Quality Control in Targeted GC-MS for Amino Acid-OMICS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
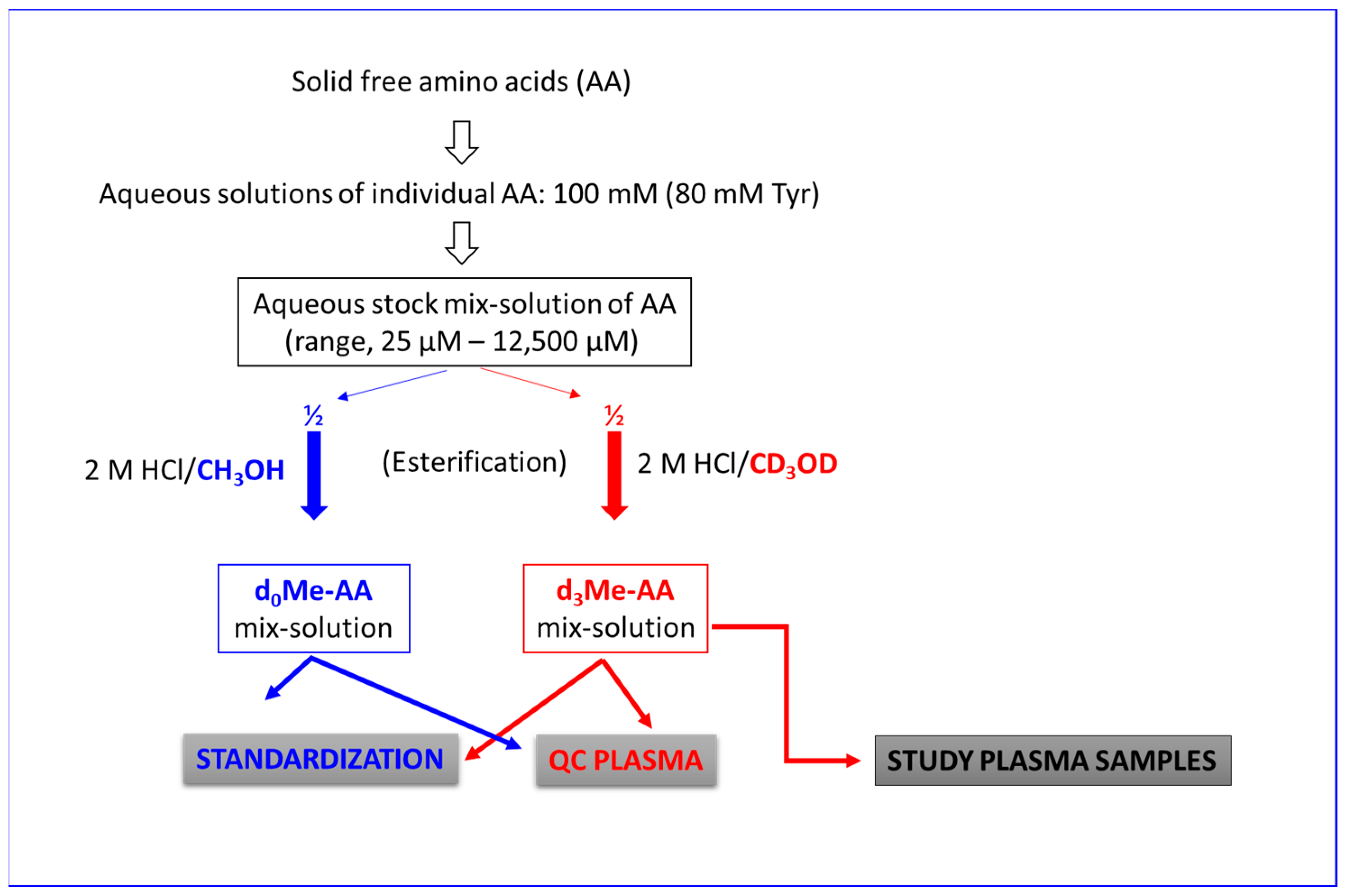
2.2. Standardization of Amino Acid Concentrations in Deionized Water
2.3. Preparation and GC-MS Analysis of Human Plasma Quality Control Samples
2.4. Procedure for the GC-MS Analysis of Amino Acids in Human Plasma Samples
2.5. Order of Analysis of the Study Human Plasma Samples and QC Samples
2.6. GC-MS Analyses
2.7. Data Handling–Statistics
3. Results
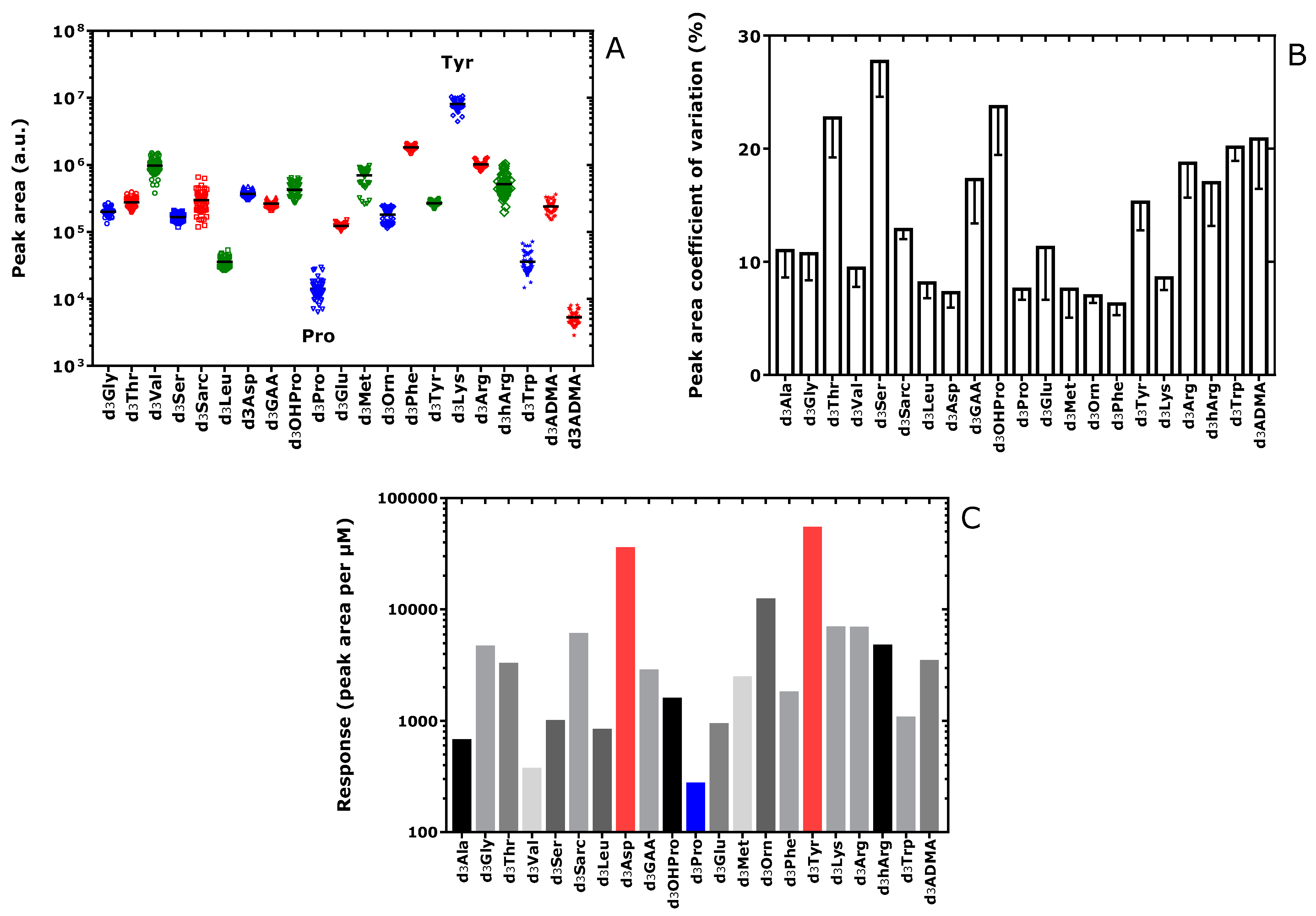
3.1. Standardization of Amino Acid Concentrations in Deionized Water Solutions
3.2. Amino Acids in Quality Control and Study Plasma Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hušek, P.; Macek, K. Gas chromatography of amino acids. J. Chromatogr. A 1975, 113, 139–230. [Google Scholar]
- Hušek, P.; Švagera, Z.; Hanzlíková, D.; Řimnáčová, L.; Zahradníčková, H.; Opekarová, I.; Šimek, P. Profiling of urinary amino-carboxylic metabolites by in-situ heptafluorobutyl chloroformate mediated sample preparation and gas-mass spectrometry. J. Chromatogr. A 2016, 1443, 211–232. [Google Scholar] [CrossRef]
- Ferré, S.; González-Ruiz, V.; Guillarme, D.; Rudaz, S. Analytical strategies for the determination of amino acids: Past, present and future trends. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1132, 121819. [Google Scholar] [CrossRef]
- Xu, W.; Zhong, C.; Zou, C.; Wang, B.; Zhang, N. Analytical methods for amino acid determination in organisms. Amino Acids 2020, 52, 1071–1088. [Google Scholar] [CrossRef]
- Tsikas, D. De novo synthesis of trideuteromethyl esters of amino acids for use in GC-MS and GC-tandem MS exemplified for ADMA in human plasma and urine: Standardization, validation, comparison and proof of evidence for their aptitude as internal standards. J. Chromatogr. B 2009, 877, 2308–2320. [Google Scholar] [CrossRef]
- Hanff, E.; Ruben, S.; Kreuzer, M.; Bollenbach, A.; Kayacelebi, A.A.; Das, A.M.; von Versen-Höynck, F.; von Kaisenberg, C.; Haffner, D.; Ückert, S.; et al. Development and validation of GC-MS methods for the comprehensive analysis of amino acids in plasma and urine and applications to the HELLP syndrome and pediatric kidney transplantation: Evidence of altered methylation, transamidination, and arginase activity. Amino Acids 2019, 51, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Baskal, S.; Bollenbach, A.; Tsikas, D. Two-Step Derivatization of Amino Acids for Stable-Isotope Dilution GC-MS Analysis: Long-Term Stability of Methyl Ester-Pentafluoropropionic Derivatives in Toluene Extracts. Molecules 2021, 26, 1726. