Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. Treatment Schedule and Method
2.3. Circadian Rhythm Sleep Disruption Method
2.4. Plasma Sample Preparation
2.5. Mass Spectrometric Analyses
2.6. Chromatography Conditions
2.7. Statistical Analysis
- Mass-to-charge ratio- m/z represents mass divided by charge number and the horizontal axis in a mass spectrum is expressed in units of m/z;
- (STDEV (five replicates of peak area for each metabolite)/AVERAGE (five replicates peak area for each metabolite)) ∗ 100.
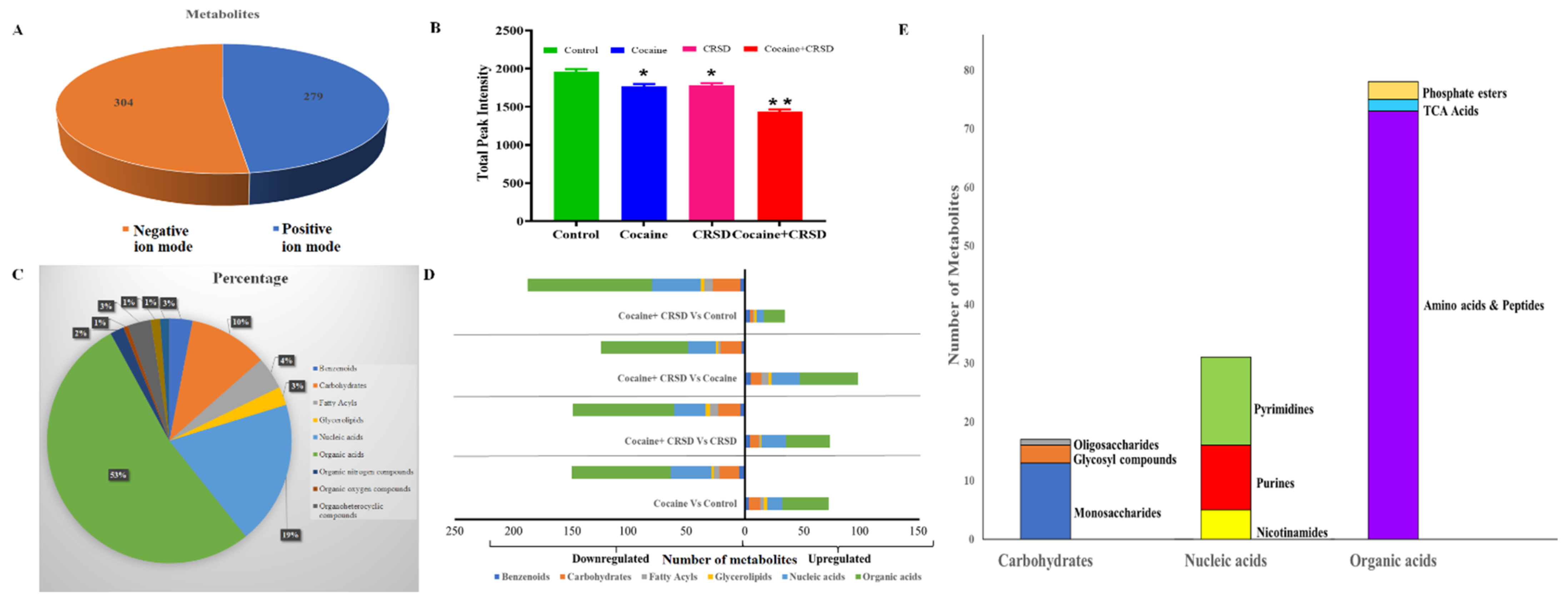
3. Results
