Gamma-ray Irradiation of Rodent Diets Alters the Urinary Metabolome in Rats with Chemically Induced Mammary Cancer
Abstract
:1. Introduction
2. Experimental Design
Metabolomics Study
3. Results
3.1. Cancer Chemoprevention
3.2. Multivariate Statistical Analysis
3.3. Pathway Analysis
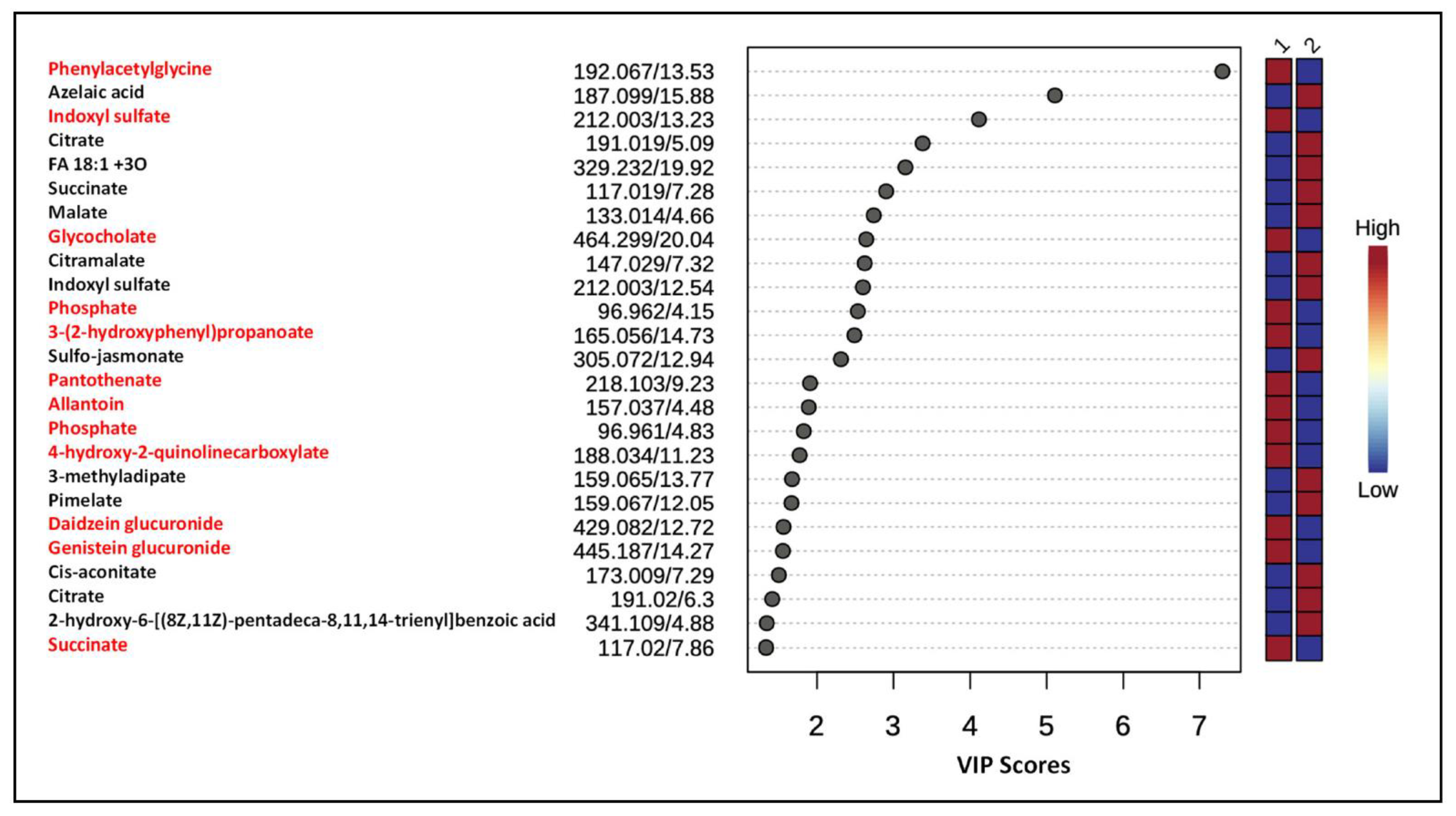
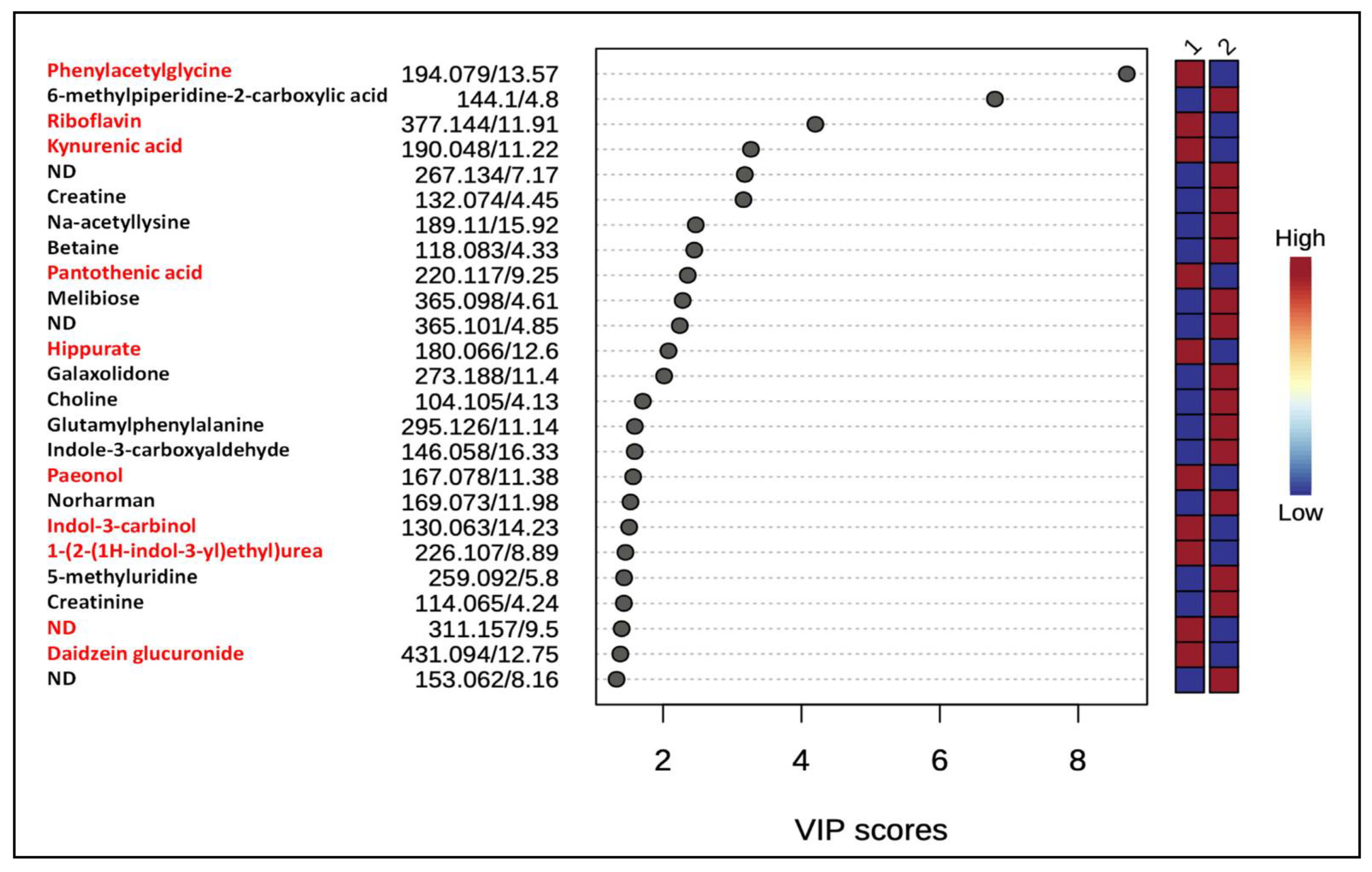
3.4. Differentially Expressed Metabolites
3.5. Metabolite Annotation
- Glycine conjugates
- m/z 194.079 [M + H]+, 192.065 [M-H]−, Rt 13.5 min
- MS/MS of m/z 194.079 contained intense fragment ions m/z 119.050 and 91.054 due to the neutral losses of 75.029 (NH2CH2COOH) and 103.025 Da (CONH2CH2COOH), respectively (Supplemental Figure S4). In addition, the characteristic product ion m/z 76.040 further indicated the presence of the glycine moiety. Based on the accurate mass and product ion pattern analyses, this metabolite was annotated as phenylacetylglycine. It is the most significantly altered and upregulated urinary metabolite among the top 25 identified by PLS-DA following chronic exposure to gID in animals.