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ni, Y.; Su, M.; Li, H.; Dong, F.; Chen, W.; Wei, R.; Zhang, L.; Guiraud, S.P.; Martin, F.P.; et al. High Throughput and Quantitative Measurement of Microbial Metabolome by Gas Chromatography/Mass Spectrometry Using Automated Alkyl Chloroformate Derivatization. Anal. Chem. 2017, 89, 5565–5577. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.G.; Theodoridis, G.A.; Earll, M.; Wilson, I.D. A QC approach to the determination of day-to-day reproducibility and robustness of LC-MS methods for global metabolite profiling in metabonomics/metabolomics. Bioanalysis 2012, 4, 2239–2247. [Google Scholar] [CrossRef]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. LC-MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. J. Chromatogr. B 2014, 966, 1–6. [Google Scholar] [CrossRef]
- Gika, H.G.; Zisi, C.; Theodoridis, G.; Wilson, I.D. Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1008, 15–25. [Google Scholar] [CrossRef]
- Begou, O.; Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Quality Control and Validation Issues in LC-MS Metabolomics. Methods Mol. Biol. 2018, 1738, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.L. Accreditation and Quality Assurance for Clinical Liquid Chromatography-Mass Spectrometry Laboratories. Clin. Lab. Med. 2018, 38, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Lippa, K.A.; Aristizabal-Henao, J.J.; Beger, R.D.; Bowden, J.A.; Broeckling, C.; Beecher, C.; Clay Davis, W.; Dunn, W.B.; Flores, R.; Goodacre, R. Reference materials for MS-based untargeted metabolomics and lipidomics: A review by the metabolomics quality assurance and quality control consortium (mQACC). Metabolomics 2022, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.A.; Gika, H.; Beger, R.D.; Bearden, D.; Dunn, W.B.; Goodacre, R.; Theodoridis, G.; Witting, M.; Yu, L.R.; Wilson, I.D. Quality Assurance and Quality Control Consortium (mQACC). Quality assurance and quality control reporting in untargeted metabolic phenotyping: mQACC recommendations for analytical quality management. Metabolomics 2022, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Sens, A.; Rischke, S.; Hahnefeld, L.; Dorochow, E.; Schäfer, S.M.; Thomas, D.; Köhm, M.; Geisslinger, G.; Behrens, F.; Gurke, R. Pre-analytical sample handling standardization for reliable measurement of metabolites and lipids in LC-MS-based clinical research. J. Mass Spectrom. Adv. Clin. Lab. 2023, 28, 35–46. [Google Scholar] [CrossRef]
- Tsikas, D. Methods of quantitative analysis of the nitric oxide metabolites nitrite and nitrate in human biological fluids. Free Radic. Res. 2005, 39, 797–815. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Schubert, B.; Gutzki, F.M.; Sandmann, J.; Frölich, J.C. Quantitative determination of circulating and urinary asymmetric dimethylarginine (ADMA) in humans by gas chromatography-tandem mass spectrometry as methyl ester tri(N-pentafluoropropionyl) derivative. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 798, 87–99. [Google Scholar] [CrossRef]
