Mechanism Analysis of Metabolic Fatty Liver on Largemouth Bass (Micropterus salmoides) Based on Integrated Lipidomics and Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Diets
2.2. Experimental Fish, Feeding and Sampling
2.3. Hematological and Liver Homogenate Parameters
2.4. Histopathological Examination of the Liver Tissue
2.5. Proteomics Analysis
2.5.1. Protein Extraction
2.5.2. Trypsin Digestion
2.5.3. TMT Labeling
2.5.4. LC-MS/MS Analysis
2.5.5. Protein Identification
2.6. Lipidomics Analysis
2.6.1. Lipid Extraction
2.6.2. UPLC-MS/MS Analysis
2.6.3. Lipid Identification and Quantitation
2.7. Statistical Analysis
3. Results
3.1. High-Starch Diet (HSD) Induced Fatty Liver of Largemouth
3.2. Proteomics Analysis
3.3. Lipidomics Analysis
3.4. Integrative Proteomic and Lipidomic Analysis
4. Discussion
4.1. Proteomic Analysis
4.2. Lipidomic Analysis
4.3. Integrative Proteomic and Lipidomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, C.F.; Li, J.; Dong, J.J.; Niu, Y.C.; Hu, J.; Lian, J.M.; Li, W.H.; Li, J.; Tian, Y.Y.; Shi, Q.; et al. Chromosome-level genome assembly for the largemouth bass Micropterus salmoides provides insights into adaptation to fresh and brackish water. Mol. Ecol. Resour. 2020, 21, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Zheng, S.X.; Ma, X.K.; Cheng, K.M.; Wu, G.Y. Effects of dietary protein and lipid levels on the growth performance, feed utilization, and liver histology of largemouth bass (Micropterus salmoides). Amino Acids 2020, 52, 1043–1061. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C. Largemouth bass Micropterus salmoides production in China. In Largemouth Bass Aquaculture; Tidwell, J.H., Coyle, S.D., Bright, L.A., Eds.; 5M Publishing Ltd., Benchmark House: Sheffield, UK, 2019. [Google Scholar]
- Bai, J.; Li, S. Development of Largemouth Bass (Micropterus salmoides) Culture; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018. [Google Scholar]
- Yu, H.H.; Liang, X.F.; Chen, P.; Wu, X.F.; Zheng, Y.H.; Luo, L.; Qin, Y.C.; Long, X.C.; Xue, M. Dietary supplementation of Grobiotic®-A increases short-term inflammatory responses and improves long-term growth performance and liver health in largemouth bass (Micropterus salmoides). Aquaculture 2018, 500, 327–337. [Google Scholar] [CrossRef]
- Chen, Y.F.; Sun, Z.Z.; Liang, Z.M.; Xie, Y.D.; Tan, X.H.; Su, J.L.; Luo, Q.L.; Zhu, J.Y.; Liu, Q.Y.; Wang, A.L. Addition of L-carnitine to formulated feed improved growth performance, antioxidant status and lipid metabolism of juvenile largemouth bass, Micropterus salmoides. Aquaculture 2019, 518, 734434. [Google Scholar] [CrossRef]
- Yu, L.L.; Yu, H.H.; Liang, X.F.; Li, N.; Wang, X.; Li, F.H.; Wu, X.F.; Zheng, Y.H.; Xue, M.; Liang, X.F. Dietary butylated hydroxytoluene improves lipid metabolism, antioxidant and anti-apoptotic response of largemouth bass (Micropterus salmoides). Fish Shellfish. Immunol. 2018, 72, 220–229. [Google Scholar] [CrossRef]
- Amoah, A.; Coyle, S.D.; Webster, C.D.; Durborow, R.M.; Bright, L.A.; Tidwell, J.H. Effects of graded levels of carbohydrate on growth and survival of largemouth bass, Micropterus salmoides. J. World Aquac. Soc. 2008, 39, 397–405. [Google Scholar] [CrossRef]
- Lin, S.M.; Shi, C.M.; Mu, M.M.; Chen, Y.J.; Luo, L. Effect of high dietary starch levels on growth, hepatic glucose metabolism, oxidative status and immune response of juvenile largemouth bass, Micropterus salmoides. Fish Shellfish. Immunol. 2018, 78, 121–126. [Google Scholar] [CrossRef]
- Asaoka, Y.; Terai, S.; Sakaida, I.; Nishina, H. The expanding role of fish models in understanding non-alcoholic fatty liver disease. Dis. Models Mech. 2013, 6, 905–914. [Google Scholar] [CrossRef]
- Han, T.; Li, X.Y.; Wang, J.T.; Hu, S.X.; Jiang, Y.D.; Zhong, X.D. Effect of dietary lipid level on growth, feed utilization and body composition of juvenile giant croaker Nibea japonica. Aquaculture 2014, 434, 145–150. [Google Scholar] [CrossRef]
- Shi, C.M.; Zhao, H.; Zhai, X.L.; Chen, Y.J.; Lin, S.M. Linseed oil can decrease liver fat deposition and improve antioxidant ability of juvenile largemouth bass, Micropterus salmoides. Fish Physiol. Biochem. 2019, 45, 1513–1521. [Google Scholar] [CrossRef]
- Goodwin, A.E.; Lochmann, R.T.; Tieman, D.M.; Mitchell, A.J. Massive hepatic necrosis and nodular regeneration in largemouth bass fed diets high in available carbohydrate. J. World Aquac. Soc. 2010, 33, 466–477. [Google Scholar] [CrossRef]
- Xu, X.T.; Chen, N.S.; Liu, Z.K.; Gou, S.P.; Jia, Y. Effects of dietary starch sources and levels on liver histology in largemouth bass, Micropterus salmoide. J. Shanghai Ocean. Univ. 2016, 25, 61–70. [Google Scholar]