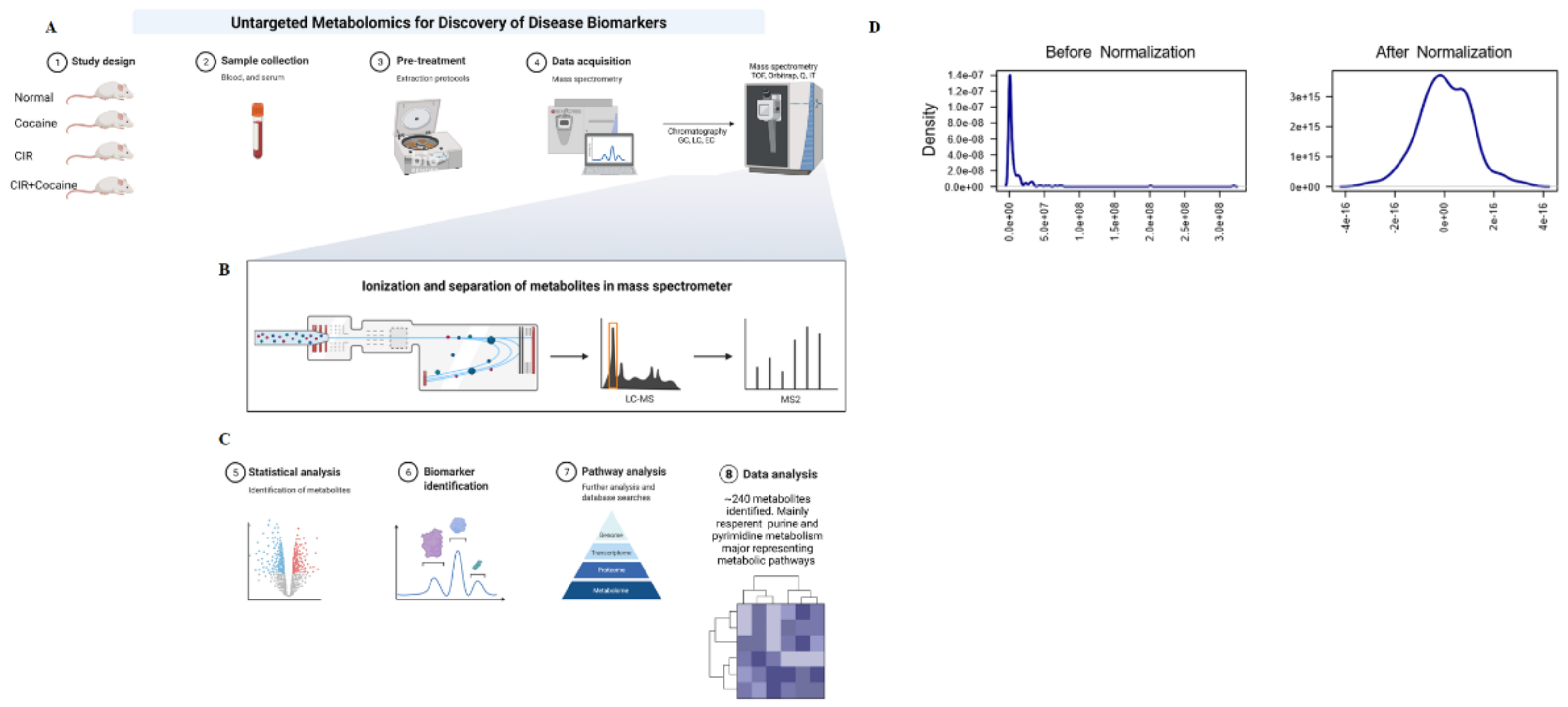
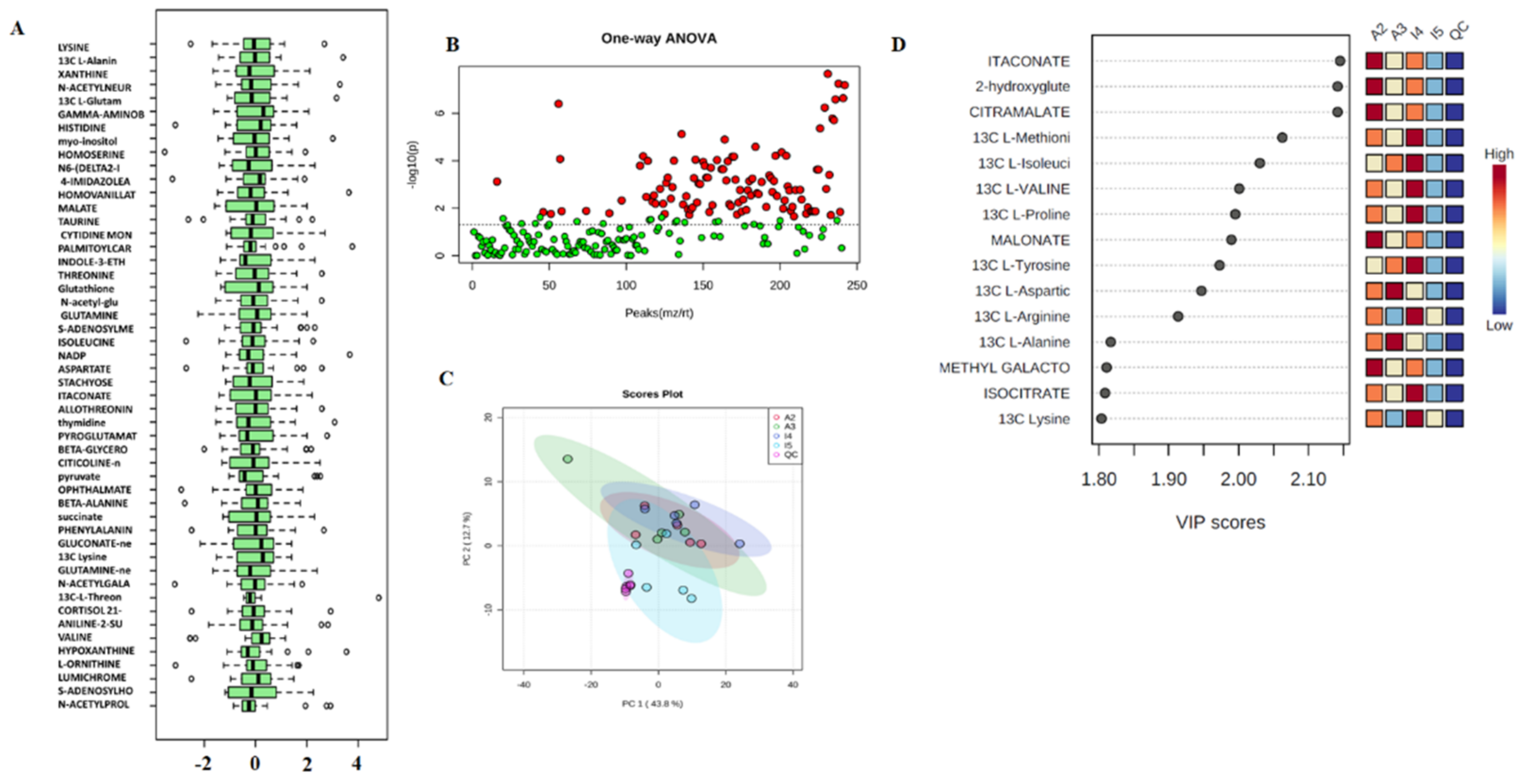
3.1. Extraction and Preprocessing
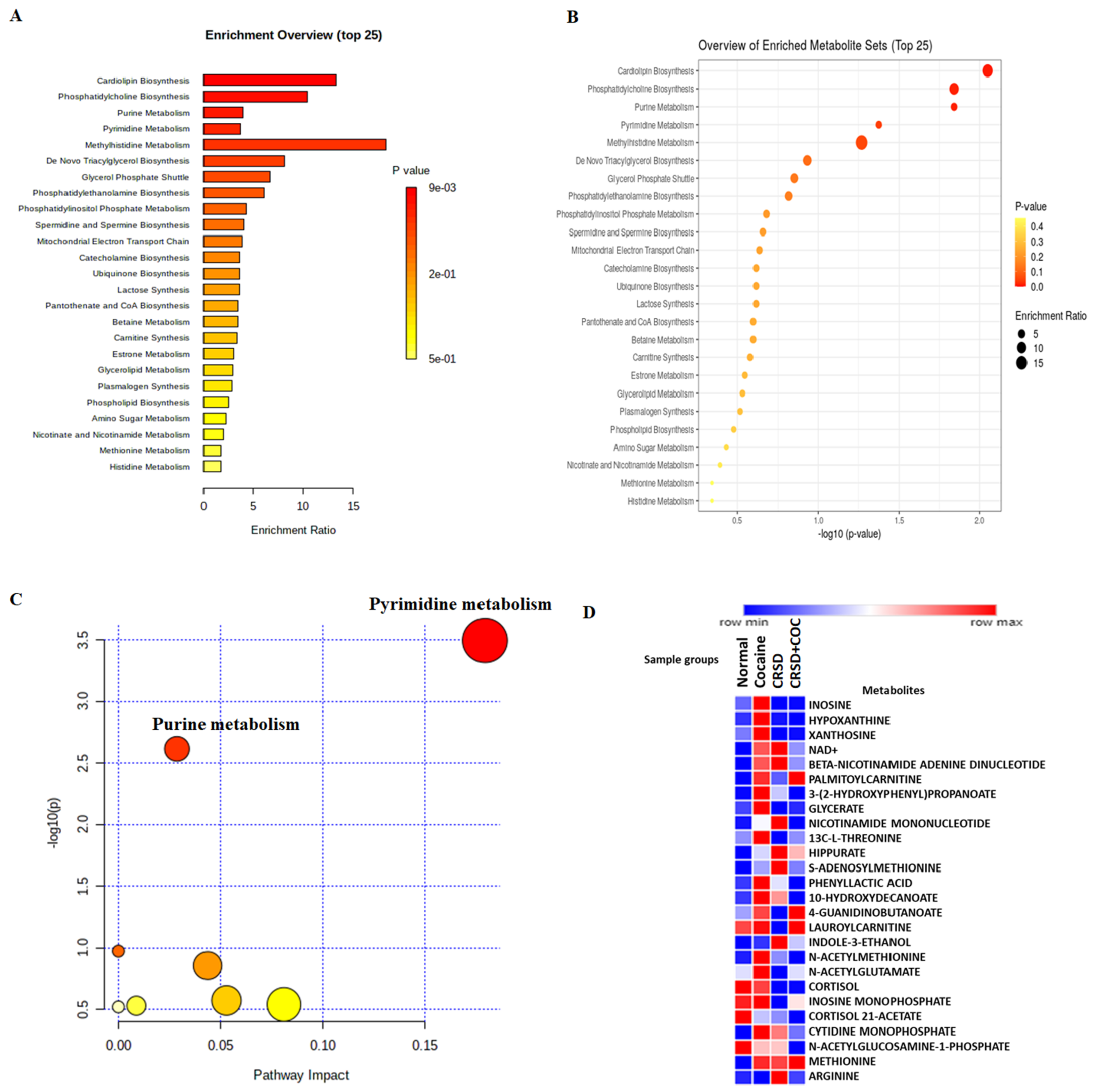
3.2. Analysis
4. Discussion
5. Conclusions and Future Direction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pickel, L.; Sung, H.K. Feeding Rhythms and the Circadian Regulation of Metabolism. Front. Nutr. 2020, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Wu, D.; Ptáček, L.J.; Fu, Y.H. Human Genetics and Sleep Behavior. Curr. Opin. Neurobiol. 