- Begou, O.; Drabert, K.; Theodoridis, G.; Tsikas, D. GC-NICI-MS analysis of acetazolamide and other sulfonamide (R-SO2-NH2) drugs as pentafluorobenzyl derivatives [R-SO2-N(PFB)2] and quantification of pharmacological acetazolamide in human urine. J. Pharm. Anal. 2020, 10, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.L.; Markus, C.; Lim, C.Y.; Tan, R.Z.; Sethi, S.K.; Loh, T.P.; IFCC Working Group on Method Evaluation Protocols. Calibration Practices in Clinical Mass Spectrometry: Review and Recommendations. Ann. Lab. Med. 2023, 43, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Cynober, L.; Blonde, F.; Nguyen Dinh, F.; Gerbet, D.; Giboudeau, J. Measurement of plasma and urinary amino acids with gas chromatography in healthy subjects. Variations as a function of age and sex. Ann. Biol. Clin. 1983, 41, 33–38. [Google Scholar]
- Bancel, E.; Strubel, D.; Bellet, H.; Polge, A.; Peray, P.; Magnan de Bornier, B. Effect of the age and the sex on plasma concentration of amino acids. Ann. Biol. Clin. 1994, 52, 667–670. [Google Scholar]
- Schwedhelm, E.; Xanthakis, V.; Maas, R.; Sullivan, L.M.; Schulze, F.; Riederer, U.; Benndorf, R.A.; Böger, R.H.; Vasan, R.S. Asymmetric dimethylarginine reference intervals determined with liquid chromatography-tandem mass spectrometry: Results from the Framingham offspring cohort. Clin. Chem. 2009, 55, 1539–1545. [Google Scholar] [CrossRef]
- Kouchiwa, T.; Wada, K.; Uchiyama, M.; Kasezawa, N.; Niisato, M.; Murakami, H.; Fukuyama, K.; Yokogoshi, H. Age-related changes in serum amino acids concentrations in healthy individuals. Clin. Chem. Lab. Med. 2012, 50, 861–870. [Google Scholar] [CrossRef]
- Yamamoto, H.; Kondo, K.; Tanaka, T.; Muramatsu, T.; Yoshida, H.; Imaizumi, A.; Nagao, K.; Noguchi, Y.; Miyano, H. Reference intervals for plasma-free amino acid in a Japanese population. Ann. Clin. Biochem. 2016, 53 Pt 3, 357–364. [Google Scholar] [CrossRef]
- Tsikas, D. A proposal for comparing methods of quantitative analysis of endogenous compounds in biological systems by using the relative lower limit of quantification (rLLOQ). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 2244–2251. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D. Bioanalytical method validation of endogenous substances according to guidelines by the FDA and other organizations: Basic need to specify concentration ranges. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1093–1094, 80–81. [Google Scholar] [CrossRef]
- Märtens, A.; Holle, J.; Mollenhauer, B.; Wegner, A.; Kirwan, J.; Hiller, K. Instrumental Drift in Untargeted Metabolomics: Optimizing Data Quality with Intrastudy QC Samples. Metabolites 2023, 13, 665. [Google Scholar] [CrossRef]