- Ma, H.J.; Mou, M.M.; Pu, D.C.; Lin, S.M.; Chen, Y.J.; Luo, L. Effect of dietary starch level on growth, metabolism enzyme and oxidative status of juvenile largemouth bass, Micropterus salmoides. Aquaculture 2019, 498, 482–487. [Google Scholar] [CrossRef]
- Wang, Z.; Kavdia, K.; Dey, K.K.; Pagala, V.R.; Kodali, K.; Liu, D.T.; Lee, D.G.; Sun, H.; Chepyala, S.R.; Cho, J.H. High-throughput and deep-proteome profiling by 16-plex tandem mass tag labeling coupled with two-dimensional chromatography and mass spectrometry. J. Vis. Exp. 2020, 162, e61684. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.W.; Ge, R.; Liu, W.L.; Liu, Q.M.; Xia, X.; Lai, M.; Liang, L.Z.; Li, C.; Song, L.; Zhen, B.; et al. Differential proteomics profiling identifies LDPs and biological functions in high-fat diet-induced fatty livers. J. Lipid Res. 2017, 58, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Willians, E.G.; Wu, Y.B.; Jha, P.; Dubuis, S.; Blattmann, P.; Argmann, C.A.; Houten, S.M.; Amariuta, T.; Wolski, W.; Zamboni, N.; et al. Systems proteomics of liver mitochondria function. Science 2016, 352, aad0189. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Geyer, P.E.; Albrechtsen, N.J.W.; Gluud, L.L.; Santos, A.; Doll, S.; Treit, P.V.; Holst, J.J.; Knop, F.K.; Vilsbol, T.; et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol. Syst. Biol. 2019, 15, e8793. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhan, C.; Chen, G.W.; Sun, J.Y. Labelfree quantitative proteomics and bioinformatics analyses of alcoholic liver disease in a chronic and binge mouse model. Mol. Med. Rep. 2018, 18, 2079–2087. [Google Scholar]
- Assini, J.M.; Mulvihill, E.E.; Huff, M.W. Citrus flavonoids and lipid metabolism. Curr. Opin. Lipidol. 2013, 24, 34–40. [Google Scholar] [CrossRef]
- Feng, K.L.; Lan, Y.Q.; Zhu, X.A.; Li, J.; Chen, T.; Huang, Q.R.; Ho, C.T.; Chen, Y.J.; Cao, Y. Hepatic Lipidomics analysis reveals the antiobesity and cholesterol-lowering effects of tangeretin in high-fat diet-fed rats. J. Agric. Food Chem. 2020, 68, 6142–6153. [Google Scholar] [CrossRef]
- Yang, K.; Han, X. Lipidomics: Techniques, applications, and outcomes related to biomedical sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Han, X.L.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wei, F.; Dong, X.Y.; Xiang, J.Q.; Quek, S.Y.; Wang, X.M. Lipidomics in food science. Curr. Opin. Food Sci. 2017, 16, 80–87. [Google Scholar] [CrossRef]
- Li, Y.X.; Luo, Z.P.; Wu, X.L.; Zhu, J.; Yu, K.; Jin, Y.; Zhang, Z.W.; Zhao, S.H.; Zhou, L. Proteomic analyses of cysteine redox in high-fat-fed and fasted mouse livers: Implications for liver metabolic homeostasis. J. Proteome Res. 2017, 14, 129–140. [Google Scholar] [CrossRef]
- Kwon, O.K.; Kim, S.J.; Lee, Y.M.; Lee, Y.H.; Bae, Y.S.; Kim, J.Y.; Peng, X.J.; Cheng, Z.Y.; Zhao, Y.M.; Lee, S. Global analysis of phosphoproteome dynamics in embryonic development of zebrafish (Danio rerio). Proteomics 2016, 16, 136–149. [Google Scholar] [CrossRef]
- Enes, P.; Panserat, S.; Kaushik, S.; Oliva-Teles, A. Nutritional regulation of hepatic glucose metabolism in fish. Fish Physiol. Biochem. 2008, 35, 519–539. [Google Scholar] [CrossRef]
- Hemre, G.I.; Mommsen, T.P.; Krogdahl, A. Carbohydrates in fish nutrition: Effects on growth, glucose metabolism and hepatic enzymes. Aquac. Nutr. 2002, 8, 175–194. [Google Scholar] [CrossRef]
- Liang, X.F.; Chen, P.; Wu, X.L.; Xing, S.J.; Morais, S.; He, M.L.; Gu, X.; Xue, M. Effects of High Starch and Supplementation of an Olive Extract on the Growth Performance, Hepatic Antioxidant Capacity and Lipid Metabolism of Largemouth Bass. Antioxidants 2022, 11, 577. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Xie, S.W.; Wei, H.L.; Zheng, L.; Liu, Z.L.; Fang, H.H.; Xie, J.J.; Liao, S.Y.; Tian, L.X.; Liu, Y.J. High dietary starch impaired growth performance, liver histology and hepatic glucose metabolism of Juvenile largemouth bass, Micropterus salmoides. Aquac. Nutr. 2020, 26, 1083–1095. [Google Scholar] [CrossRef]
- Yu, H.H.; Zhang, L.L.; Chen, P.; Liang, X.F.; Cao, A.Z.; Han, J.; Wu, X.F.; Zheng, Y.H.; Qin, Y.C.; Xue, M. Dietary Bile Acids Enhance Growth, and Alleviate Hepatic Fibrosis Induced by a High Starch Diet via AKT/FOXO1 and cAMP/AMPK/SREBP1 Pathway in Micropterus salmoides. Front. Physiol. 2019, 10, 1430. [Google Scholar] [CrossRef]
- English, A.R.; Voeltz, G.K. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013, 5, a013227. [Google Scholar] [CrossRef] [PubMed]