2017, 44, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Mignot, E. Genetics of Sleep and Sleep Disorders. Cell 2011, 146, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Hasler, B.P.; Smith, L.J.; Cousins, J.C.; Bootzin, R.R. Circadian Rhythms, Sleep, and Substance Abuse. Sleep Med. Rev. 2012, 16, 67–81. [Google Scholar] [CrossRef]
- Roehrs, T.A.; Roth, T. Sleep Disturbance in Substance Use Disorders. Psychiatr. Clin. N. Am. 2015, 38, 793–803. [Google Scholar] [CrossRef]
- Angarita, G.A.; Emadi, N.; Hodges, S.; Morgan, P.T. Sleep Abnormalities Associated with Alcohol, Cannabis, Cocaine, and Opiate Use: A Comprehensive Review. Addict. Sci. Clin. Pract. 2016, 11, 9. [Google Scholar] [CrossRef]
- Verwey, M.; Dhir, S.; Amir, S. Circadian Influences on Dopamine Circuits of the Brain: Regulation of Striatal Rhythms of Clock Gene Expression and Implications for Psychopathology and Disease. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Strum, J. Cocaine & Sleep: Circadian Rhythms, Insomnia, and More|The Recovery Village. Available online: https://www.therecoveryvillage.com/cocaine-addiction/faq/cocaines-effects-on-sleep/ (accessed on 18 February 2022).