- m/z 180.066 [M + H]+, Rt 12.6 min
- MS/MS spectrum of the precursor ion m/z 180.066 contained a product ion m/z 162.055 due to the neutral loss of H2O. The observation of an intense product ion m/z 105.031 with a neutral 75.035 Da loss from the precursor ion indicated the presence of glycine residue [24]. Based on these data, this metabolite ion was annotated as hippurate.
- m/z 170.041 [M + H]+, Rt 9.7 min
- Upon MS/MS fragmentation, the protonated precursor ion m/z 170.041 produced several product ions characteristic of N-acylglycine. A neutral loss of 75.029 Da from the precursor ion generated an intense product ion m/z 95.012. In addition, it also showed product ions m/z 152.034 and 124.039 due the losses of 18 Da (H2O) and 46 Da (HCOOH), respectively. Based on these data, this metabolite ion was annotated as N(2-furoyl)glycine. Glycine N-acyltransferase catalyzes the transfer of acyl group from the acyl-CoA to glycine in the mitochondria [25]. Although glycine can be endogenously formed, composition of an animal diet (irradiated vs. non-irradiated) may influence their levels.
- Glycine betaine
- m/z 118.083, Rt 4.3 min
- The precursor ion m/z 118.083 in the positive ion mode generated major product ions, m/z 58.069 and 59.076, with almost equal intensities (Supplemental Figure S5). These are characteristic product ions of betaine and correspond to C3H8N+ and (CH3)3N+ ions, respectively [26]. This metabolite was elevated in the gID group.
- Glycocholate
- m/z 464.299 [M−H]−, Rt 20.0 min
- MS/MS fragmentation of m/z 464.299 generated the product ions m/z 402.297 and 74.027, after neutral losses of 62.00 Da (H2O + CO2) and glycine moiety, respectively. Comparing these data with those of a published report [27], this metabolite was annotated as a glycine conjugate of cholic acid.
- Riboflavin (vitamin B2)
- m/z 377.146 [M + H]+, Rt 11.9 min
- MS/MS fragmentation of m/z 377.146 generated the product ions m/z 359.138, 341.124 and 243.087, characteristic of riboflavin (Supplemental Figure S6) [28]. Riboflavin was elevated in the urines of the gID group compared to nonID group.
- Pantothenic acid (vitamin B5)
- m/z 220.117 [M + H]+, Rt 9.25 min
- Its product ion m/z 184.095 occurs after the sequential losses of two H2O molecules. Product ions m/z 142.084, 124.074, 90.055 and 72.046 were also observed in the MS/MS spectrum (Supplemental Figure S7). The observed characteristic product ions m/z 90.055 and 72.046 corresponded to the β-alanine residue [29]. Based on this evidence, the structure of this metabolite is annotated as pantothenic acid. Bacteria in the gut can produce this vitamin and its increased levels in gID fed animals may be associated with breast cancer prevention [30].
- Glucuronide conjugates
- m/z 431.094 [M + H]+, Rt 12.7 min
- MS/MS spectrum of the precursor ion m/z 431.094 contained an intense product ion m/z 255.065 due to a neutral loss of 176.025 Da, indicating the presence of a glucuronic acid moiety [31]. In addition, the presence of product ions m/z 177.050 and 149.074 enabled annotation of this metabolite ion as daidzein glucuronide (cal. m/z 431.097). The NIH 7001 diet contains soy protein and therefore contributed expected glucuronide metabolites of the isoflavones, daidzein and genistein.