- Juan, R.C.; Sarah, E.B.; Laurie, H.G. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell 2017, 168, 692–706. [Google Scholar]
- Michael, J. Endoplasmic reticulum stress in nonalcoholic fatty liver disease. Annu. Rev. Nutr. 2012, 32, 17–33. [Google Scholar]
- Christopher, L.; Melinda, A.; Michael, J. Fatty acids and the endoplasmic reticulum in nonalcoholic fatty liver disease. BioFactors 2011, 37, 8–16. [Google Scholar]
- Pan, Y.; Cao, F.; Guo, A.; Chang, W.; Ma, W.; Gao, X.; Guo, S.; Fu, C.; Zhu, J. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br. J. Cancer 2015, 113, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y.; Yang, Y.F.; Wu, A.T.; Yang, C.J.; Liu, Y.P.; Jan, Y.H.; Lee, C.H.; Hsiao, Y.W.; Yeh, C.T.; Shen, C.N. Endoplasmic reticulum ribosome-binding protein1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene 2013, 32, 4921–4931. [Google Scholar] [CrossRef]
- Liang, X.S.; Sun, S.S.; Zhang, X.Y.; Wu, H.; Tao, W.Y.; Liu, T.; Wei, W.; Geng, J.S.; Pang, D. Expression of ribosome-binding protein 1 correlates with shorter survival in Her-2 positive breast cancer. Cancer Sci. 2015, 106, 740–746. [Google Scholar] [CrossRef]
- Xiong, L.; Zhou, X.R.; Liu, D.; Liu, G.C. Effect of overexpression of RRBP1 on metastasis and invasion of liver cancer cells. J. Chengdu Med. Coll. 2021, 16, 5. [Google Scholar]
- Li, H.Y.; Wang, Y.Z.; Yang, H.G.; Zhang, Y.D.; Xing, L.; Wang, J.Q.; Zheng, N. Furosine, a maillard reaction product, triggers necroptosis in hepatocytes by regulating the RIPK1/RIPK3/MLKL pathway. Int. J. Mol. Sci. 2019, 20, 2388. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Y.Z. Transport Transporter on the Endoplasmic Reticulum-TRAP. Chin. J. Biochem. Mol. Biol. 2008, 24, 95–100. [Google Scholar]
- Lu, Y.C.; Chang, C.C.; Wang, C.P.; Hung, W.C.; Tsai, I.T.; Tang, W.H.; Wu, C.C.; Wei, C.T.; Chung, F.M.; Lee, Y.J. Circulating fatty acid-binding protein 1 (FABP1) and nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus. Int. J. Med. Sci. 2020, 17, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Shen, H.; Ganesh, R.; Roberts, M.S.; Gong, Y.W.; Jiang, P.; Burczynski, F. Expression and antioxidant function of liver fatty acid binding protein in normal and bile-duct ligated rats. Eur. J. Pharmacol. 2007, 560, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Waikar, S.S.; Bonventre, J.V. Biomarkers for the diagnosis of acute kidney injury. Nephron Clin. Pract. 2008, 109, c192–c197. [Google Scholar] [CrossRef] [PubMed]
- Carla, G.; Marta, B.; Sandra, P.V.; Marta, M.M.; Victoria, G.M.; M.Luz, M.C.; Javer, G.G.; Jose, V.C.; Sonia, S.C.; Ramiro, J. The human liver fatty acid binding protein (FABP1) gene is activated by FOXA1 and PPARα; and repressed by C/EBPα: Implications in FABP1 down-regulation in nonalcoholic fatty liver disease. Biochim. Biophys. Acta 2013, 1831, 803–818. [Google Scholar]
- Zhang, Y. Anti-Hepatic Fibrosis Effects of Bifendate in Rat and Its Relationship with CBR1 and FABP1. Master’s Thesis, Hebei Medical University, Shijiazhuang, China, 2015. [Google Scholar]
- Akbal, E.; Köklü, S.; Koçak, E.; Cakal, B.; Gunes, F.; Basar, O.; Tuna, Y.; Senes, M. Liver fatty acid-binding protein is a diagnostic marker to detect liver injury due to chronic hepatitis C infection. Arch. Med. Res. 2013, 44, 34–38. [Google Scholar] [CrossRef]
- Ozenirler, S.; Degertekin, C.K.; Erkan, G.; Elbeg, S.; Tuncer, C.; Kandilci, U.; Akyol, G. Serum liver fatty acid binding protein shows good correlation with liver histology in NASH. Hepatogastroenterology 2013, 60, 1095–1100. [Google Scholar]
- Akbal, E.; Koçak, E.; Akyürek, Ö.; Koklu, S.; Batgi, H.; Senes, M. Liver fatty acid-binding protein as a diagnostic marker for non-alcoholic fatty liver disease. Wien. Klin. Wochenschr. 2016, 128, 48–52. [Google Scholar] [CrossRef]
- Hussain, M.M.; Shi, J.; Dreizen, P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J. Lipid Res. 2003, 44, 22–32. [Google Scholar] [CrossRef]
- Khatun, I.; Walsh, M.T.; Hussain, M.M. Loss of both phospholipid and triglyceride transfer activities of microsomal triglyceride transfer protein in abetalipoproteinemia. J. Lipid Res. 2013, 54, 1541–1549. [Google Scholar] [CrossRef]
- Love, J.D.; Suzuki, T.; Robinson, D.B.; Harris, C.M.; Johnson, J.E.; Mohler, P.J.; Jerome, W.G.; Swift, L.L. Microsomal triglyceride transfer protein (MTP) associates with cytosolic lipid droplets in 3T3-L1 adipocytes. PLoS ONE 2015, 10, e0135598. [Google Scholar] [CrossRef]
