes. Limnol. Oceanogr. 2016, 61, 1065–1077. [Google Scholar] [CrossRef]
- Ozuolmez, D.; Moore, E.K.; Hopmans, E.C.; Sinninghe Damsté, J.S.; Stams, A.J.M.; Plugge, C.M. Butyrate Conversion by Sulfate-Reducing and Methanogenic Communities from Anoxic Sediments of Aarhus Bay, Denmark. Microorganisms 2020, 8, 606. [Google Scholar] [CrossRef]
- Boyer, G.M.; Schubotz, F.; Summons, R.E.; Woods, J.; Shock, E.L. Carbon Oxidation State in Microbial Polar Lipids Suggests Adaptation to Hot Spring Temperature and Redox Gradients. Front. Microbiol. 2020, 11, 229. [Google Scholar] [CrossRef]
- Wörmer, L.; Gajendra, N.; Schubotz, F.; Matys, E.D.; Evans, T.W.; Summons, R.E.; Hinrichs, K.-U. A micrometer-scale snapshot on phototroph spatial distributions: mass spectrometry imaging of microbial mats in Octopus Spring, Yellowstone National Park. Geobiology 2020, 18, 742–759. [Google Scholar] [CrossRef]
- Jauhiainen, J.; Takahashi, H.; Heikkinen, J.E.P.; Martikainen, P.J.; Vasander, H. Carbon fluxes from a tropical peat swamp forest floor. Glob. Chang. Biol. 2005, 11, 1788–1797. [Google Scholar] [CrossRef]
- Hirano, T.; Segah, H.; Kusin, K.; Limin, S.; Takahashi, H.; Osaki, M. Effects of disturbances on the carbon balance of tropical peat swamp forests. Glob. Chang. Biol. 2012, 18, 3410–3422. [Google Scholar] [CrossRef]
- Kanokratana, P.; Uengwetwanit, T.; Rattanachomsri, U.; Bunterngsook, B.; Nimchua, T.; Tangphatsornruang, S.; Plengvidhya, V.; Champreda, V.; Eurwilaichitr, L. Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 2011, 61, 518–528. [Google Scholar] [CrossRef]
- Bowen, J.L.; Morrison, H.G.; Hobbie, J.E.; Sogin, M.L. Salt marsh sediment diversity: A test of the variability of the rare biosphere among environmental replicates. ISME J. 2012, 6, 2014–2023. [Google Scholar] [CrossRef]
- Osburn, C.L.; Mikan, M.P.; Etheridge, J.R.; Burchell, M.R.; Birgand, F. Seasonal variation in the quality of dissolved and particulate organic matter exchanged between a salt marsh and its adjacent estuary. J. Geophys. Res. Biogeosciences 2015, 120, 1430–1449. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, J.; Cui, L.; Feng, W.; Wang, Y.; Zhang, J. Soil organic carbon stabilization mechanisms in a subtropical mangrove and salt marsh ecosystems. Sci. Total Environ. 2019, 673, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Marsh, L.M.; Polcin, S. Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments. Nature 1982, 296, 143–145. [Google Scholar] [CrossRef]
- Metcalf, J.L.; Xu, Z.Z.; Bouslimani, A.; Dorrestein, P.; Carter, D.O.; Knight, R. Microbiome Tools for Forensic Science. Trends Biotechnol. 2017, 35, 814–823. [Google Scholar] [CrossRef]
- Langley, N.R.; Wood, P.; Herling, P.; Steadman, D.W. Forensic Postmortem Interval Estimation from Skeletal Muscle Tissue: A Lipidomics Approach. Forensic Anthropol. 2019, 2, 152–157. [Google Scholar] [CrossRef]
- Pechal, J.L.; Crippen, T.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Tomberlin, J.K. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Legal Med. 2014, 128, 193–205. [Google Scholar] [CrossRef]
- Howard, G.T.; Duos, B.; Watson-Horzelski, E.J. Characterization of the soil microbial community associated with the decomposition of a swine carcass. Int. Biodeterior. Biodegrad. 2010, 64, 300–304. [Google Scholar] [CrossRef]
- Cobaugh, K.L.; Schaeffer, S.M.; DeBruyn, J.M. Functional and Structural Succession of Soil Microbial Communities below Decomposing Human Cadavers. PLoS ONE 2015, 10, e0130201. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moore, E.K. Trimethylornithine Membrane Lipids: Discovered in Planctomycetes and Identified in Diverse Environments. Metabolites 2021, 11, 49. https://doi.org/10.3390/metabo11010049
Moore EK. Trimethylornithine Membrane Lipids: Discovered in Planctomycetes and Identified in Diverse Environments. Metabolites. 2021; 11(1):49. https://doi.org/10.3390/metabo11010049
Chicago/Turabian StyleMoore, Eli K. 2021. "Trimethylornithine Membrane Lipids: Discovered in Planctomycetes and Identified in Diverse Environments" Metabolites 11, no. 1: 49. https://doi.org/10.3390/metabo11010049
APA StyleMoore, E. K. (2021). Trimethylornithine Membrane Lipids: Discovered in Planctomycetes and Identified in Diverse Environments. Metabolites, 11(1), 49. https://doi.org/10.3390/metabo11010049