- Creatine
- m/z 132.074 [M + H]+, Rt 4.4 min
- In an MS/MS experiment, the precursor ion m/z 132.074 generated prominent product ions m/z 114.064 and 90.055 due to neutral losses of H2O and NHCNH (42.02 Da), respectively (Supplemental Figure S8). Comparison of these product ion features with previously published data [32] enabled the annotation of this metabolite ion as creatine (cal. m/z 132.077).
- Creatinine
- m/z 114.065 [M + H]+, Rt 4.2 min
- This precursor ion m/z 114.065 had the product ion m/z 86.073 after the neutral loss of CO (27.995 Da). In addition, the product ion m/z 72.046 is characteristic of metabolites with a guanidino group [32]. These data allowed annotation of this metabolite as creatinine (cal. m/z 114.066).
- Kynurenic acid
- m/z 190.047 [M + H]+, Rt 11.2 min
- The precursor ion 190.047 generated the product ions m/z 172.027 and 162.055 following the neutral losses of H2O and CO, respectively. The intense product ion m/z 144.042 was due to loss of 46.005 Da (H2O + CO2). Other product ions included m/z 116.051, 89.040, and 70.070 (Supplemental Figure S9). These characteric features in the MS/MS spectrum of m/z 190.047 allowed annotation of this metabolite ion as kynurenic acid. In this study, gID-fed animals had increased levels of this metabolite in their urine.
- Indoxyl-sulfate
- m/z 212.003 [M−H]−, 13.2 min
- Its precursor ion generated the major product ions m/z 132.046, 112.040, and 79.958. The product ion m/z 79.958 corresponds to a sulfate group attached to aromatic ring in the molecule. Indoxyl-sulfate is a gut microbial metabolite of tryptophan produced from sulfonation of indole by hepatic sulfotransferase [33].
- Citramalate
- m/z 147.029 [M−H]−, Rt 7.3 min
- Upon dissociation, it had a series of product ions m/z 129.019, 101.026, 103.054, and 85.030 due to neutral losses of H2O, HCOOH, CO2, and H2O + CO2, respectively. Based on accurate mass measurement, and comparison of experimental MS/MS fragmentation spectrum with the published report [34], the peak Rt 7.3 min with m/z 147.029 is annotated as citramalate (Supplemental Figure S10). The level of citramalate is higher in samples derived from animals exposed to gID, compared to non-irradiated diet fed animals.
- TCA cycle intermediates
- MS/MS of the precursor ion m/z 117.018 ([M−H]−, Rt 7.2 min) contained the prominent product ion m/z 73.031 along with m/z 99.010. The product ion m/z 73.031 is considered as a characteristic product ion of succinate.
- The precursor ion m/z 133.013 ([M−H]−, Rt 4.6 min) produced the major product ions m/z 115.003, 89.024, 71.016, and 59.019. The product ions m/z 115.003 and 89.024 are due to neutral losses of 18.010 (H2O) and 43.990 (CO2), respectively. These fragmention patterns correspond to malate.
- Similarly, MS/MS of the precursor ion m/z 191.021 ([M−H]−, 6.3 min) contained the product ions m/z 173.008, 147.030, 129.019, 111.009, 87.010, 67.021. Based on these data, this metabolite was annotated as citrate (Figure 3).
4. Discussion
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gautam, S.; Tripathi, J. Food Processing by Irradiation—An effective technology for food safety and security. Indian J. Exp. Biol. 2016, 54, 700–707. [Google Scholar] [PubMed]
- Hussain, P.R.; Wani, I.A.; Suradkar, P.P.; Dar, M.A. Gamma irradiation induced modification of bean polysaccharides: Impact on physicochemical, morphological and antioxidant properties. Carbohydr. Polym. 2014, 110, 183–194. [Google Scholar] [CrossRef]
- Food Safety and Irradiation: Protecting the Public from Foodborne Infections. Available online: https://wwwnc.cdc.gov/eid/article/7/7/01-7706_article (accessed on 13 October 2022).
- Safety and Nutritional Adequacy of Irradiated Food. Available online: https://apps.who.int/iris/bitstream/handle/10665/39463/9241561629-eng.pdf?sequence=4 (accessed on 18 September 2022).