- Iqbal, J.; Walsh, M.T.; Hammad, S.M.; Cuchel, M.; Tarugi, P.; Hegele, R.A.; Davidson, N.O.; Rader, D.J.; Klein, R.L.; Hussain, M.M. Microsomal triglyceride transfer protein transfers and determines plasma concentrations of ceramide and sphingomyelin but not glycosylceramide. J. Biol. Chem. 2015, 290, 25863–25875. [Google Scholar] [CrossRef] [PubMed]
- Ueshima, K.; Akihisa-Umeno, H.; Nagayoshi, A.; Takakura, S.; Matsuo, M.; Mutoh, S. Implitapide, a microsomal triglyceride transfer protein inhibitor, reduces progression of atherosclerosis in apolipoprotein E knockout mice fed a Western-type diet: Involvement of the inhibition of postprandial triglyceride elevation. Biol. Pharm. Bull. 2005, 28, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.H. Nonalcoholic fatty liver disease: Molecular mechanisms for the hepatic steatosis. Clin. Mol. Hepatol. 2013, 19, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Cohen, D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 434–441. [Google Scholar] [CrossRef]
- Chang, X.X.; Yan, H.M.; Fei, J.; Jiang, M.H.; Zhu, H.G.; Lu, D.R.; Gao, X. Berberine reduces methylation of the MTTP promoter and alleviates fatty liver induced by a high-fat diet in rats. J. Lipid Res. 2010, 51, 2504–2515. [Google Scholar] [CrossRef]
- Liu, M.; Chung, S.; Shelness, G.S.; Parks, J.S. Hepatic ABCA1 and VLDL triglyceride production. Biochim. Biophys. Acta 2012, 1821, 770–777. [Google Scholar] [CrossRef]
- Vaisman, B.L.; Lambert, G.; Amar, M.; Joyce, C.; Ito, T.; Shamburek, R.D.; Cain, W.J.; Fruchart, N.J.; Neufeld, E.D.; Remaley, A.T. ABCA1 overexpression leads to hyperalphalipoproteinemia and increased biliary cholesterol excretion in transgenic mice. J. Clin. Investig. 2001, 108, 303–309. [Google Scholar] [CrossRef]
- Vega-Badillo, J.; Gutiérrez-Vidal, R.; Hernández-Pérez, H.A.; Villanil, R.Z.; Leon, M.P.; Sanchez, M.F.; Moran, R.S.; Larriieta, C.E.; Fernandez, S.I.; Mendez, S.N.; et al. Hepatic miR-33a/miR-144 and their target gene ABCA1 are associated with steatohepatitis in morbidly obese subjects. Liver Int. 2016, 36, 1383–1391. [Google Scholar] [CrossRef]
- Ma, K.L.; Ruan, X.Z.; Powis, S.H.; Chen, Y.; Moorhead, J.F.; Varghese, Z. Inflammatory stress exacerbates lipid accumulation in hepatic cells and fatty livers of apolipoprotein E knockout mice. Hepatology 2008, 48, 770–778. [Google Scholar] [CrossRef]
- Fukunaga, K.; Imachi, H.; Lyu, J.; Dong, T.; Sato, S.; Ibata, T.; Kobayashi, T.; Yoshimoto, T.; Yonezaki, K.; Matsunaga, T. IGF1 suppresses cholesterol accumulation in the liver of growth hormone-deficient mice via the activation of ABCA1. Am. J. Physiol. Endocrinol. Metab. 2018, 315, 1232–1241. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.S.; Lu, L.L.; Liao, S.L.; Yue, H.Y.; Dong, Q.J.; Xin, Y.N.; Xuan, S.Y. Association Between Four ABCA1 gene Polymorphisms and Risk of Non-Alcoholic Fatty Liver Disease in a Chinese Han Population. Hepat. Mon. 2018, 6, e66149. [Google Scholar] [CrossRef]
- Farrell, G.C.; Larter, C.Z. Nonalcoholic fatty liver disease: From steatosis to cirrhosis. Hepatology 2006, 43, S99–S112. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.M.; Gan, L.; Yang, C.S.; Liu, A.B.; Lu, W.Y.; Shao, P.; Dai, Z.Q.; Sun, P.L.; Luo, Z.S. Effects of Stigmasterol and β-Sitosterol on Nonalcoholic Fatty Liver Disease in a Mouse Model: A Lipidomic Analysis. J. Agric. Food Chem. 2018, 66, 3417–3425. [Google Scholar] [CrossRef] [PubMed]
- Perla, F.M.; Prelati, M.; Lavorato, M.; Visicchio, D.; Anania, C. The role of lipid and lipoprotein metabolism in non-alcoholic fatty liver disease. Children 2017, 4, 46. [Google Scholar] [CrossRef]
- Zhai, R.N.; Feng, L.; Zhang, Y.; Liu, W.; Li, S.L.; Hu, Z.Y. Combined Transcriptomic and Lipidomic Analysis Reveals Dysregulated Genes Expression and Lipid Metabolism Profiles in the Early Stage of Fatty Liver Disease in Rats. Front. Nutr. 2021, 8, 733197. [Google Scholar] [CrossRef]
- Goldberg, I.J.; Ginsberg, H.N. Ins and outs modulating hepatic triglyceride and development of nonalcoholic fatty liver disease. Gastroenterology 2006, 130, 1343–1346. [Google Scholar] [CrossRef]
- Ma, L.N.; Feng, M.; Zhou, Y.Z. The relationship between nonalcoholic fatty liver disease and insulin resistance. J. Clin. Hepatol. 2010, 26, 173–175. [Google Scholar]
- Zhang, C.H.; Zhou, B.G.; Sheng, J.Q.; Chen, Y.; Cao, Y.Q.; Chen, C. Molecular mechanisms of hepatic insulin resistance in nonalcoholic fatty liver disease and potential treatment strategies. Pharmacol. Res. 2020, 159, 104984. [Google Scholar] [CrossRef]