- Pickett, W.C.; Murphy, R.C. Enzymatic preparation of carboxyl oxygen-18 labeled prostaglandin F2 alpha and utility for quantitative mass spectrometry. Anal. Biochem. 1981, 111, 115–121. [Google Scholar] [CrossRef] [PubMed]
- de Leenheer, A.P.; Thienpont, L.M. Applications of isotope dilution-mass spectrometry in clinical chemistry, pharmacokinetics, and toxicology. Mass Spectrom. Rev. 1992, 11, 249–307. [Google Scholar] [CrossRef]
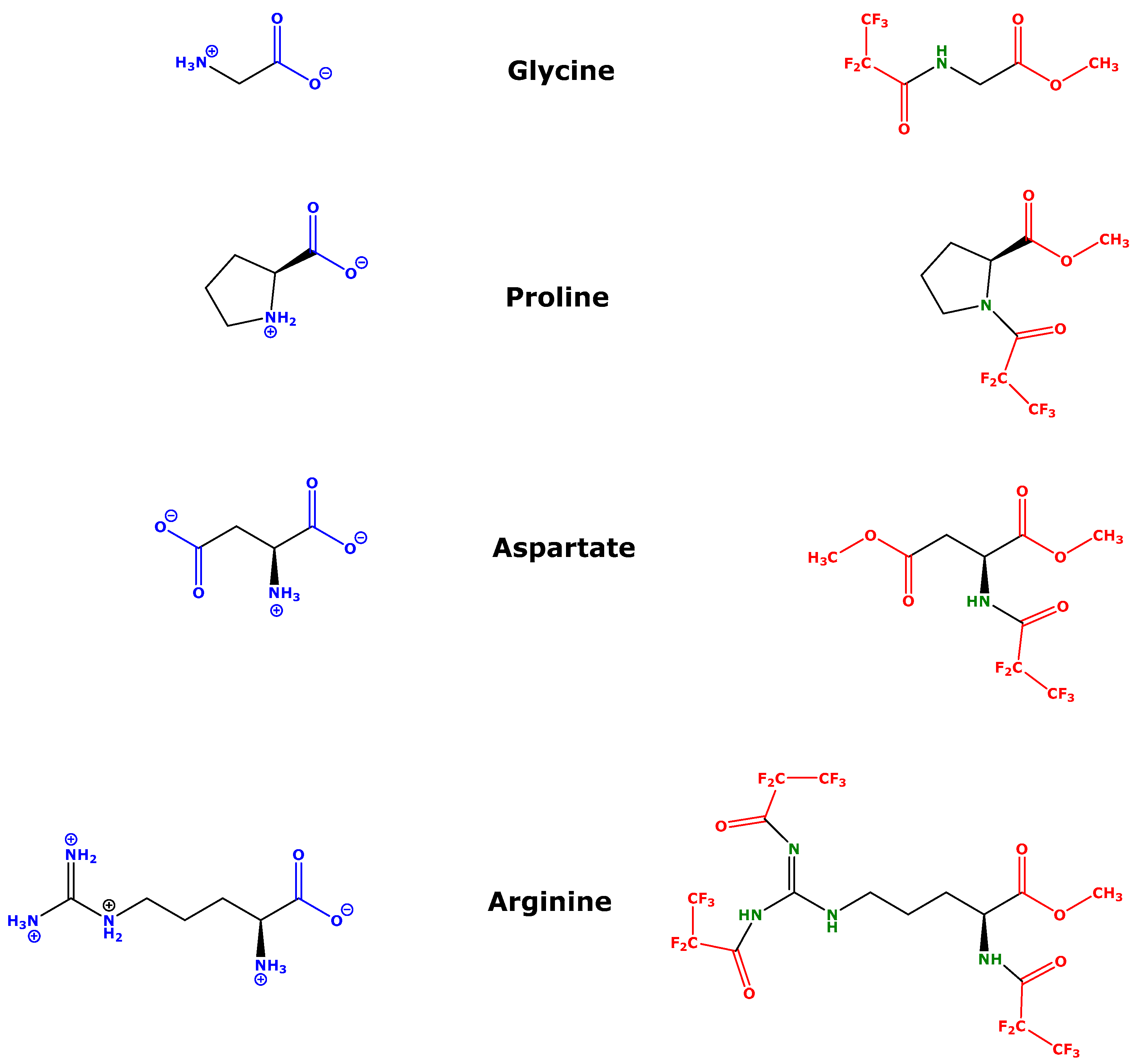


| AA | STD1; QC1 (µM) | STD2; QC2 (µM) | STD3; QC3 (µM) | STD4; QC4 (µM) | IS (µM) | Stock Solution (µM) | m/z of AA | m/z of IS | Dwell Time (ms) | Time Window (min) |
|---|---|---|---|---|---|---|---|---|---|---|
| Ala | 0 | 75 | 150 | 300 | 200 | 5000 | 229 | 232 | 100 | 3.20 |
| Thr | 0 | 15 | 30 | 60 | 40 | 1000 | 259 | 262 | 50 | 3.65 |
| Gly | 0 | 75 | 150 | 300 | 200 | 5000 | 215 | 218 | 50 | 3.65 |
| Val | 0 | 112 | 224 | 448 | 300 | 7500 | 257 | 260 | 50 | 3.65 |
| Ser | 0 | 75 | 150 | 300 | 200 | 5000 | 207 | 210 | 50 | 3.65 |
| Sarc | 0 | 1.5 | 3.0 | 6.0 | 4 | 100 | 229 | 232 | 50 | 4.32 |
| Leu/Ile | 0 | 112 | 224 | 448 | 300 | 7500 | 271 | 274 | 100 | 5.10 |
| GAA | 0 | 1.9 | 3.8 | 7.6 | 50 | 125 | 383 | 386 | 50 | 5.85 |
| Asp/Asn | 0 | 37 | 74 | 148 | 100 | 2500 | 287 | 293 | 50 | 5.85 |
| OH-Pro | 0 | 22.5 | 45 | 90 | 6 | 1500 | 397 | 400 | 50 | 5.85 |
| Pro | 0 | 112 | 224 | 448 | 30 | 7500 | 255 | 258 | 100 | 6.52 |
| Glu/Gln | 0 | 187 | 375 | 750 | 500 | 1250 | 301 | 307 | 100 | 7.10 |
| Met | 0 | 19 | 38 | 76 | 50 | 12,500 | 289 | 292 | 100 | 7.10 |
| Orn/Cit | 0 | 37 | 74 | 148 | 100 | 2500 | 418 | 421 | 50 | 7.80 |
| Phe | 0 | 37 | 74 | 148 | 100 | 2500 | 305 | 308 | 50 | 7.80 |
| Tyr | 0 | 37 | 74 | 148 | 100 | 2500 | 233 | 236 | 100 | 8.35 |
| Lys | 0 | 37 | 74 | 148 | 100 | 2500 | 432 | 435 | 50 | 8.80 |
| Arg | 0 | 19 | 38 | 76 | 50 | 1250 | 586 | 589 | 50 | 8.80 |
| hArg | 0 | 1.9 | 3.8 | 7.6 | 5 | 125 | 600 | 603 | 100 | 9.75 |
| Trp | 0 | 56 | 112 | 224 | 150 | 3750 | 233 | 236 | 50 | 10.40 |
| ADMA | 0 | 0.37 | 0.74 | 1.48 | 1.0 | 25 | 634 | 637 | 100 | 10.40 |
| AA | STD1 (µM) | STD2 (µM) | STD3 (µM) | STD4 (µM) | [IS]nom (µM) | Regression Equation (y= a + b × x) (y = PAR, x = [STD]) | [IS]std (µM) | [IS]nom /[IS]std |
|---|---|---|---|---|---|---|---|---|
| Ala | 0 | 75 | 150 | 300 | 200 | y = −0.0007 + 0.0043 × x, r2 = 0.9995 | 232 | 0.86 |
| (CV, %) | 7.7 | 2.9 | 1.7 | 2.5 | ||||
| Thr | 0 | 15 | 30 | 60 | 40 | y = −0.0067 + 0.03187 × x, r2 = 0.9999 | 31 | 1.29 |
| (CV, %) | 30 | 5.7 | 2.8 | 2.2 | ||||
| Gly | 0 | 75 | 150 | 300 | 200 | y = −0.00024 + 0.0041 × x, r2 = 0.9999 | 243 | 0.82 |