- Musiek, E.S. Circadian Clock Disruption in Neurodegenerative Diseases: Cause and Effect? Front. Pharmacol. 2015, 6, 29. [Google Scholar] [CrossRef]
- Du, X.; Xu, W.; Li, X.; Zhou, D.; Han, C. Sleep Disorder in Drug Addiction: Treatment With Transcranial Magnetic Stimulation. Front. Psychiatry 2019, 10, 848. [Google Scholar] [CrossRef]
- Schulte, M.T.; Hser, Y.I. Substance Use and Associated Health Conditions throughout the Lifespan. Public Health Rev. 2014, 35, 3. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, X.; Qi, D.; Sun, M.; Hou, Q.; Li, Y.; Jiang, H. Establishment of the Circadian Metabolic Phenotype Strategy in Spontaneously Hypertensive Rats: A Dynamic Metabolomics Study. J. Transl. Med. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Videnovic, A.; Zee, P.C. Consequences of Circadian Disruption on Neurologic Health. Sleep Med. Clin. 2015, 10, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, R.; Sumner, S. Using Metabolomics to Investigate Biomarkers of Drug Addiction. Trends Mol. Med. 2018, 24, 197–205. [Google Scholar] [CrossRef]
- Patkar, A.A.; Rozen, S.; Mannelli, P.; Matson, W.; Pae, C.U.; Krishnan, K.R.; Kaddurah-Daouk, R. Alterations in Tryptophan and Purine Metabolism in Cocaine Addiction: A Metabolomic Study. Psychopharmacology 2009, 206, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, N.; Zhao, T.Y.; Li, J. The Potential Biomarkers of Drug Addiction: Proteomic and Metabolomics Challenges. Biomarkers 2016, 21, 678–685. [Google Scholar] [CrossRef]
- bin Heyat, M.B.; Akhtar, F.; Khan, A.; Noor, A.; Benjdira, B.; Qamar, Y.; Abbas, S.J.; Lai, D. A Novel Hybrid Machine Learning Classification for the Detection of Bruxism Patients Using Physiological Signals. Appl. Sci. 2020, 10, 7410. [Google Scholar] [CrossRef]
- bin Heyat, M.B.; Akhtar, F.; Khan, M.H.; Ullah, N.; Gul, I.; Khan, H.; Lai, D. Detection, Treatment Planning, and Genetic Predisposition of Bruxism: A Systematic Mapping Process and Network Visualization Tech-nique. CNS Neurol. Disord. Drug Targets 2020, 20, 755–775. [Google Scholar] [CrossRef]
- bin Heyat, M.B.; Akhtar, F.; Ansari, M.A.; Khan, A.; Alkahtani, F.; Khan, H.; Lai, D. Progress in Detection of Insomnia Sleep Disorder: A Comprehensive Review. Curr. Drug Targets 2020, 22, 672–684. [Google Scholar] [CrossRef]
- Lewis, G.D.; Wei, R.; Liu, E.; Yang, E.; Shi, X.; Martinovic, M.; Farrell, L.; Asnani, A.; Cyrille, M.; Ramanathan, A.; et al. Metabolite Profiling of Blood from Individuals Undergoing Planned Myocardial Infarction Reveals Early Markers of Myocardial Injury. J. Clin. Investig. 2008, 118, 3503–3512. [Google Scholar] [CrossRef]
- Zhang, J.; Bowers, J.; Liu, L.; Wei, S.; Gowda, G.A.N.; Hammoud, Z.; Raftery, D. Esophageal Cancer Metabolite Biomarkers Detected by LC-MS and NMR Methods. PLoS ONE 2012, 7, e30181. [Google Scholar] [CrossRef]
- Psychology Today Staff Alcohol Use Disorder | Psychology Today. Available online: https://www.psychologytoday.com/us/conditions/cocaine-use-disorder (accessed on 21 February 2022).