- Marieke, D.V.; Westerink, J.; EL-Morabit, F.; Kaasjager, H.A.H.; De Valk, H.W. Prevalence of non-alcoholic fatty liver disease (NAFLD) and its association with surrogate markers of insulin resistance in patients with type 1 diabetes. Diabetes Res. Clin. Pract. 2022, 186, 109827. [Google Scholar]
- Byrne, C.D. Dorothy Hodgkin Lecture: Non-alcoholic fatty liver disease, insulin resistance and ectopic fat: A new problem in diabetes management. Diabet. Med. 2019, 29, 1098–1107. [Google Scholar] [CrossRef]
- Marjot, T.; Moolla, A.; Cobbold, J.F.; Hodson, L.; Tomlinson, J.W. Nonalcoholic Fatty Liver Disease in Adults: Current Concepts in Etiology, Outcomes, and Management. Endocr. Rev. 2020, 41, 66–117. [Google Scholar] [CrossRef] [PubMed]
- Holland, W.L.; Summers, S.A. Sphingolipids, insulin resistance, and metabolic disease: New insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 2008, 29, 381–402. [Google Scholar] [CrossRef] [PubMed]
- Kotronen, A.; Seppänen-Laakso, T.; Westerbacka, J.; Kiviluoto, T.; Arola, J.; Ruskeepaa, A.L.; Yki-Jarvinen, H.; Oresic, M. Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity 2010, 18, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Llacuna, L.; Marí, M.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; Morales, A. Critical role of acidic sphingomyelinase in murine hepatic ischemia-reperfusion injury. Hepatology 2006, 44, 561–572. [Google Scholar] [CrossRef]
- Regnier, M.; Polizzi, A.; Guillou, H.; Loiseau, N. Sphingolipid metabolism in non-alcoholic fatty liver diseases. Biochimie 2019, 1590, 9–22. [Google Scholar] [CrossRef]
- Chocian, G.; Chabowski, A.; Żendzian-Piotrowska, M.E.; Harasim, E.; Lukaszuk, B.; Gorski, J. High fat diet induces ceramide and sphingomyelin formation in rat’s liver nuclei. Mol. Cell. Biochem. 2010, 340, 125–131. [Google Scholar] [CrossRef]
- Brown, E.M. Fatty liver? Microbiome sphingolipids to the rescue. Cell Host Microbe 2022, 30, 755–757. [Google Scholar] [CrossRef]
- Dowman, J.K.; Tomlinson, J.W.; Newsome, P.N. Pathogenesis of non-alcoholic fatty liver disease. Int. J. Med. 2010, 103, 71–83. [Google Scholar] [CrossRef]
- Amrutkar, M.; Cansby, E.; Chursa, U.; Nunez-Duran, E.; Chanclon, B.; Stahlman, M.; Friden, V.; Manneras-Holm, L.; Wickman, A.; Smith, U.; et al. Genetic disruption of protein kinase STK25 ameliorates metabolic defects in a diet-induced type 2 diabetes model. Diabetes 2015, 64, 2791–2804. [Google Scholar] [CrossRef]
- Martin, G.G.; Danneberg, H.; Kumar, L.S.; Atshaves, B.P.; Erol, E.; Bader, M.; Schroeder, F.; Binas, B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J. Biol. Chem. 2003, 278, 21429–21438. [Google Scholar] [CrossRef]
- Atshaves, B.P.; McIntosh, A.M.; Lyuksyutova, O.I.; Zipfel, W.; Webb, W.W.; Schroeder, F. Liver fatty acid-binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J. Biol. Chem. 2004, 279, 30954–30965. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Egawa, M.; Takeuchi, T.; Yamashita, H.; Kusudo, T. Silencing of FABP1 ameliorates hepatic steatosis, inflammation, and oxidative stress in mice with nonalcoholic fatty liver disease. FEBS Open Bio 2017, 7, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Newberry, E.P.; Xie, Y.; Kennedy, S.M.; Luo, J.; Davidson, N.O. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology 2006, 44, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.G.; Atshaves, B.P.; Huang, H.; Mclntosh, A.L.; Williams, B.J.; Pai, P.J.; Russell, D.H.; Kier, A.B.; Schroeder, F. Hepatic phenotype of liver fatty acid binding protein gene-ablated mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 297, G1053–G1065. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Ioannou, G.N. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol. Metab. 2016, 27, 84–95. [Google Scholar] [CrossRef]
- Sozen, E.; Ozer, N.K. Impact of high cholesterol and endoplasmic reticulum stress on metabolic diseases: An updated mini-review. Redox Biol. 2017, 12, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Oram, J.F.; Heinecke, J.W. ATP-binding cassette transporter A1: A cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 2005, 85, 1343–1372. [Google Scholar] [CrossRef]
- Oram, J.F. The ins and outs of ABCA. J. Lipid Res. 2008, 49, 1150–1151. [Google Scholar] [CrossRef]
- Tang, C.R.; Liu, Y.H.; Yang, W.; Storey, C.; McMillen, T.S.; Houston, B.A.; Heinecke, J.W.; LeBoeuf, R.C. Hematopoietic ABCA1 deletion promotes monocytosis and worsens diet-induced insulin resistance in mice. J. Lipid Res. 2016, 57, 100–108. [Google Scholar] [CrossRef]