| (CV, %) | 30 | 6.1 | 3.3 | 2.6 | ||||
| Val | 0 | 112 | 224 | 448 | 300 | y = −0.02417 + 0.0530 × x, r2 = 0.9993 | 188 | 1.60 |
| (CV, %) | 53 | 5.0 | 2.4 | 4.6 | ||||
| Ser | 0 | 75 | 150 | 300 | 200 | y = 0.01259 + 0.00466 × x, r2 = 0.9999 | 215 | 0.93 |
| (CV, %) | 28 | 4.5 | 3.6 | 2.7 | ||||
| Sarc | 0 | 1.5 | 3.0 | 6.0 | 4 | y = 0.00472 + 0.2022 × x, r2 = 1.0000 | 5 | 0.80 |
| (CV, %) | 39 | 2.0 | 2.4 | 8.0 | ||||
| Leu/Ile | 0 | 112 | 224 | 448 | 300 | y = 0.02317 + 0.00259 × x, r2 = 0.9998 | 386 | 0.78 |
| (CV, %) | 19 | 4.7 | 1.7 | 1.9 | ||||
| GAA | 0 | 1.9 | 3.8 | 7.6 | 50 | y = 0.01701 + 0.01816 × x, r2 = 0.9974 | 55 | 0.91 |
| (CV, %) | 30 | 65 | 11.3 | 38 | ||||
| Asp/Asn | 0 | 37 | 74 | 148 | 100 | y = 0.02115 + 0.00982 × x, r2 = 0.9989 | 102 | 0.98 |
| (CV, %) | 0 | 4.7 | 2.6 | 2.9 | ||||
| OH-Pro | 0 | 22.5 | 45 | 90 | 6 | y = 0.02633 + 0.1322 × x, r2 = 0.9999 | 8 | 0.75 |
| (CV, %) | 0 | 4.8 | 4.9 | 9.2 | ||||
| Pro | 0 | 112 | 224 | 448 | 30 | y = 0.0845 + 0.02901 × x, r2 = 0.9955 | 35 | 0.86 |
| (CV, %) | 17.1 | 5.7 | 3.3 | 4.4 | ||||
| Glu/Gln | 0 | 187 | 375 | 750 | 500 | y = 0.04569 + 0.00192 × x, r2 = 0.9947 | 520 | 0.96 |
| (CV, %) | 0 | 6.3 | 2.2 | 2.8 | ||||
| Met | 0 | 19 | 38 | 76 | 50 | y = 0.0792 + 0.0158 × x, r2 = 0.9940 | 63 | 0.79 |
| (CV, %) | 7.8 | 2.6 | 1.9 | 4.7 | ||||
| Orn/Cit | 0 | 37 | 74 | 148 | 100 | y = −0.0131 + 0.00957 × x, r2 = 0.9995 | 105 | 0.95 |
| (CV, %) | 53 | 5.6 | 2.4 | 2.3 | ||||
| Phe | 0 | 37 | 74 | 148 | 100 | y = −0.0044 + 0.00884 × x, r2 = 0.9997 | 113 | 0.88 |
| (CV, %) | 11.1 | 5.0 | 1.6 | 2.4 | ||||
| Tyr | 0 | 37 | 74 | 148 | 100 | y = −0.0076 + 0.00789 × x, r2 = 0.9996 | 127 | 0.79 |
| (CV, %) | 28.4 | 5.5 | 2.7 | 1.8 | ||||
| Lys | 0 | 37 | 74 | 148 | 100 | y = 0.0472 + 0.00865 × x, r2 = 0.9931 | 116 | 0.86 |
| (CV, %) | 44 | 5.3 | 2.3 | 3.0 | ||||
| Arg | 0 | 19 | 38 | 76 | 50 | y = 0.0104 + 0.01776 × x, r2 = 0.9979 | 56 | 0.89 |
| (CV, %) | 28 | 12.3 | 13.2 | 9.1 | ||||
| hArg | 0 | 1.9 | 3.8 | 7.6 | 5 | y = 0.0156 + 0.2167 × x, r2 = 0.9982 | 4.6 | 1.09 |
| (CV, %) | 28.4 | 10.5 | 17.6 | 6.6 | ||||
| Trp | 0 | 56 | 112 | 224 | 150 | y = −0.0036 + 0.0056 × x, r2 = 0.9998 | 179 | 0.84 |
| (CV, %) | 38.5 | 6.1 | 4.2 | 4.1 | ||||
| ADMA | 0 | 0.37 | 0.74 | 1.48 | 1.0 | y = 0.0558 + 0.8995 × x, r2 = 0.9998 | 1.1 | 0.91 |
| (CV, %) | 74.2 | 5.5 | 9.9 | 7.3 |
| AA | tR | IE | δ(H/D) | Regression Equation | |||
|---|---|---|---|---|---|---|---|
| min (CV, %) | (CV, %) | (s) | y-axis Intercept (a) | Slope (b) | r2 | [IS]std (µM) | |
| Ala | 3.382 (0.51) | 1.005 (0.15) | 0.96 | 430 | 0.91 | 0.9980 | 232 |
| Thr | 3.799 (0.23) | 1.004 (0.15) | 0.90 | 196 | 0.89 | 0.9968 | 31 |
| Gly | 3.794 (0.33) | 1.005 (0.07) | 1.22 | 247 | 0.91 | 0.9982 | 243 |
