Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Broadly Targeted Metabolomic Analysis
- Sample preparation and extraction
- UPLC conditions
- ESI-Q TRAP-MS/MS
- Metabolite characterization and quantification
2.3. RNA Extraction and Transcriptome Analysis
2.4. Data Processing and Statistical Analysis
3. Results
3.1. Dynamic Metabolic Profiles at Five Hawthorn Developmental Stages Revealed via UPLC-MS/MS Analysis
3.2. Characteristics of Dynamic Changes in Primary Metabolites during Hawthorn Fruit Development
3.2.1. Metabolism of Sugars and Alcohols
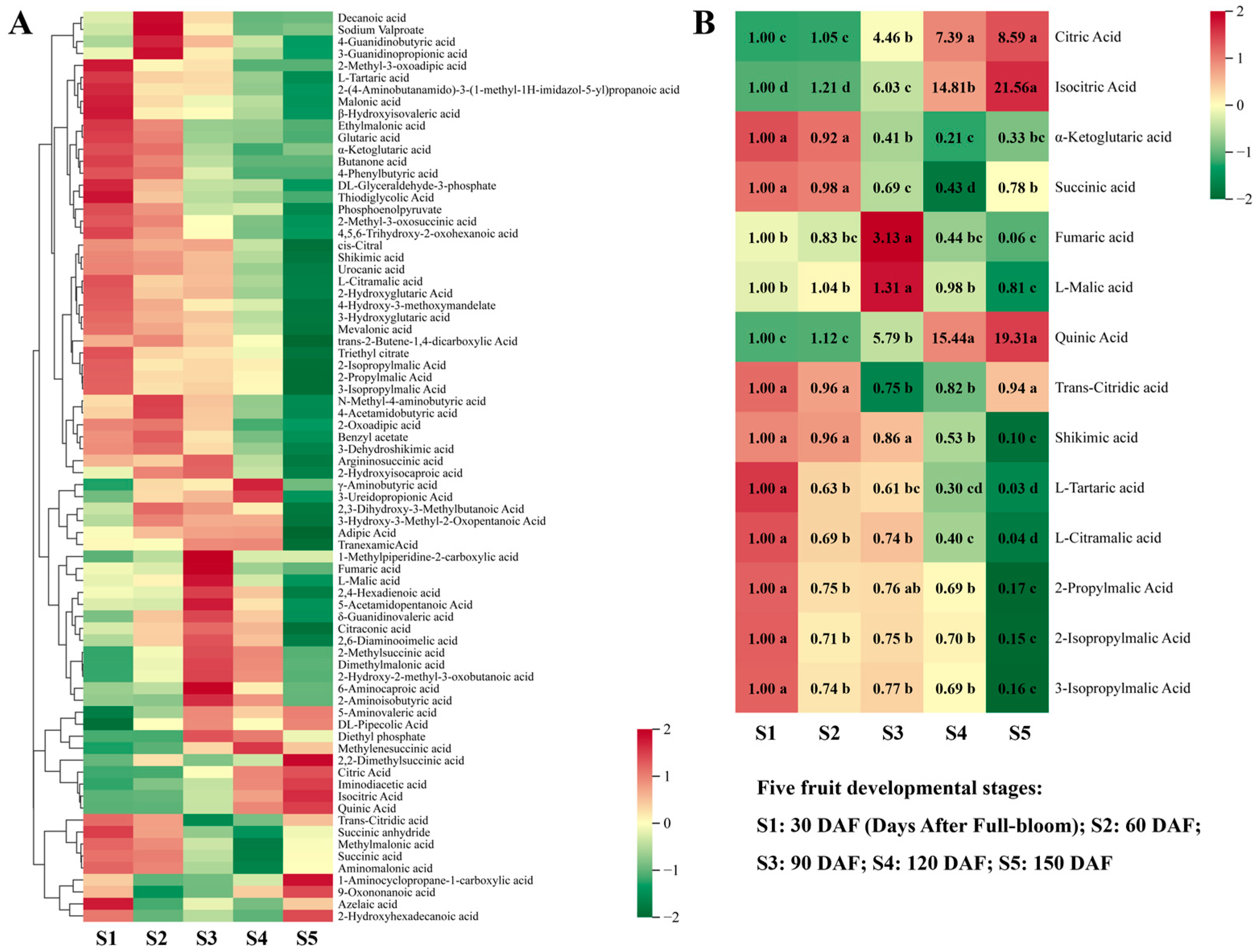
3.2.2. Organic Acid Metabolism during Hawthorn Development