- Kolovou, V.; Marvaki, A.; Boutsikou, M.; Vasilopoulos, G.; Degiannis, D.; Marvaki, C.; Kolovou, G. Effect of ATP-binding Cassette Transporter A1 (ABCA1) Gene Polymorphisms on Plasma Lipid Variables and Common Demographic Parameters in Greek Nurses. Open Cardiovasc. Med. J. 2016, 10, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Clee, S.M.; Kastelein, J.J.; van Dam, M.; Marcil, M.; Roomp, K.; Zwarts, K.Y.; Collins, J.A.; Roelants, R.; Tamasawa, N.; Stulc, T.; et al. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Investig. 2000, 106, 1263–1270. [Google Scholar] [CrossRef]
- Lee, K.; Kerner, J.; Hoppel, C.L. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J. Biol. Chem. 2011, 286, 25655–25662. [Google Scholar] [CrossRef] [PubMed]
- Tonazzi, A.; Giangregorio, N.; Console, L.; Indiveri, C. Mitochondrial carnitine/acylcarnitine translocase: Insights in structure/ function relationships. Basis for drug therapy and side effects prediction. Mini Rev. Med. Chem. 2015, 15, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Pittala, S.; Krelin, Y.; Kuperman, Y.; Shoshan-Barmatz, V. A Mitochondrial VDAC1-Based Peptide Greatly Suppresses Steatosis and NASH-Associated Pathologies in a Mouse Model. Mol. Ther. 2019, 27, 1848–1862. [Google Scholar] [CrossRef] [PubMed]
- Turkaly, P.; Kerner, J.; Hoppel, C. A 22 kDa polyanion inhibits carnitine-dependent fatty acid oxidation in rat liver mitochondria. FEBS Lett. 1999, 460, 241–245. [Google Scholar] [CrossRef]
- Zhong, Z.; Lemasters, J.J. A Unifying Hypothesis Linking Hepatic Adaptations for Ethanol Metabolism to the Proinflammatory and Profibrotic Events of Alcoholic Liver Disease. Alcohol. Clin. Exp. Res. 2018, 42, 2072–2089. [Google Scholar] [CrossRef]
- Holmuhamedov, E.L.; Czerny, C.; Beeson, C.C.; Lemasters, J.J. Ethanol suppresses ureagenesis in rat hepatocytes: Role of acetaldehyde. J. Biol. Chem. 2012, 287, 7692–7700. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Krelin, A.Y.; Shteinfer-Kuzmine, T. Voltage-Dependent Anion Channel 1 As an Emerging Drug Target for Novel Anti-Cancer Therapeutics. Front. Oncol. 2017, 7, 154. [Google Scholar] [CrossRef]
- Maldonado, E.N.; Shoshan-Barmatz, V.; Krelin, Y. VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell Stress 2017, 1, 11–36. [Google Scholar]
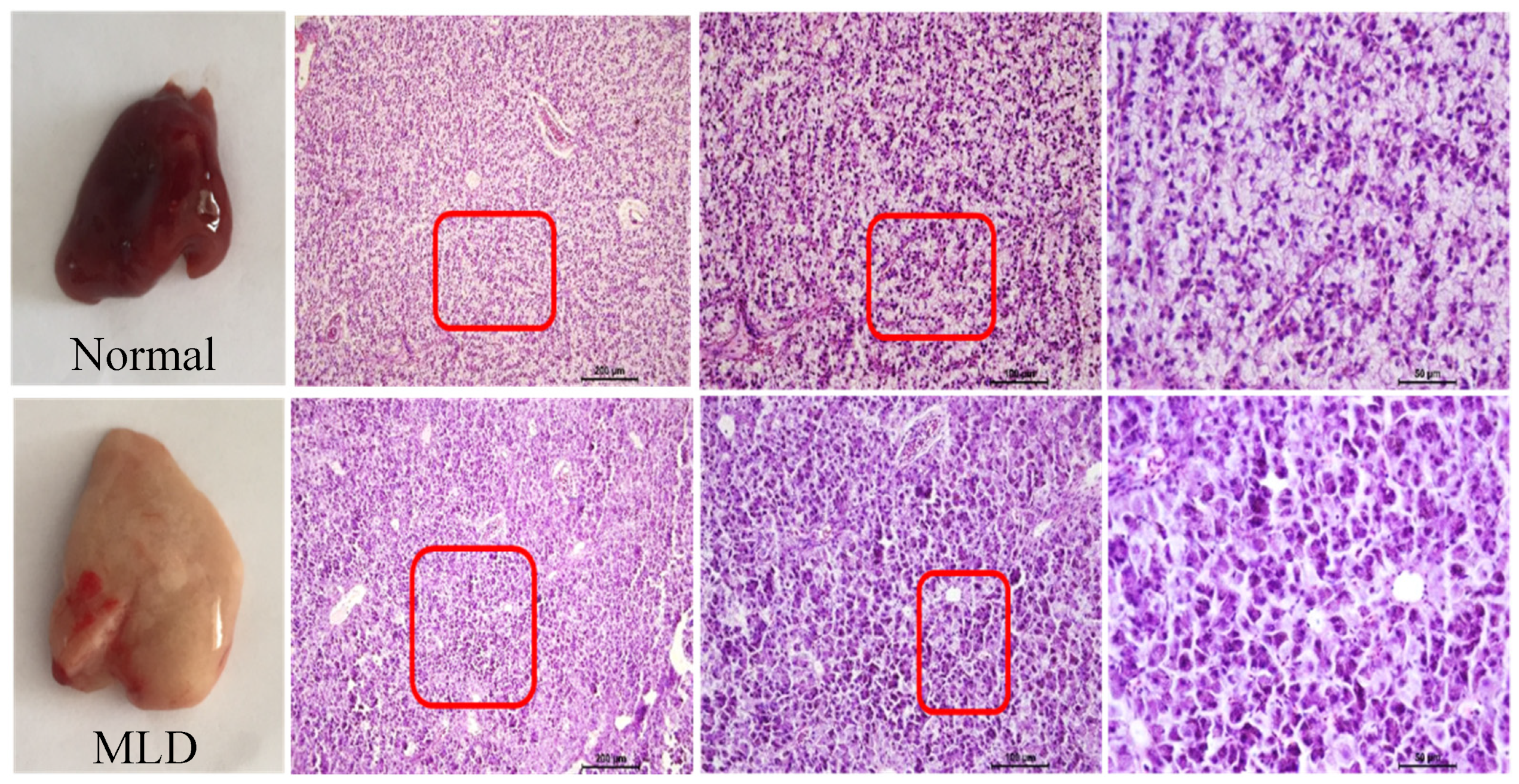
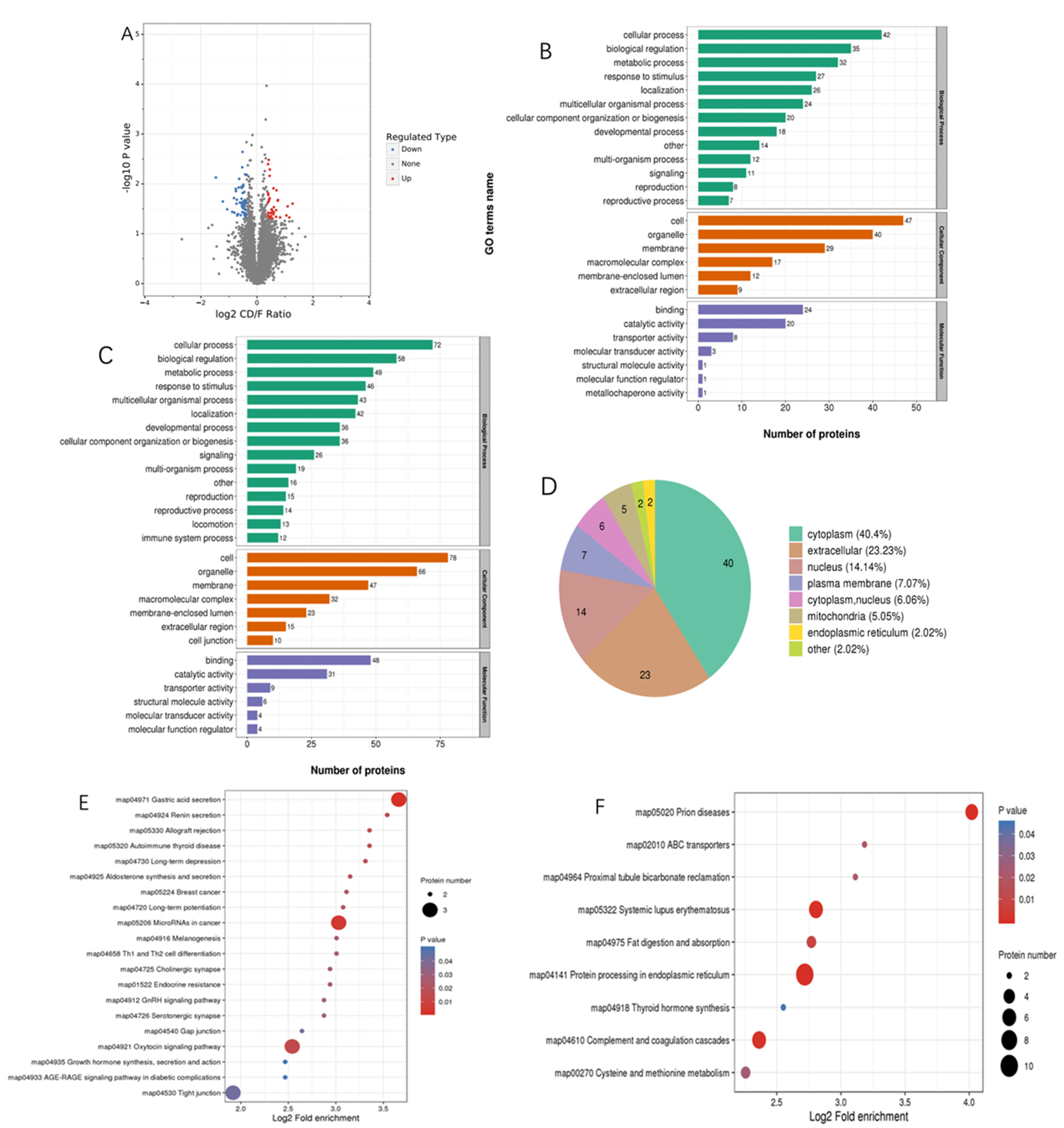
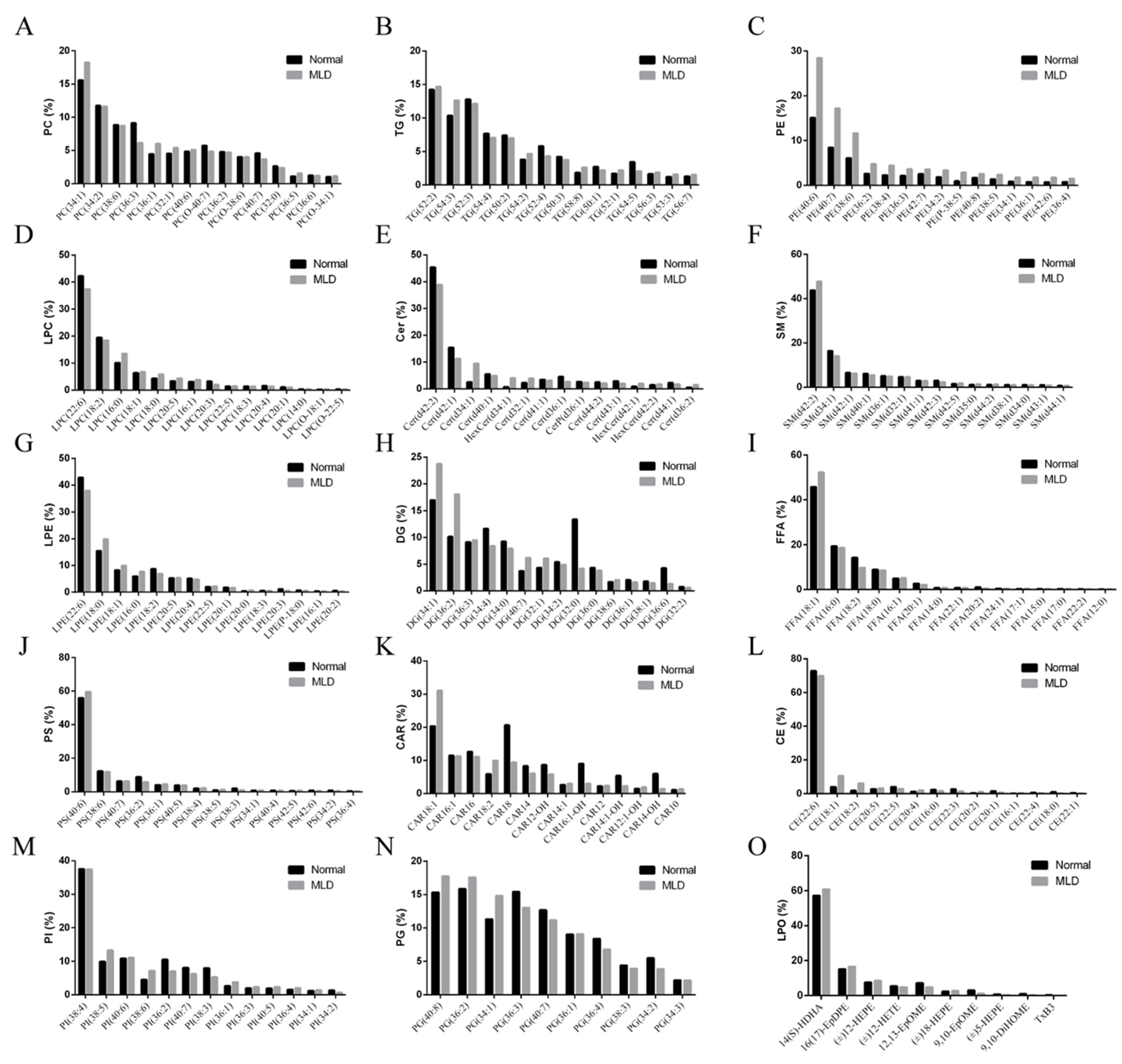

| Ingredients (in Dry Matter Basis, %) | Normal Group | MLD Group |
|---|---|---|
| a Fish meal | 30.0 | 30.0 |
| Tapioca starch | 5 | 5 |
| Wheat flour | 9.0 | 18 |
| Microbial protein | 4 | 4 |
| Cottonseed concentrate protein | 23.5 | 22.6 |
| Wheat gluten meal | 4 | 4 |
| Soybean meal | 2 | - |
| Spay-dried blood cell powder | 4 | 4 |
| α-cellulose | 4.6 | - |
| Ca(H2PO4)2 | 1.7 | 1.7 |
| Lecithin oil | 2.0 | 2.0 |
| Fish oil | 3.5 | 3.5 |
| Soybean oil | 3.5 | 3.5 |
| Kelp powder | 1.5 | 0 |
| L-Threonine | 0.1 | 0.1 |
| DL-Methionine | 0.2 | 0.2 |
| b Vitamin and mineral premix | 1.4 | 1.4 |
| Total | 100 | 100 |
| Nutrients compositions | ||
| Crude protein | 50.83 | 51.15 |
| Ether extract | 12.36 | 12.33 |
| Crude ash | 10.08 | 10.04 |
| Moisture | 6.10 | 7.43 |
| Gross energy (MJ/Kg) | 20.45 | 20.15 |
| Lipids | Abbreviation | CAS | Standards Type | |
|---|---|---|---|---|
| 1 | Lysophosphatidyl choline | LPC (13:0) | 20559-17-5 | Internal |
| 2 | Cholesterol heptadecanoate | CE (17:0) | 24365-37-5 | Internal |
| 3 | Ceramide C4 | Cer (d18:1/4:0) | 74713-58-9 | Internal |
| 4 | Diester of glycerol dodecanoate | DG (12:0/12:0) | 60562-15-4 | Internal |
| 5 | Lysophosphatidyl ethanolamine | LPE (14:0) | 123060-40-2 | Internal |
| 6 | Phosphatidylcholine | PC (13:0/13:0) | 71242-28-9 | Internal |
| 7 | Phosphatidyl ethanolamine | PE (12:0/12:0) | 59752-57-7 | Internal |
| 8 | Diphosphatidyl glycerol | PG (12:0/12:0) | 322647-27-8 | Internal |
| 9 | Phosphatidylserin | PS (14:0/14:0) | 105405-50-3 | Internal |
| 10 | Triglyceride dodecyl | TG (12:0/12:0/12:0) | 555-44-2 | Internal |
| 11 | Phosphatidyl inositol | PI (16:0/16:0) | 34290-57-8 | Internal |
| 12 | Palmitic acid -d31 | FFA (16:0)-d31 | 39756-30-4 | Internal |
| 13 | Cholesterol linoleate | CE (18:2) | 604-33-1 | External |
| 14 | Ceramide C17 | Cer (d18:1/17:0) | 67492-16-4 | External |
| 15 | Diester of glycerol hexadecanoate | DG (16:0/16:0) | 30334-71-5 | External |
| 16 | Lysophosphatidyl choline | LPC (17:0) | 50930-23-9 | External |
| 17 | Lysophosphatidyl ethanolamine | LPE (16:0) | 53862-35-4 | External |
| 18 | Phosphatidyl choline | PC (17:0/17:0) | 70897-27-7 | External |
| 19 | Phosphatidyl ethanolamine | PE (17:0/17:0) | 140219-78-9 | External |