- Sharma, S.; Kavuru, M. Sleep and Metabolism: An Overview. Int. J. Endocrinol. 2010, 2010, 270832. [Google Scholar] [CrossRef] [PubMed]
- Logan, R.W.; Williams, W.P.; McClung, C.A. Circadian Rhythms and Addiction: Mechanistic Insights and Future Directions. Behav. Neurosci. 2014, 128, 387–412. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.; Maertzdorf, J.; Sutherland, J.S.; Duffy, F.J.; Thompson, E.; Suliman, S.; McEwen, G.; Thiel, B.; Parida, S.K.; Zyla, J.; et al. Metabolite Changes in Blood Predict the Onset of Tuberculosis. Nat. Commun. 2018, 9, 5208. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.J. Fundamentals of Biochemistry: Life at the Molecular Level (Third Edition) by D. Voet, J. Voet, and C. W. Pratt. Biochem. Mol. Biol. Educ. 2008, 36, 319–320. [Google Scholar] [CrossRef]
- Winkler, J.K.; Suttle, D.P. Analysis of UMP Synthase Gene and MRNA Structure in Hereditary Orotic Aciduria Fibroblasts. Am. J. Hum. Genet. 1988, 43, 86–94. [Google Scholar] [PubMed]
- Myers, C.S.; Napolitano, M.; Fisher, H.; Wagner, G.C. Uridine and Stimulant-Induced Motor Activity. Proc. Soc. Exp. Biol. Med. 1993, 204, 49–53. [Google Scholar] [CrossRef]
- Ocalan, B.; Cakir, A.; Koc, C.; Suyen, G.G.; Kahveci, N. Uridine Treatment Prevents REM Sleep Deprivation-Induced Learning and Memory Impairment. Neurosci. Res. 2019, 148, 42–48. [Google Scholar] [CrossRef]
- Ma, Q.F.; Kenyon, G.L.; Markham, G.D. Specificity of S-Adenosylmethionine Synthetase for ATP Analogues Mono- and Disubstituted in Bridging Positions of the Polyphosphate Chain. Biochemistry 1990, 29, 1412–1416. [Google Scholar] [CrossRef]
- Kubrusly, R.C.C.; Bhide, P.G. Cocaine Exposure Modulates Dopamine and Adenosine Signaling in the Fetal Brain. Neuropharmacology 2010, 58, 436–443. [Google Scholar] [CrossRef]
- Ferré, S.; Fredholm, B.B.; Morelli, M.; Popoli, P.; Fuxe, K. Adenosine-Dopamine Receptor-Receptor Interactions as an Integrative Mechanism in the Basal Ganglia. Trends Neurosci. 1997, 20, 482–487. [Google Scholar] [CrossRef]
- Fiorillo, C.D.; Williams, J.T. Selective Inhibition by Adenosine of MGluR IPSPs in Dopamine Neurons after Cocaine Treatment. J. Neurophysiol. 2000, 83, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Loidl, C.F.; Herrera-Marschitz, M.; Andersson, K.; You, Z.B.; Goiny, M.; O’Connor, W.T.; Silveira, R.; Rawal, R.; Bjelke, B.; Chen, Y.; et al. Long-Term Effects of Perinatal Asphyxia on Basal Ganglia Neurotransmitter Systems Studied with Microdialysis in Rat. Neurosci. Lett. 1994, 175, 9–12. [Google Scholar] [CrossRef]
- Doke, M.; Kashanchi, F.; Khan, M.A.; Samikkannu, T. HIV-1 Tat and Cocaine Impact Astrocytic Energy Reservoir Influence on MiRNA Epigenetic Regulation. Genomics 2021, 113, 3461–3475. [Google Scholar] [CrossRef] [PubMed]
- Doke, M.; Jeganathan, V.; McLaughlin, J.P.; Samikkannu, T. HIV-1 Tat and Cocaine Impact Mitochondrial Epigenetics: Effects on DNA Methylation. Epigenetics 2020. [Google Scholar] [CrossRef]