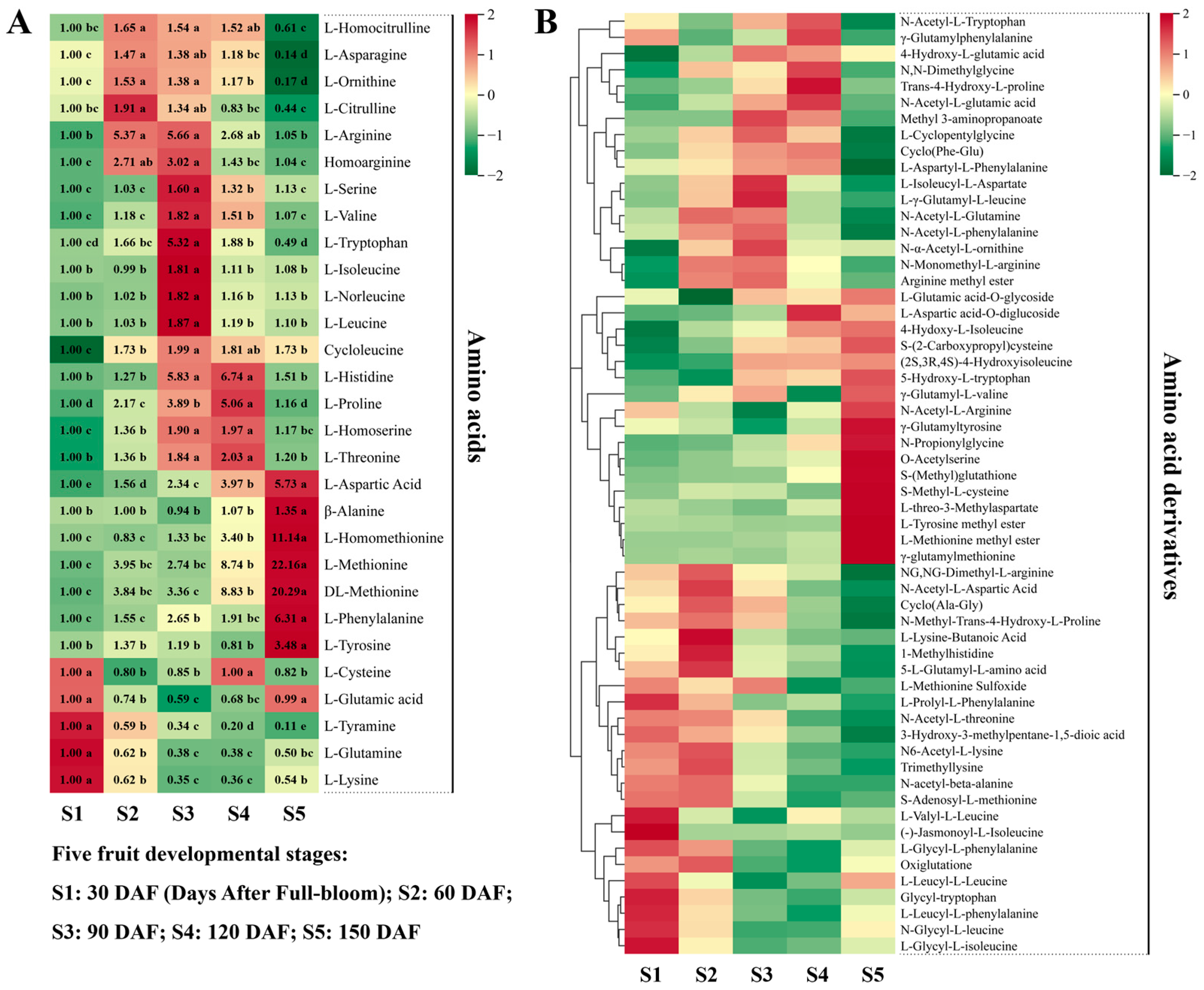
3.2.3. Accumulation Curves for Other Primary Metabolites
3.3. Accumulation of Secondary Metabolites during Hawthorn Fruit Development
3.3.1. Metabolism of Phenolic Acid Compounds
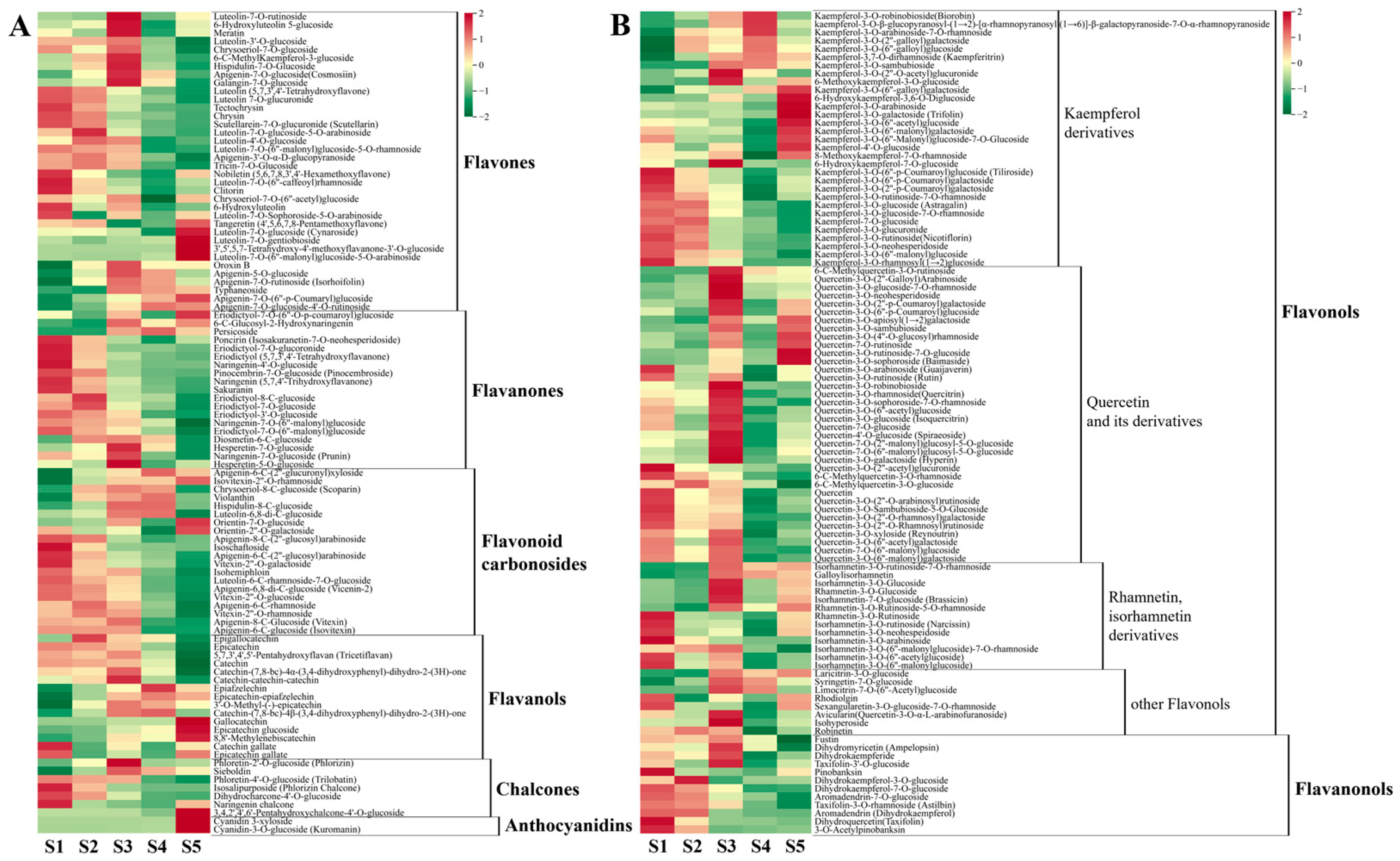
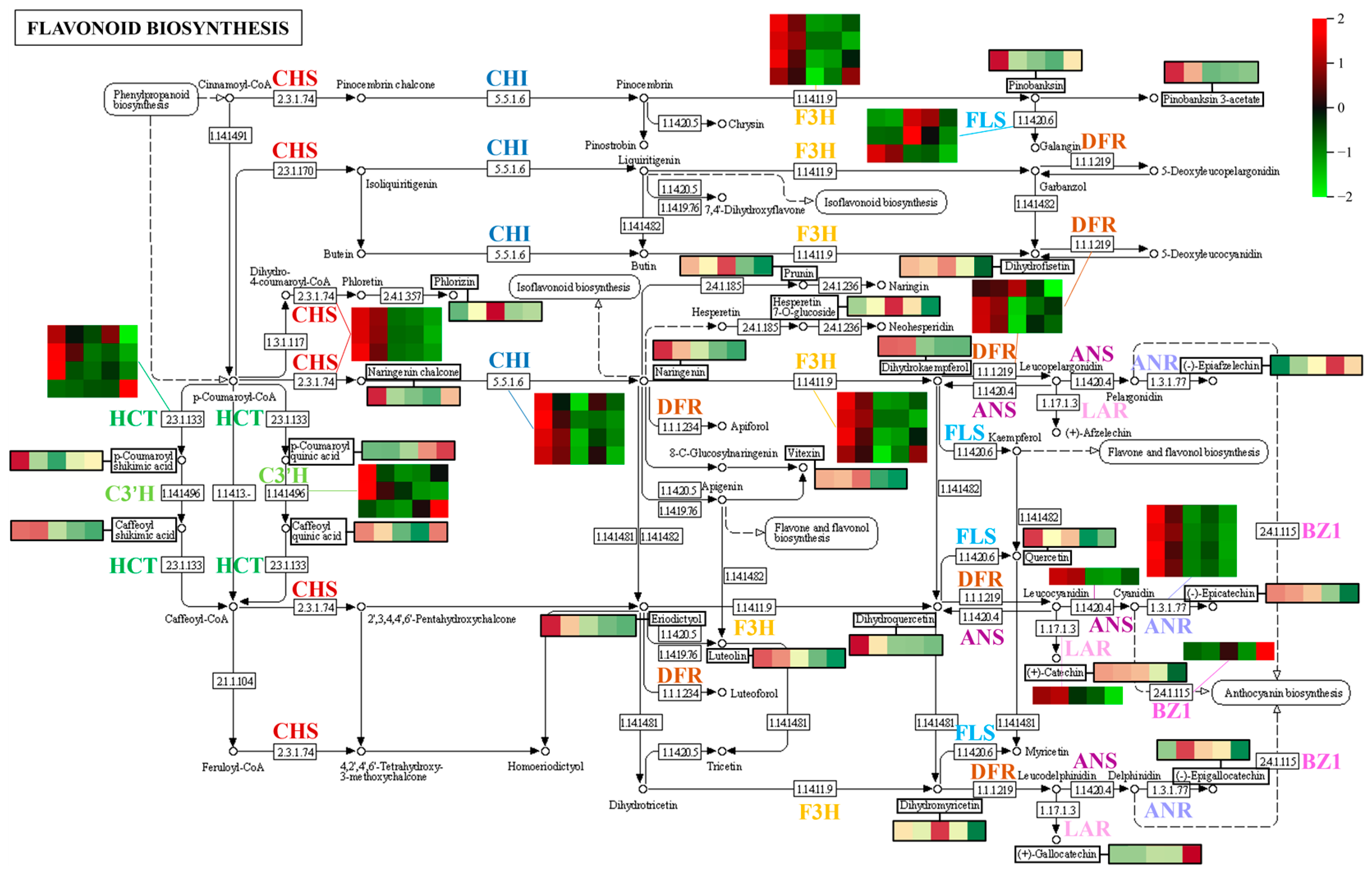
3.3.2. Metabolism of Flavonoids
- Flavonoids
- Flavanones
- Flavonoid carbonosides
- Flavanols
- Chalcones
- Anthocyanidins
- Flavonols
- Flavanonols
3.3.3. Accumulation of Other Secondary Metabolites (Excluding Flavonoids and Phenolic Acids)
- Phytohormones
- Lignans and coumarins
- Terpenoids
- Stilbenes
- Tannins
- Alkaloids and other nitrogen-containing metabolites