| 20 | Phosphatidyl glycerol | PG (17:0/17:0) | 799268-52-3 | External |
| 21 | Phosphatidylserine | PS (17:0/17:0) | 799268-51-2 | External |
| 22 | Sphingomyelin | SM (d18:1/17:0) | 121999-64-2 | External |
| 23 | Triglyceride heptadecanoate | TG (17:0/17:0/17:0) | 2438-40-6 | External |
| 24 | Phosphatidyl inositol | PI (16:0/18:1) | 50730-13-7 | External |
| 25 | Palmitic acid | FFA (16:0) | 57-10-3 | External |
| Items | Normal | MLD |
|---|---|---|
| CF (g/cm3) | 2.03 ± 0.09 | 2.01 ± 0.08 |
| VSI (%) | 7.17 ± 1.86 | 7.36 ± 0.20 |
| HSI (%) | 1.66 ± 0.08 b | 2.36 ± 1.17 a |
| FBW (g) | 105.83 ± 1.68 b | 95.54 ± 1.30 a |
| SGR | 2.00 ± 0.31 b | 1.80 ± 0.03 a |
| FCR | 0.98 ± 0.01 | 1.00 ± 0.18 |
| HL | 6.54 ± 0.29 b | 7.39 ± 0.10 a |
| Items | Normal | MLD |
|---|---|---|
| TP (g/L) | 16.49 ± 0.47 | 15.69 ± 0.50 |
| GLU (mmol/L) | 5.71 ± 0.35 b | 4.25 ± 0.45 a |
| TG (mmol/L) | 5.64 ± 1.18 | 5.48 ± 0.45 |
| TC (mmol/L) | 7.70 ± 0.79 | 8.16 ± 0.62 |
| HDL-C (mmol/L) | 1.75 ± 0.38 | 1.64 ± 0.32 |
| LDL-C (mmol/L) | 2.00 ± 0.20 b | 2.24 ± 0.13 a |
| AKP (U/L) | 49.10 ± 5.56 b | 77.34 ± 5.13 a |
| AST (U/L) | 5.87 ± 1.05 b | 11.15 ± 1.88 a |
| ALT (U/L) | 5.87 ± 1.04 b | 12.65 ± 1.97 a |
| TBA (umol/L) | 74.41 ± 1.12 b | 78.11 ± 4.49 a |
| Items | Normal | MLD |
|---|---|---|
| TG (mmol/g·prot) | 0.17 ± 0.01 b | 0.21 ± 0.05 a |
| TC (mmol/g·prot) | 0.15 ± 0.02 | 0.15 ± 0.01 |
| TBA (umol/mg·prot) | 2.33 ± 0.28 | 2.33 ± 0.65 |
| LDL-C (umol/g·prot) | 30.03 ± 2.92 b | 43.92 ± 4.89 a |
| LDL-C/TC | 0.22 ± 0.03 | 0.29 ± 0.03 |
| Items | Normal | MLD |
|---|---|---|
| ROS (U/mg·prot) | 66.78 ± 4.92 b | 88.14 ± 4.85 a |
| T-AOC (umol/g·prot) | 76.78 ± 5.96 | 89.14 ± 9.02 |
| CAT (U/mg·prot) | 46.62 ± 2.07 a | 31.13 ± 3.42 b |
| GSH-Px (U/ug·prot) | 4.07 ± 0.51 | 4.38 ± 0.57 |
| SOD (U/mg·prot) | 182.40 ± 9.45 | 168.75 ± 19.81 |
| MDA (nmol/mg·prot) | 2.69 ± 0.71 | 3.00 ± 0.38 |
| KEGG Pathway | Related Proteins (Up-Regulated) | Related Proteins (Down-Regulated) |
|---|---|---|
| Protein processing in endoplasmic reticulum | RRBP1 | TRAPα, PDIA4 |
| Fat digestion and absorption | FABP1 | ABCA1, MTTP |
| ABC transporters | ABCA1 | |
| Cholesterol metabolism | ABCA1, VDAC1, AK1R1D1 | |
| PPAR signal pathway | HRAs, FABP1 | PEPCK |
| FoxO signaling pathway; | HRAS | PEPCK |
| mTOR signaling pathway | HRAs | |
| Glycolysis/Gluconeogenesis | PEPCK | |
| Phosphatidylinositol signalingsystem | PI4Kβ | |
| Metabolic pathways | FA-CoA, UGT, lipocalin | PEPCK, CBS, L2HGDH, B3GNT3, PI4Kβ, CYP2U1, AK1R1D1 |
| Insulin signaling pathway | HRAs, GNAQ | PEPCK |
| Thyroid hormone synthesis | GNAQ | PDIA4 |
| Primary bile acids synthesis | AK1R1D1 | |
| Themogenesis | HRAs | NPR-A |
| Classification | Subclass | Composition |
|---|---|---|
| Fatty acyl | Free fatty acid | FFA(18:1), FFA(16:0), FFA(18:2), FFA(18:0), FFA(16:1), FFA(20:1), FFA(14:0), FFA(22:1), FFA(20:2), FFA(24:1) |
| Acyl carnitine | CAR18:1, CAR16:1, CAR16, CAR18:2, CAR18, CAR14, CAR12-OH, CAR14:1, CAR18:1-OH, CAR12 | |
| Glyveride | Diacylglycerol | DG(34:1), DG(36:2), DG(36:3), DG(34:4), DG(34:0), DG(40:7), DG(32:1), DG(34:2), DG(32:0), DG(36:0) |
| Triacylglycerol | TG(52:2), TG(54:3), TG(52:3), TG(54:5), TG(50:2), TG(54:2), TG(52:4), TG(50:3), TG(58:8), TG(50:1) | |
| Glyceryl phosphatide | Lysophosphatidyl choline | LPC(22:6), LPC(18:2), LPC(16:0), LPC(18:1), LPC(18:0), LPC(20:5), LPC(16:1), LPC(20:3), LPC(22:5), LPC(18:3) |
| Lysophosphatidyl ethanolamine | PE(40:6), PE(40:7), PE(38:6), PE(36:2), PE(38:4), PE(36:3), PE(42:7), PE(34:2), PE(P-38:5), PE(40:8), LPE(22:6), LPE(18:0), LPE(18:1), LPE(16:0), LPE(18:2), LPE(20:5), LPE(20:4), LPE(22:5), LPE(20:1), LPE(20:0) | |
| Phosphatidyl choline | PC(34:1), PC(34:2), PC(38:6), PC(36:3), PC(36:1), PC(32:1), PC(40:6), PC(O-40:7), PC(36:2), PC(O-38:6) | |
| Phosphatidyl glycerol | PG(40:8), PG(36:2), PG(34:1), PG(36:3), PG(40:7), PG(36:1), PG(36:4), PG(38:3), PG(34:2), PG(34:3) | |
| Phosphatidylinositol | PI(38:4), PI(38:5), PI(40:6), PI(38:6), PI(36:2), PI(40:7), PI(38:3), PI(36:1), PI(36:3), PI(40:5) | |
| Phosphatidylserine | PS(40:6), PS(38:6), PS(40:7), PS(36:2), PS(36:1), PS(40:5), PS(38:4), PS(38:5), PS(38:3), PS(34:1) | |
| Sphingolipid | Sphingomyelin | SM(d42:2), SM(d34:1), SM(d42:1), SM(d40:1), SM(d36:1), SM(d32:1), SM(d41:1), SM(d42:3), SM(d42:5), SM(d35:0) |
| Ceramide | Cer(d42:2), Cer(d42:1), Cer(d34:1), Cer(d40:1), HerCer(d34:1), Cer(d32:1), Cer(d41:1), Cer(d36:1), CerP(d36:1), Cer(d44:2) | |
| Cholesterol | Cholesteryl ester | CE(22:6), CE(18:0), CE(18:1), CE(18:2), CE(20:5), CE(22:5), CE(20:4), CE(16:0), CE(22:3), CE(20:2), CE(20:1) |