- Doke, M.; Kashanchi, F.; Khan, M.A.; Samikkannu, T. HIV-1 Tat and Cocaine Coexposure Impacts PiRNAs to Affect Astrocyte Energy Metabolism. Epigenomics 2022. [Google Scholar] [CrossRef] [PubMed]
- Mugabo, Y.; Zhao, S.; Seifried, A.; Gezzar, S.; Al-Mass, A.; Zhang, D.; Lamontagne, J.; Attane, C.; Poursharifi, P.; Iglesias, J.; et al. Identification of a Mammalian Glycerol-3-Phosphate Phosphatase: Role in Metabolism and Signaling in Pancreatic β-Cells and Hepatocytes. Proc. Natl. Acad. Sci. USA 2016, 113, E430–E439. [Google Scholar] [CrossRef]
- Pecinová, A.; Alán, L.; Brázdová, A.; Vrbacký, M.; Pecina, P.; Drahota, Z.; Houštěk, J.; Mráček, T. Role of Mitochondrial Glycerol-3-Phosphate Dehydrogenase in Metabolic Adaptations of Prostate Cancer. Cells 2020, 9, 1764. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Schroeder, T.; Mueller-Klieser, W. Lactate in Solid Malignant Tumors: Potential Basis of a Metabolic Classification in Clinical Oncology. Curr. Med. Chem. 2012, 11, 2195–2204. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite Profiling of Human Colon Carcinoma—Deregulation of TCA Cycle and Amino Acid Turnover. Mol. Cancer 2008, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]



| Samples | Features (Positive) | Missing/Zero | Features (Processed) |
|---|---|---|---|
| A2-CIR-1 | 243 | 0 | 242 |
| A2-CIR-2 | 243 | 0 | 242 |
| A2-CIR-3 | 243 | 0 | 242 |
| A2-CIR-4 | 243 | 0 | 242 |
| A2-CIR-5 | 243 | 0 | 242 |
| A3-COC-CIR-1 | 243 | 0 | 242 |
| A3-COC-CIR-2 | 243 | 0 | 242 |
| A3-COC-CIR-3 | 243 | 0 | 242 |
| A3-COC-CIR-4 | 243 | 0 | 242 |
| A3-COC-CIR-5 | 243 | 0 | 242 |
| I4-Control-1 | 243 | 0 | 242 |
| I4-Control-2 | 243 | 0 | 242 |
| I4-Control-3 | 243 | 0 | 242 |
| I4-Control-4 | 243 | 0 | 242 |
| I4-Control-5 | 243 | 0 | 242 |
| I5-COCAINE-1 | 243 | 0 | 242 |
| I5-COCAINE-2 | 243 | 0 | 242 |
| I5-COCAINE-3 | 243 | 0 | 242 |
| I5-COCAIN-4 | 243 | 0 | 242 |
| I5-COCAIN-5 | 243 | 0 | 242 |
| QC-start-run-1 | 243 | 0 | 242 |
| QC-start-run-2 | 243 | 0 | 242 |
| QC-start-run-3 | 243 | 0 | 242 |
| QC-end-run-1 | 243 | 0 | 242 |
| QC-end-run-2 | 243 | 0 | 242 |
| QC-end-run-3 | 243 | 0 | 242 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doke, M.; McLaughlin, J.P.; Baniasadi, H.; Samikkannu, T. Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures. Metabolites 2022, 12, 869. https://doi.org/10.3390/metabo12090869
Doke M, McLaughlin JP, Baniasadi H, Samikkannu T. Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures. Metabolites. 2022; 12(9):869. https://doi.org/10.3390/metabo12090869
Chicago/Turabian StyleDoke, Mayur, Jay P. McLaughlin, Hamid Baniasadi, and Thangavel Samikkannu. 2022. "Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures" Metabolites 12, no. 9: 869. https://doi.org/10.3390/metabo12090869
APA StyleDoke, M., McLaughlin, J. P., Baniasadi, H., & Samikkannu, T. (2022). Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures. Metabolites, 12(9), 869. https://doi.org/10.3390/metabo12090869