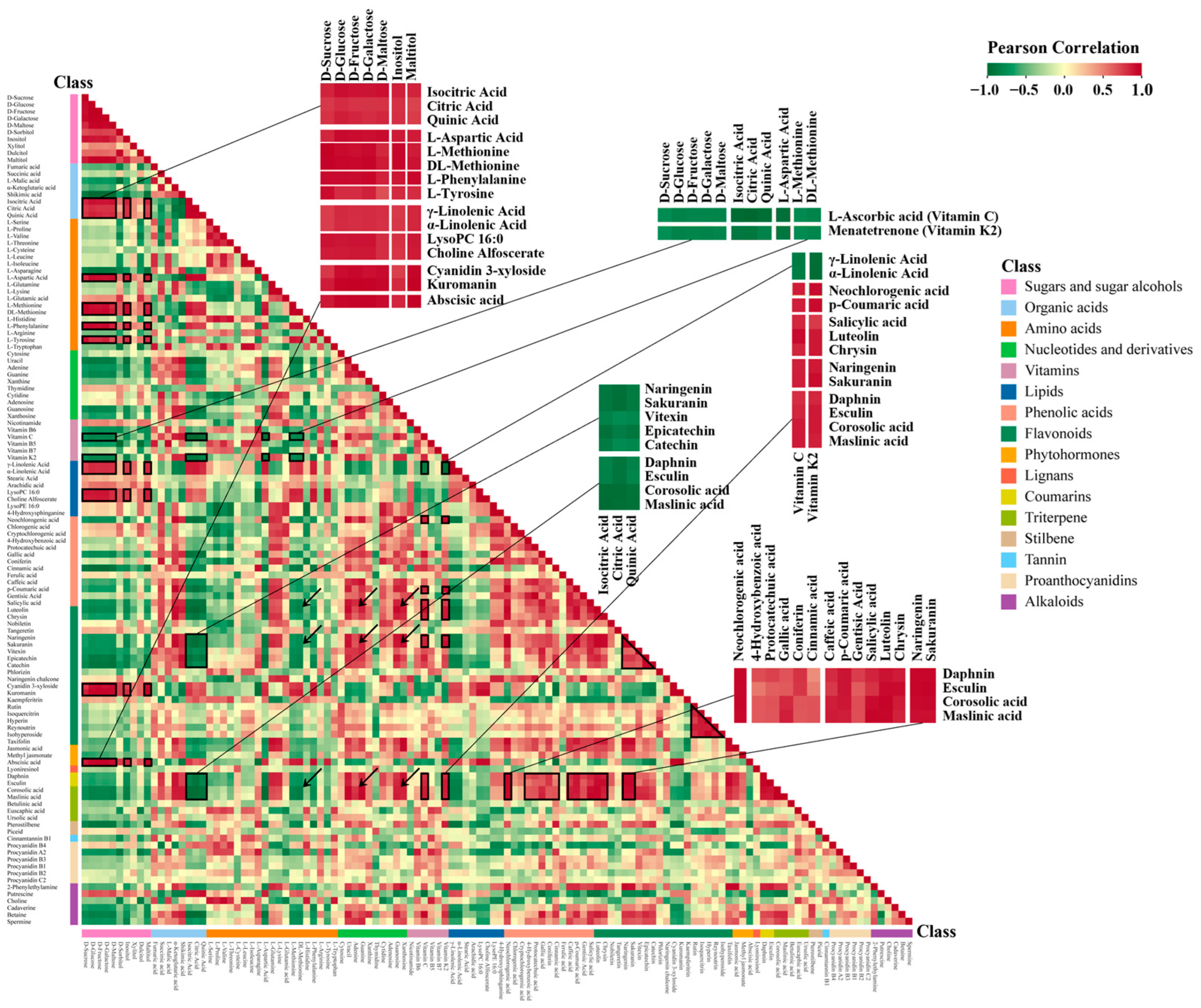
3.4. Metabolite-Metabolite Correlations during Hawthorn Development and Ripening
3.5. Differential Accumulation of Metabolites among Stages of Fruit Development
3.6. Expression Patterns of Genes Associated with Various Metabolic Pathways during Fruit Development and Ripening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nazhand, A.; Lucarini, M.; Durazzo, A.; Zaccardelli, M.; Cristarella, S.; Souto, S.B.; Silva, A.M.; Severino, P.; Souto, E.B.; Santini, A. Hawthorn (Crataegus spp.): An Updated Overview on Its Beneficial Properties. Forests 2020, 11, 564. [Google Scholar] [CrossRef]