| Val | 4.017 (0.22) | 1.005 (0.13) | 1.17 | 367 | 0.92 | 0.9992 | 188 |
| Ser | 4.139 (0.16) | 1.006 (0.15) | 1.37 | 149 | 0.95 | 0.9990 | 215 |
| Sarc | 4.491 (0.17) | 1.005 (0.08) | 1.28 | 1.49 | 0.94 | 0.9984 | 5 |
| Leu/Ile | 4.644 (0.14) | 1.004 (0.07) | 1.17 | 196 | 0.96 | 0.9996 | 386 |
| GAA | 6.258 (0.09) | 1.004 (0.08) | 1.61 | 6.93 | 0.87 | 0.9986 | 55 |
| Asp/Asn | 6.200 (0.05) | 1.006 (0.04) | 2.40 | 60 | 0.99 | 0.9995 | 102 |
| OH-Pro | 6.400 (0.03) | 1.003 (0.03) | 1.18 | 10.1 | 1.08 | 0.9976 | 8 |
| Pro | 6.602 (0.06) | 1.003 (0.05) | 1.28 | 214 | 1.01 | 0.9998 | 35 |
| Glu/Gln | 7.383 (0.06) | 1.005 (0.05) | 2.42 | 985 | 1.25 | 0.9949 | 520 |
| Met | 7.393 (0.06) | 1.006 (0.06) | 2.56 | 86 | 0.97 | 0.9999 | 63 |
| Orn/Cit | 8.099 (0.07) | 1.002 (0.00) | 1.20 | 179 | 0.96 | 0.9988 | 105 |
| Phe | 8.154 (0.06) | 1.002 (0.00) | 1.20 | 78 | 0.99 | 0.9996 | 113 |
| Tyr | 8.578 (0.06) | 1.002 (0.04) | 1.14 | 96 | 0.92 | 0.9995 | 127 |
| Lys | 9.002 (0.07) | 1.002 (0.05) | 1.22 | 181 | 1.01 | 0.9995 | 116 |
| Arg | 9.230 (0.05) | 1.002 (0.04) | 1.20 | 62 | 0.86 | 0.9992 | 56 |
| hArg | 10.03 (0.20) | 1.003 (0.06) | 1.61 | 1.58 | 0.79 | 0.9997 | 4.6 |
| Trp | 10.92 (0.04) | 1.002 (0.04) | 1.58 | 31 | 1.23 | 0.9993 | 179 |
| ADMA | 11.08 (0.06) | 1.002 (0.03) | 1.12 | 0.609 | 0.80 | 0.9924 | 1.1 |
| AA | QC1 | QC2 | QC3 | QC4 | Mean QC |
|---|---|---|---|---|---|
| Ala | 4.12 (3.95) | 5.94 (3.58) | 3.61 (3.44) | 3.64 (3.94) | 4.33 (1.10) |
| Thr | 4.51 (4.72) | 2.59 (2.04) | 1.74 (1.69) | 3.33 (3.94) | 3.04 (1.17) |
| Gly | 1.89 (1.16) | 3.48 (2.21) | 2.34 (1.57) | 1.60 (1.69) | 2.33 (0.83) |
| Val | 2.90 (1.87) | 2.94 (1.54) | 2.79 (2.46) | 2.39 (2.12) | 2.75 (0.25) |
| Ser | 2.04 (1.54) | 2.10 (1.57) | 3.04 (1.84) | 1.82 (1.82) | 2.25 (0.54) |
| Sarc | 1.59 (1.17) | 4.29 (3.28) | 4.00 (2.69) | 1.98 (1.89) | 2.96 (1.37) |
| Leu/Ile | 2.56 (1.79) | 3.43 (1.71) | 2.34 (2.19) | 1.25 (0.59) | 2.40 (0.90) |
| GAA | 1.53 (1.19) | 3.45 (1.74) | 3.44 (3.66) | 2.08 (1.75) | 2.63 (0.97) |
| Asp/Asn | 1.91 (1.00) | 2.44 (1.86) | 1.72 (2.35) | 3.18 (3.07) | 2.31 (0.65) |
| OH-Pro | 5.39 (9.04) | 3.96 (3.74) | 3.70 (3.66) | 2.78 (1.05) | 3.96 (1.08) |
| Pro | 3.16 (2.15) | 4.64 (2.44) | 3.87 (2.67) | 2.97 (2.36) | 3.61 (0.67) |
| Glu/Gln | 4.28 (2.69) | 3.41 (3.25) | 4.87 (4.71) | 2.99 (3.33) | 3.88 (0.85) |
| Met | 3.69 (6.00) | 1.77 (2.02) | 2.66 (2.27) | 1.90 (1.50) | 2.50 (0.88) |
| Orn/Cit | 2.07 (2.03) | 4.00 (2.90) | 2.98 (3.26) | 1.22 (1.34) | 2.57 (1.19) |
| Phe | 1.92 (1.20) | 2.85 (1.67) | 3.36 (3.07) | 2.17 (1.54) | 2.58 (0.66) |
| Tyr | 3.06 (2.72) | 3.97 (3.18) | 3.43 (2.38) | 1.39 (1.15) | 2.96 (1.11) |
| Lys | 2.88 (1.19) | 2.12 (1.70) | 1.93 (1.22) | 1.32 (0.81) | 2.06 (0.64) |
| Arg | 2.17 (1.62) | 3.57 (2.32) | 3.35 (3.07) | 1.64 (0.73) | 2.68 (0.93) |