- Sundaralingam, R. A Debate on Safety of the Irradiated Food. Int. J. Res. Stud. Microbiol. Biotechnol. 2017, 3, 4–5. [Google Scholar]
- Rao, C.V. Do Irradiated Foods Causes or Promote Colon Cancer? Available online: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.504.5316&rep=rep1&type=pdf (accessed on 18 September 2022).
- Magaki, M.; Ishii, H.; Yamasaki, A.; Kitai, Y.; Kametani, S.; Nakai, R.; Dabid, A.; Tsuda, H.; Ohnishi, T. A high-fat diet increases the incidence of mammary cancer inc-Ha- ras proto-oncogene transgenic rats. J. Toxicol. Pathol. 2017, 30, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Willett, W.C. Diet, nutrition, and avoidable cancer. Environ. Health Perspect. 1995, 103 (Suppl. 8), 165–170. [Google Scholar]
- Doll, R. An overview of the epidemiological evidence linking diet and cancer. Proc. Nutr. Soc. 1990, 49, 119–131. [Google Scholar] [CrossRef]
- Barnes, S.; Grubbs, C.; Setchell, K.D.; Carlson, J. Soybeans inhibit mammary tumors in models of breast cancer. Prog. Clin. Biol. Res. 1990, 347, 239–253. [Google Scholar]
- Messina, M.; Barnes, S. The role of soy products in reducing risk of cancer. J. Natl. Cancer Inst. 1991, 83, 541–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badger, T.M.; Ronis, M.J.; Simmen, R.C.; Simmen, F.A. Soy protein isolate and protection against cancer. J. Am. Coll. Nutr. 2005, 24, 146S–149S. [Google Scholar] [CrossRef]
- Prasain, J.K.; Wilson, L.S.; Arabshahi, A.; Grubbs, C.; Barnes, S. Mass spectrometric evidence for the modification of small molecules in a cobalt-60-irradiated rodent diet. J. Mass Spectrom. 2017, 52, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Yu, J.; Wang, N.; Zhao, S.; Han, W.; Chen, D.; Li, L.; Li, L. High-Coverage Metabolome Analysis Reveals Significant Diet Effects of Autoclaved and Irradiated Feed on Mouse Fecal and Urine Metabolomics. Mol. Nutr. Food Res. 2021, 65, e2100110. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Spinelli, J.J.; Dummer, T.J.B.; Bhatti, P.; Playdon, M.C.; Levitt, J.O.; Hauner, B.; Moore, S.C.; Murphy, R.A. Metabolomics and cancer preventive behaviors in the BC Generations Project. Sci. Rep. 2021, 11, 12094. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, C.J.; Lubet, R.A.; Atigadda, V.R.; Christov, K.; Deshpande, A.M.; Tirmal, V.; Xia, G.; Bland, K.I.; Eto, I.; Brouillette, W.J.; et al. Efficacy of new retinoids in the prevention of mammary cancers and correlations with short-term biomarkers. Carcinogenesis 2006, 27, 1232–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aminu, M.; Ahmad, N.A. Complex Chemical Data Classification and Discrimination Using Locality Preserving Partial Least Squares Discriminant Analysis. ACS Omega 2020, 5, 26601–26610. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, D.; Guan, H.; Madhivanan, P.; Mathee, K.; Narasimhan, G. So you think you can PLS-DA? BMC Bioinform. 2020, 21 (Suppl. 1), 2. [Google Scholar] [CrossRef]
- Deininger, S.O.; Cornett, D.S.; Paape, R.; Becker, M.; Pineau, C.; Rauser, S.; Walch, A.; Wolski, E. Normalization in MALDI-TOF imaging datasets of proteins: Practical considerations. Anal. Bioanal. Chem. 2011, 401, 167–181. [Google Scholar] [CrossRef] [Green Version]
- Wulff, J.E.; Mitchell, M.W. A Comparison of Various Normalization Methods for LC/MS Metabolomics Data. Adv. Biosci. Biotechnol. 2018, 9, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 13. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Lewis-Stanislaus, A.E.; Li, L. A method for comprehensive analysis of urinary acylglycines by using ultra-performance liquid chromatography quadrupole linear ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 2105–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badenhorst, C.P.; van der Sluis, R.; Erasmus, E.; van Dijk, A.A. Glycine conjugation: Importance in metabolism, the role of glycine N-acyltransferase, and factors that influence interindividual variation. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1139–1153. [Google Scholar] [CrossRef] [PubMed]