- Zhang, J.; Chai, X.; Zhao, F.; Hou, G.; Meng, Q. Food Applications and Potential Health Benefits of Hawthorn. Foods 2022, 11, 2861. [Google Scholar] [CrossRef]
- Jurikova, T.; Sochor, J.; Rop, O.; Mlcek, J.; Balla, S.; Szekeres, L.; Adam, V.; Kizek, R. Polyphenolic Profile and Biological Activity of Chinese Hawthorn (Crataegus pinnatifida BUNGE) Fruits. Molecules 2012, 17, 14490–14509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Arya, V.; Bhat, Z.A.; Khan, N.A.; Prasad, D.N. The genus Crataegus: Chemical and pharmacological perspectives. Rev. Bras. Farmacogn 2012, 22, 1187–1200. [Google Scholar] [CrossRef]
- Orhan, E.I. Phytochemical and Pharmacological Activity Profile of Crataegus oxyacantha L. (Hawthorn)—A Cardiotonic Herb. Curr. Med. Chem. 2018, 25, 4854–4865. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Perrone, A.; Yousefi, S.; Papini, A.; Castiglione, S.; Guarino, F.; Cicatelli, A.; Aelaei, M.; Arad, N.; Gholami, M.; et al. Botanical, Phytochemical, Anti-Microbial and Pharmaceutical Characteristics of Hawthorn (Crataegus monogyna Jacq.), Rosaceae. Molecules 2021, 26, 7266. [Google Scholar] [CrossRef]
- Liu, J.; Xu, D.; Chen, S.; Yuan, F.; Mao, L.; Gao, Y. Superfruits in China: Bioactive phytochemicals and their potential health benefits-A Review. Food Sci. Nutr. 2021, 9, 6892–6902. [Google Scholar] [CrossRef]
- Li, T.P.; Fu, S.Y.; Huang, X.; Zhang, X.S.; Cui, Y.M.; Zhang, Z.Y.; Ma, Y.; Zhang, X.; Yu, Q.H.; Yang, S.N.; et al. Biological properties and potential application of hawthorn and its major functional components: A review. J. Funct. Foods 2022, 90, 104988. [Google Scholar] [CrossRef]
- Zheng, G.Q.; Deng, J.; Wen, L.R.; You, L.J.; Zhao, Z.G.; Zhou, L. Release of phenolic compounds and antioxidant capacity of Chinese hawthorn “Crataegus pinnatifida” during, in vitro, digestion. J. Funct. Foods 2018, 40, 76–85. [Google Scholar] [CrossRef]
- European Medicines Agency. 2016. Available online: https://www.ema.europa.eu/en/medicines/herbal/crataegi-folium-cum-flore (accessed on 19 September 2022).