| hArg | 1.53 (1.55) | 1.91 (1.89) | 3.24 (3.97) | 1.07 (0.94) | 1.94 (0.93) |
| Trp | 2.44 (1.85) | 3.68 (1.99) | 2.99 (3.20) | 2.65 (1.24) | 2.94 (0.54) |
| ADMA | 1.79 (0.73) | 2.30 (1.10) | 3.23 (3.84) | 1.22 (1.04) | 2.13 (0.86) |
| Mean QC | 2.73 (1.10) | 3.27 (1.02) | 3.08 (0.78) | 2.12 (0.79) |
| AA | tR(H) (min) | tR(D) (min) | IE | δ(H/D) (s) | tR(H) (min) | tR(D) (min) | IE | δ(H/D) (s) |
|---|---|---|---|---|---|---|---|---|
|
| |||||||
| Ala | 3.369 (0.5) | 3.355 (0.6) | 1.005 (0.2) | 0.92 (44) | 3.372 (0.5) | 3.36 (0.7) | 1.004 (0.3) | 0.90 (56) |
| (n) | 353 | 352 | 340 | 340 | 64 | 62 | 59 | 59 |
| Thr | 3.790 (0.3) | 3.773 (0.5) | 1.004 (0.3) | 0.98 (77) | 3.790 (0.3) | 3.776 (0.3) | 1.004 (0.2) | 0.84 (58) |
| (n) | 353 | 353 | 349 | 349 | 64 | 62 | 62 | 62 |
| Gly | 3.787 (0.3) | 3.767 (0.5) | 1.006 (0.2) | 1.34 (27) | 3.788 (0.3) | 3.768 (0.6) | 1.006 (0.2) | 1.35 (30) |
| (n) | 353 | 353 | 341 | 341 | 64 | 62 | 60 | 60 |
| Val | 4.007 (0.3) | 3.984 (0.5) | 1.005 (0.2) | 1.23 (33) | 4.007 (0.3) | 3.983 (0.7) | 1.006 (0.7) | 1.43 (90) |
| (n) | 353 | 352 | 344 | 344 | 64 | 62 | 62 | 62 |
| Ser | 4.128 (0.2) | 4.107 (0.4) | 1.005 (0.2) | 1.17 (35) | 4.128 (0.2) | 4.105 (0.4) | 1.006 (0.4) | 1.39 (72) |
| (n) | 353 | 353 | 344 | 323 | 64 | 61 | 61 | 54 |
| Sarc | 4.485 (0.2) | 4.463 (0.3) | 1.005 (0.1) | 1.26 (19) | 4.484 (0.2) | 4.461 (0.3) | 1.005 (0.3) | 1.40 (59) |
| (n) | 353 | 352 | 344 | 344 | 64 | 62 | 62 | 62 |
| Leu/Ile | 4.634 (0.2) | 4.614 (0.3) | 1.004 (0.1) | 1.17 (25) | 4.632 (0.2) | 4.611 (0.3) | 1.004 (0.2) | 1.24 (49) |
| (n) | 353 | 353 | 345 | 345 | 64 | 61 | 61 | 61 |
| GAA | 6.285 (0.1) | 6.255 (0.1) | 1.006 (0.1) | 2.34 (50) | 6.250 (0.1) | 6.229 (0.1) | 1.003 (0.1) | 1.28 (22) |
| (n) | 352 | 352 | 297 | 297 | 63 | 61 | 59 | 59 |
| Asp/Asn | 6.191 (0.1) | 6.150 (0.1) | 1.007 (0.1) | 2.49 (16) | 6.187 (0.1) | 6.146 (0.1) | 1.007 (0.1) | 2.45 (18) |
| (n) | 353 | 353 | 351 | 351 | 64 | 62 | 62 | 62 |
| OH-Pro | 6.393 (0.1) | 6.375 (0.1) | 1.003 (0.1) | 1.11 (26) | 6.400 (0.1) | 6.381 (0.1) | 1.003 (0.1) | 1.19 (23) |
| (n) | 353 | 353 | 350 | 350 | 64 | 62 | 61 | 61 |
| Pro | 6.594 (0.1) | 6.574 (0.1) | 1.003 (0.1) | 1.23 (21) | 6.593 (0.1) | 6.573 (0.1) | 1.003 (0.1) | 1.21 (21) |
| (n) | 353 | 352 | 347 | 347 | 64 | 62 | 61 | 61 |
| Glu/Gln | 7.372 (0.1) | 7.332 (0.2) | 1.006 (0.1) | 2.44 (15) | 7.373 (0.1) | 7.329 (0.2) | 1.006 (0.1) | 2.64 (24) |
| (n) | 353 | 352 | 352 | 352 | 64 | 62 | 62 | 62 |
| Met | 7.380 (0.1) | 7.340 (0.1) | 1.005 (0.1) | 2.42 (16) | 7.383 (0.1) | 7.342 (0.2) | 1.006 (0.2) | 2.46 (31) |
| (n) | 353 | 352 | 352 | 352 | 64 | 62 | 62 | 62 |
| Orn/Cit | 8.086 (0.1) | 8.067 (0.2) | 1.002 (0.1) | 1.12 (19) | 8.108 (0.2) | 8.088 (0.2) | 1.003 (0.1) | 1.22 (24) |