| Metabolites | Class | VIP | p-Value | Log2FC | Type |
|---|---|---|---|---|---|
| TG (51:0) | TGs | 1.22 | N/A | 4.50 | up |
| TG (51:1) | TGs | 1.22 | 0.09 | 3.44 | up |
| TG (54:1) | TGs | 1.11 | 0.09 | 2.93 | up |
| TG (50:0) | TGs | 1.25 | 0.07 | 2.79 | up |
| TG (52:0) | TGs | 1.19 | 0.13 | 2.64 | up |
| TG (49:1) | TGs | 1.29 | 0.03 | 2.62 | up |
| TG (54:0) | TGs | 1.09 | 0.16 | 2.53 | up |
| TG (52:1) | TGs | 1.17 | 0.06 | 2.49 | up |
| TG (48:0) | TGs | 1.32 | 0.03 | 2.43 | up |
| TG (46:0) | TGs | 1.34 | 0.02 | 2.39 | up |
| TG (56:0) | TGs | 1.09 | 0.12 | 2.37 | up |
| TG (52:7) | TGs | 1.52 | 0.02 | 2.33 | up |
| TG (53:2) | TGs | 1.07 | 0.06 | 2.26 | up |
| TG (58:7) | TGs | 1.32 | 0.04 | 2.21 | up |
| TG (44:0) | TGs | 1.18 | 0.03 | 2.14 | up |
| TG (56:9) | TGs | 1.21 | 0.07 | 2.02 | up |
| TG (58:10) | TGs | 1.14 | 0.05 | 1.92 | up |
| DG (38:6) | DGs | 1.33 | 0.00 | 2.03 | up |
| PI (38:5) | PIs | 1.54 | 0.00 | 2.25 | up |
| PE (P-40:5) | PEs | 1.47 | 0.02 | 2.37 | up |
| LPC (20:2) | LPCs | 1.32 | 0.03 | 0.96 | down |
| Cer (d34:1) | Cers | 1.16 | 0.05 | 0.72 | down |
| PE (P-34:2) | PEs | 1.17 | 0.05 | 1.38 | down |
| Pathways | ko_ID | Unique Compound |
|---|---|---|
| Metabolic pathways | ko01100 | 99 |
| Insulin resistance | ko04931 | 36 |
| Sphingolipid signaling pathway | ko04071 | 21 |
| Fat digestion and absorption | ko04975 | 29 |
| Cholesterol metabolism | ko04979 | 29 |
| Glycerophospholipid metabolism | ko00564 | 50 |
| Vitamin digestion and absorption | ko04977 | 29 |
| Regulation of lipolysis in adipocytes | ko04923 | 28 |
| Necroptosis | ko04217 | 21 |
| Neurotrophin signaling pathway | ko04722 | 8 |
| Adipocytokine signaling pathway | ko04920 | 8 |
| Sphingolipid metabolism | ko00600 | 25 |
| AGE-RAGE signaling pathway in diabetic complications | ko04933 | 8 |
| Leishmaniasis | ko05140 | 8 |
| Glycerolipid metabolism | ko00561 | 33 |
| Inositol phosphate metabolism | ko00562 | 5 |
| Phosphatidylinositol signaling system | ko04070 | 5 |
| Long-term depression | ko04730 | 5 |
| Choline metabolism in cancer | ko05231 | 32 |
| Arachidonic acid metabolism | ko00590 | 27 |
| Linoleic acid metabolism | ko00591 | 27 |
| alpha-Linolenic acid metabolism | ko00592 | 27 |
| Retrograde endocannabinoid signaling | ko04723 | 40 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | ko00563 | 13 |
| Autophagy—other | ko04136 | 13 |
| Autophagy | ko04140 | 13 |
| Pathogenic Escherichia coli infection | ko05130 | 13 |
| Kaposi sarcoma-associated herpesvirus infection | ko05167 | 13 |
| Thermogenesis | ko04714 | 28 |
| Pathway | Number of Lipids | Number of Proteins | Differential Lipids | Differential Proteins (Up-Regulated) | Differential Proteins (Down-Regulated) |
|---|---|---|---|---|---|
| Themogenesis | 27 | 2 | TG | NPR-A, HRAs | |
| Fat digestion and absorption | 27 | 2 | TG | FABP1 | ABCA1 |
| Cholesterol metabolism | 27 | 2 | TG | ABCA1, VDCA1 | |
| Metabolic pathways | 48 | 17 | TG | NPR-A, FA-CoA, UGT | PI4Kβ, AK1R1D1, PEPCK, L2HGDH, CBS |
| Arachidonic acid metabolism | 11 | 2 | TG | CYP2U1 | |
| Inositol phosphate metabolism | 3 | 2 | TG | ITPK1 | PI4Kβ |
| Phosphatidylinositol signaling system | 3 | 2 | TG | ITPK1 | PI4Kβ |
| Long-term depression | 3 | 2 | TG | GNAQ, HRAs | |
| Sphingolipid signaling pathway | 2 | 2 | SM | GNAQ, HRAs |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, M.; Yao, T.; Xue, M.; Francis, F.; Qin, Y.; Jia, M.; Li, J.; Gu, X. Mechanism Analysis of Metabolic Fatty Liver on Largemouth Bass (Micropterus salmoides) Based on Integrated Lipidomics and Proteomics. Metabolites 2022, 12, 759. https://doi.org/10.3390/metabo12080759
Xue M, Yao T, Xue M, Francis F, Qin Y, Jia M, Li J, Gu X. Mechanism Analysis of Metabolic Fatty Liver on Largemouth Bass (Micropterus salmoides) Based on Integrated Lipidomics and Proteomics. Metabolites. 2022; 12(8):759. https://doi.org/10.3390/metabo12080759
Chicago/Turabian StyleXue, Moyong, Ting Yao, Min Xue, Frédéric Francis, Yuchang Qin, Ming Jia, Junguo Li, and Xu Gu. 2022. "Mechanism Analysis of Metabolic Fatty Liver on Largemouth Bass (Micropterus salmoides) Based on Integrated Lipidomics and Proteomics" Metabolites 12, no. 8: 759. https://doi.org/10.3390/metabo12080759
APA StyleXue, M., Yao, T., Xue, M., Francis, F., Qin, Y., Jia, M., Li, J., & Gu, X. (2022). Mechanism Analysis of Metabolic Fatty Liver on Largemouth Bass (Micropterus salmoides) Based on Integrated Lipidomics and Proteomics. Metabolites, 12(8), 759. https://doi.org/10.3390/metabo12080759