- Xu, J.; Yan, J.; Li, W.; Wang, Q.; Wang, C.; Guo, J.; Geng, D.; Guan, Q.; Ma, F. Integrative Analyses of Widely Targeted Metabolic Profiling and Transcriptome Data Reveals Molecular Insight into Metabolomic Variations during Apple (Malus domestica) Fruit Development and Ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.P.; Tang, L.; Mei, X.Y.; Liu, H.Z.; Luo, H.R.; Deng, Y.M.; Su, J.L. Single-molecule long-read sequencing of the full-length transcriptome of Rhododendron lapponicum L. Sci. Rep. 2020, 10, 6755. [Google Scholar] [CrossRef] [Green Version]
- Nabavi, S.F.; Habtemariam, S.; Ahmed, T.; Sureda, A.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.M. Polyphenolic Composition of Crataegus monogyna Jacq.: From Chemistry to Medical Applications. Nutrients 2015, 7, 7708–7728. [Google Scholar] [CrossRef]
- Dehghani, S.; Mehri, S.; Hosseinzadeh, H. The effects of Crataegus pinnatifida (Chinese hawthorn) on metabolic syndrome: A review. Iran. J. Basic Med. Sci. 2019, 22, 460–468. [Google Scholar]
- Rocchetti, G.; Senizza, B.; Zengin, G.; Mahomodally, M.F.; Senkardes, I.; Lobine, D.; Lucini, L. Untargeted metabolomic profiling of three Crataegus species (hawthorn) and their in vitro biological activities. J. Sci. Food Agric. 2020, 100, 1998–2006. [Google Scholar] [CrossRef]
- Wen, L.R.; Guo, X.B.; Liu, R.H.; You, L.J.; Abbasi, A.M.; Fu, X. Phenolic contents and cellular antioxidant activity of Chinese hawthorn “Crataegus pinnatifida”. Food Chem. 2015, 186, 54–62. [Google Scholar] [CrossRef]
- Huang, X.X.; Xu, Y.; Bai, M.; Zhou, L.; Song, S.J.; Wang, X.B. Lignans from the seeds of Chinese hawthorn (Crataegus pinnatifida var. major N.E.Br.) against β-amyloid aggregation. Nat. Prod. Res. 2018, 32, 1706–1713. [Google Scholar] [CrossRef]
- González-Jiménez, F.E.; Salazar-Montoya, J.A.; Calva-Calva, G.; Ramos-Ramírez, E.G. Phytochemical Characterization, In Vitro Antioxidant Activity, and Quantitative Analysis by Micellar Electrokinetic Chromatography of Hawthorn (Crataegus pubescens) Fruit. J. Food Qual. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Lv, T.M.; Han, F.Y.; Lin, B.; Yao, G.D.; Wang, X.B.; Huang, X.X.; Song, S.J. Chiral resolution and neuroprotective activities of enantiomeric dihydrobenzofuranneolignans from the fruit of Crataegus pinnatifida. Bioorg. Chem. 2019, 85, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, H.; Han, F.Y.; Guo, R.; Huang, S.W.; Lin, B.; Huang, X.X.; Song, S.J. Chiral resolution and neuroprotective activities of enantiomeric 8-O-4′ neolignans from the fruits of Crataegus pinnatifida Bge. Fitoterapia 2019, 136, 104164. [Google Scholar] [CrossRef]
- Zhang, J.J.; Wang, X.; Yu, O.; Tang, J.J.; Gu, X.G.; Wan, X.C.; Fang, C.B. Metabolic profiling of strawberry (Fragaria × ananassa Duch.) during fruit development and maturation. J. Exp. Bot. 2011, 62, 1103–1118. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.T.; Wang, S.C.; Huang, Z.J.; Zhang, S.B.; Liao, Q.G.; Zhang, C.Z.; Lin, T.; Qin, M.; Peng, M.; Yang, C.K.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhao, X.; Fu, D.; Zhao, Y. Integrated Analysis of Widely Targeted Metabolomics and Transcriptomics Reveals the Effects of Transcription Factor NOR-like1 on Alkaloids, Phenolic Acids, and Flavonoids in Tomato at Different Ripening Stages. Metabolites 2022, 12, 1296. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Zhou, X.; Zhao, J.; Liu, X.; Jiang, Q.; Ren, F. Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.). Metabolites 2022, 12, 905. [Google Scholar] [CrossRef]
- Wu, S.; Tohge, T.; Cuadros-Inostroza, Á.; Tong, H.; Tenenboim, H.; Kooke, R.; Méret, M.; Keurentjes, J.B.; Nikoloski, Z.; Fernie, A.R.; et al. Mapping the Arabidopsis Metabolic Landscape by Untargeted Metabolomics at Different Environmental Conditions. Mol. Plant 2018, 11, 118–134. [Google Scholar] [CrossRef] [Green Version]
- Etienne, A.; Génard, M.; Lobit, P.; Mbeguié-A-Mbéguié, D.; Bugaud, C. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 2013, 64, 1451–1469. [Google Scholar] [CrossRef] [Green Version]