| (n) | 353 | 352 | 351 | 351 | 64 | 62 | 62 | 62 |
| Phe | 8.144 (0.1) | 8.123 (0.1) | 1.003 (0.1) | 1.27 (22) | 8.143 (0.1) | 8.124 (0.1) | 1.003 (0.1) | 1.26 (21) |
| (n) | 353 | 352 | 345 | 345 | 64 | 62 | 60 | 60 |
| Tyr | 8.566 (0.2) | 8.548 (0.1) | 1.002 (0.1) | 1.13 (22) | 8.581 (0.2) | 8.562 (0.1) | 1.002 (0.1) | 1.15 (31) |
| (n) | 353 | 353 | 350 | 350 | 63 | 62 | 61 | 61 |
| Lys | 8.988 (0.1) | 8.970 (0.1) | 1.002 (0.1) | 1.06 (30) | 8.995 (0.1) | 8.977 (0.1) | 1.002 (0.1) | 1.11 (32) |
| (n) | 353 | 353 | 351 | 351 | 64 | 62 | 62 | 62 |
| Arg | 9.284 (1.0) | 9.255 (0.8) | 1.003 (0.2) | 1.68 (54) | 9.219 (0.1) | 9.206 (0.9) | 1.002 (0.1) | 1.00 (34) |
| (n) | 353 | 350 | 324 | 324 | 63 | 62 | 59 | 60 |
| hArg | 10.13 (1.0) | 10.11 (1.0) | 1.003 (0.2) | 1.80 (55) | 10.03 (0.3) | 10.01 (0.4) | 1.003 (0.1) | 1.66 (35) |
| (n) | 351 | 347 | 314 | 310 | 64 | 61 | 59 | 59 |
| Trp | 10.91 (0.1) | 10.89 (0.1) | 1.002 (0.1) | 1.42 (24) | 10.91 (0.1) | 10.89 (0.1) | 1.002 (0.1) | 1.41 (28) |
| (n) | 353 | 352 | 348 | 348 | 64 | 62 | 62 | 62 |
| ADMA | 11.07 (0.1) | 11.06 (0.1) | 1.002 (0.1 | 1.10 (27) | 11.07 (0.1) | 11.05 (0.1) | 1.002 (0.1 | 1.08 (25) |
| (n) | 352 | 352 | 350 | 350 | 63 | 62 | 60 | 60 |
| AA | Median | 25% Percentile | 75% Percentile | CV (%) |
|---|---|---|---|---|
| Ala | 379 | 326 | 442 | 22 |
| Gly | 289 | 220 | 472 | 61 |
| Thr | 149 | 127 | 177 | 25 |
| Val | 437 | 351 | 529 | 31 |
| Ser | 121 | 107 | 143 | 37 |
| Sar | 10.7 | 8.8 | 12.2 | 29 |
| Leu/Ile | 217 | 177 | 307 | 40 |
| Asp/Asn | 54.3 | 46.7 | 61.6 | 22 |
| GAA | 6.24 | 5.4 | 7.2 | 47 |
| OH-Pro | 6.94 | 5.53 | 8.7 | 38 |
| Pro | 227 | 171 | 299 | 40 |
| Gln/Glu | 811 | 741 | 902 | 16 |
| Met | 70.2 | 65.5 | 75.2 | 11 |
| Orn/Cit | 87.9 | 73.1 | 106 | 26 |
| Phe | 71.6 | 61.0 | 84.9 | 24 |
| Tyr | 87.1 | 70.2 | 115 | 35 |
| Lys | 212 | 188 | 249 | 26 |
| Arg | 93.5 | 79.1 | 110.2 | 35 |
| hArg | 1.94 | 1.57 | 2.52 | 139 |
| Trp | 43.2 | 36.5 | 51.4 | 27 |
| ADMA | 0.57 | 0.48 | 0.64 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsikas, D.; Beckmann, B. Quality Control in Targeted GC-MS for Amino Acid-OMICS. Metabolites 2023, 13, 986. https://doi.org/10.3390/metabo13090986
Tsikas D, Beckmann B. Quality Control in Targeted GC-MS for Amino Acid-OMICS. Metabolites. 2023; 13(9):986. https://doi.org/10.3390/metabo13090986
Chicago/Turabian StyleTsikas, Dimitrios, and Bibiana Beckmann. 2023. "Quality Control in Targeted GC-MS for Amino Acid-OMICS" Metabolites 13, no. 9: 986. https://doi.org/10.3390/metabo13090986
APA StyleTsikas, D., & Beckmann, B. (2023). Quality Control in Targeted GC-MS for Amino Acid-OMICS. Metabolites, 13(9), 986. https://doi.org/10.3390/metabo13090986