- Holm, P.I.; Ueland, P.M.; Kvalheim, G.; Lien, E.A. Determination of choline, betaine, and dimethylglycine in plasma by a high-throughput method based on normal-phase chromatography-tandem mass spectrometry. Clin. Chem. 2003, 49, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Choucair, I.; Nemet, I.; Li, L.; Cole, M.A.; Skye, S.M.; Kirsop, J.D.; Fischbach, M.A.; Gogonea, V.; Brown, J.M.; Tang, W.H.W.; et al. Quantification of bile acids: A mass spectrometry platform for studying gut microbe connection to metabolic diseases. J. Lipid Res. 2020, 61, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Zhong, X.; Li, J.; Wang, X.; Ji, L.; Shang, X. Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis. Molecules 2019, 24, 3014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, A.K.; Ramarathinam, S.H.; Purcell, A.W.; O’Hair, R.A. Can alpha- and beta-alanine containing peptides be distinguished based on the CID spectra of their protonated ions? J. Am. Soc. Mass Spectrom. 2008, 19, 1743–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egnell, M.; Fassier, P.; Lécuyer, L.; Zelek, L.; Vasson, M.P.; Hercberg, S.; Latino-Martel, P.; Galan, P.; Deschasaux, M.; Touvier, M. B-Vitamin Intake from Diet and Supplements and Breast Cancer Risk in Middle-Aged Women: Results from the Prospective NutriNet-Santé Cohort. Nutrients 2017, 9, 488. [Google Scholar] [CrossRef] [Green Version]
- Hosoda, K.; Furuta, T.; Yokokawa, A.; Ishii, K. Identification and quantification of daidzein-7-glucuronide-4’-sulfate, genistein-7-glucuronide-4’-sulfate and genistein-4’,7-diglucuronide as major metabolites in human plasma after administration of kinako. Anal. Bioanal. Chem. 2010, 397, 1563–1572. [Google Scholar] [CrossRef]
- Hou, H.; Xiong, W.; Zhang, X.; Song, D.; Tang, G.; Hu, Q. LC-MS-MS Measurements of Urinary Creatinine and the Application of Creatinine Normalization Technique on Cotinine in Smokers’ 24 Hour Urine. J. Anal. Methods Chem. 2012, 2012, 245415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huć, T.; Nowinski, A.; Drapala, A.; Konopelski, P.; Ufnal, M. Indole and indoxyl sulfate, gut bacteria metabolites of tryptophan, change arterial blood pressure via peripheral and central mechanisms in rats. Pharm. Res. 2018, 130, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, B.; Feng, X.; Rubens, J.R.; Huang, R.; Hicks, L.M.; Pakrasi, H.B.; Tang, Y.J. Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology 2010, 156, 596–602. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.M.; Lu, X.; Merchant, F.M.; Iglehart, J.D.; Miron, P.L. Gene expression profiling of NMU-induced rat mammary tumors: Cross species comparison with human breast cancer. Carcinogenesis 2005, 26, 1343–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steele, V.E.; Lubet, R.A. The use of animal models for cancer chemoprevention drug development. Semin. Oncol. 2010, 37, 327–338. [Google Scholar] [CrossRef]
- Aiderus, A.; Black, M.A.; Dunbier, A.K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer 2018, 18, 805. [Google Scholar] [CrossRef] [PubMed]
- Long, J.P.; Li, X.N.; Zhang, F. Targeting metabolism in breast cancer: How far we can go? World J. Clin. Oncol. 2016, 7, 122–130. [Google Scholar] [CrossRef]