- Reuscher, S.; Fukao, Y.; Morimoto, R.; Otagaki, S.; Oikawa, A.; Isuzugawa, K.; Shiratake, K. Quantitative Proteomics-Based Reconstruction and Identification of Metabolic Pathways and Membrane Transport Proteins Related to Sugar Accumulation in Developing Fruits of Pear (Pyrus communis). Plant Cell Physiol. 2016, 57, 505–518. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Zhang, Z.Q.; Li, B.Q.; Qin, G.Z.; Tian, S.P. Molecular basis for optimizing sugar metabolism and transport during fruit development. aBIOTECH 2021, 2, 330–340. [Google Scholar] [CrossRef]
- Oikawa, A.; Otsuka, T.; Nakabayashi, R.; Jikumaru, Y.; Isuzugawa, K.; Murayama, H.; Saito, K.; Shiratake, K. Metabolic Profiling of Developing Pear Fruits Reveals Dynamic Variation in Primary and Secondary Metabolites, Including Plant Hormones. PLoS ONE 2015, 10, e0131408. [Google Scholar] [CrossRef] [PubMed]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Santana-Gálvez, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Chlorogenic Acid: Recent Advances on Its Dual Role as a Food Additive and a Nutraceutical against Metabolic Syndrome. Molecules 2017, 22, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Pei, J.; Zheng, Y.; Miao, Y.J.; Duan, B.Z.; Huang, L.F. Gallic Acid: A Potential Anti-Cancer Agent. Chin. J. Integr. Med. 2021, 28, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 29, e47. [Google Scholar] [CrossRef] [Green Version]
- Slika, H.; Mansour, H.; Wehbe, N.; Nasser, S.A.; Iratni, R.; Nasrallah, G.; Shaito, A.; Ghaddar, T.; Kobeissy, F.; Eid, A.H. Therapeutic potential of flavonoids in cancer: ROS-mediated mechanisms. Biomed. Pharmacother. 2022, 146, 112442. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- González-Mas, M.C.; Blázquez, M.A.; López-Gresa, M.P.; Mena, P.; García-Viguera, C. Flavonoids: From Biosynthesis and Metabolism to Health Benefits. Front. Plant Sci. 2021, 12, 727043. [Google Scholar] [CrossRef]
- Lou, H.; Hu, L.; Lu, H.; Wei, T.; Chen, Q. Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules 2021, 26, 4522. [Google Scholar] [CrossRef] [PubMed]
- Naik, J.; Misra, P.; Trivedi, P.K.; Pandey, A. Molecular components associated with the regulation of flavonoid biosynthesis. Plant Sci. 2022, 317, 111196. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Yang, C.K.; Tu, H.; Zhou, J.J.; Liu, X.Q.; Cheng, Y.J.; Luo, J.; Deng, X.X.; Zhang, H.Y.; Xu, J. Characterization and metabolic diversity of flavonoids in citrus species. Sci. Rep. 2017, 7, 10549. [Google Scholar] [CrossRef]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C.J. A Proteolytic Regulator Controlling Chalcone Synthase Stability and Flavonoid Biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, A.; Qi, S.; Su, K.; Guo, Y. Identification of candidate genes related to anthocyanin biosynthesis in red sarcocarp hawthorn (Crataegus pinnatifida). Sci. Hortic. 2022, 298, 110987. [Google Scholar] [CrossRef]






| Group Name | All | Down | Up |
|---|---|---|---|
| S1_vs_S2 | 106 | 39 | 67 |
| S2_vs_S3 | 197 | 78 | 119 |
| S3_vs_S4 | 177 | 128 | 49 |
| S4_vs_S5 | 262 | 156 | 106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Hao, R.; Guo, R.; Nong, H.; Qin, Y.; Dong, N. Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development. Metabolites 2023, 13, 423. https://doi.org/10.3390/metabo13030423
Wang Y, Hao R, Guo R, Nong H, Qin Y, Dong N. Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development. Metabolites. 2023; 13(3):423. https://doi.org/10.3390/metabo13030423
Chicago/Turabian StyleWang, Yan, Ruixin Hao, Rongkun Guo, Huilan Nong, Yu Qin, and Ningguang Dong. 2023. "Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development" Metabolites 13, no. 3: 423. https://doi.org/10.3390/metabo13030423
APA StyleWang, Y., Hao, R., Guo, R., Nong, H., Qin, Y., & Dong, N. (2023). Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development. Metabolites, 13(3), 423. https://doi.org/10.3390/metabo13030423