- Chetwynd, A.J.; David, A. A review of nanoscale LC-ESI for metabolomics and its potential to enhance the metabolome coverage. Talanta 2018, 182, 380–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- di Salvo, M.L.; Contestabile, R.; Paiardini, A.; Maras, B. Glycine consumption and mitochondrial serine hydroxymethyltransferase in cancer cells: The heme connection. Med. Hypotheses 2013, 80, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, X.; Wu, L.; Jiang, M.; Zhang, Z.; Wu, R.; Miao, J.; Liu, C.; Gao, S. Downregulation of GLYAT Facilitates Tumor Growth and Metastasis and Poor Clinical Outcomes Through the PI3K/AKT/Snail Pathway in Human Breast Cancer. Front. Oncol. 2021, 11, 641399. [Google Scholar] [CrossRef]
- Battista, N.; Bari, M.; Bisogno, T. N-Acyl Amino Acids: Metabolism, Molecular Targets, and Role in Biological Processes. Biomolecules 2019, 9, 822. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Gammon, M.D.; Zeisel, S.H.; Bradshaw, P.T.; Wetmur, J.G.; Teitelbaum, S.L.; Neugut, A.I.; Santella, R.M.; Chen, J. High intakes of choline and betaine reduce breast cancer mortality in a population-based study. FASEB J. 2009, 23, 4022–4028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, C.T.; Rodionov, D.A.; Osterman, A.L.; Peterson, S.N. B Vitamins and Their Role in Immune Regulation and Cancer. Nutrients 2020, 12, 3380. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; tan, Y.; Zhu, L. Dietary vitamin B2 intake and breast cancer risk: A systematic review and meta-analysis. Arch. Gynecol. Obstet. 2017, 295, 721–729. [Google Scholar] [CrossRef]
- Bolca, S.; Urpi-Sarda, M.; Blondeel, P.; Roche, N.; Vanhaecke, L.; Possemiers, S.; Al-Maharik, N.; Botting, N.; De Keukeleire, D.; Bracke, M.; et al. Disposition of soy isoflavones in normal human breast tissue. Am. J. Clin. Nutr. 2010, 91, 976–984. [Google Scholar] [CrossRef] [Green Version]
- Walczak, K.; Zurawska, M.; Kiś, J.; Starownik, R.; Zgrajka, W.; Bar, K.; Turski, W.A.; Rzeski, W. Kynurenic acid in human renal cell carcinoma: Its antiproliferative and antimigrative action on Caki-2 cells. Amino Acids 2012, 43, 1663–1670. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021, 19, 77–94. [Google Scholar] [CrossRef]
- Fernández, M.F.; Reina-Pérez, I.; Astorga, J.M.; Rodríguez-Carrillo, A.; Plaza-Díaz, J.; Fontana, L. Breast Cancer and Its Relationship with the Microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruo, S.W.; Alkayyali, T.; Win, M.; Tara, A.; Joseph, C.; Kannan, A.; Srivastava, K.; Ochuba, O.; Sandhu, J.K.; Went, T.R.; et al. Role of Gut Microbiota Dysbiosis in Breast Cancer and Novel Approaches in Prevention, Diagnosis, and Treatment. Cureus 2021, 13, e17472. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasain, J.K.; Wilson, L.S.; Grubbs, C.; Barnes, S. Gamma-ray Irradiation of Rodent Diets Alters the Urinary Metabolome in Rats with Chemically Induced Mammary Cancer. Metabolites 2022, 12, 976. https://doi.org/10.3390/metabo12100976
Prasain JK, Wilson LS, Grubbs C, Barnes S. Gamma-ray Irradiation of Rodent Diets Alters the Urinary Metabolome in Rats with Chemically Induced Mammary Cancer. Metabolites. 2022; 12(10):976. https://doi.org/10.3390/metabo12100976
Chicago/Turabian StylePrasain, Jeevan K., Landon S. Wilson, Clinton Grubbs, and Stephen Barnes. 2022. "Gamma-ray Irradiation of Rodent Diets Alters the Urinary Metabolome in Rats with Chemically Induced Mammary Cancer" Metabolites 12, no. 10: 976. https://doi.org/10.3390/metabo12100976
APA StylePrasain, J. K., Wilson, L. S., Grubbs, C., & Barnes, S. (2022). Gamma-ray Irradiation of Rodent Diets Alters the Urinary Metabolome in Rats with Chemically Induced Mammary Cancer. Metabolites, 12(10), 976. https://doi.org/10.3390/metabo12100976






