Metabolic Niches and Plasticity of Sand-Dune Plant Communities Along a Trans-European Gradient
Abstract
1. Introduction
2. Materials and Methods
2.1. Common-Garden Study (Inter and Intraspecific Metabolic Variation)
2.2. Metabolic Plasticity in the Field Using Reciprocal Transplants
2.3. Metabolite Quenching and Extraction
2.4. Metabolite Fingerprinting—Direct Injection Mass Spectrometry (DIMS)
2.5. Metabolomic Analysis
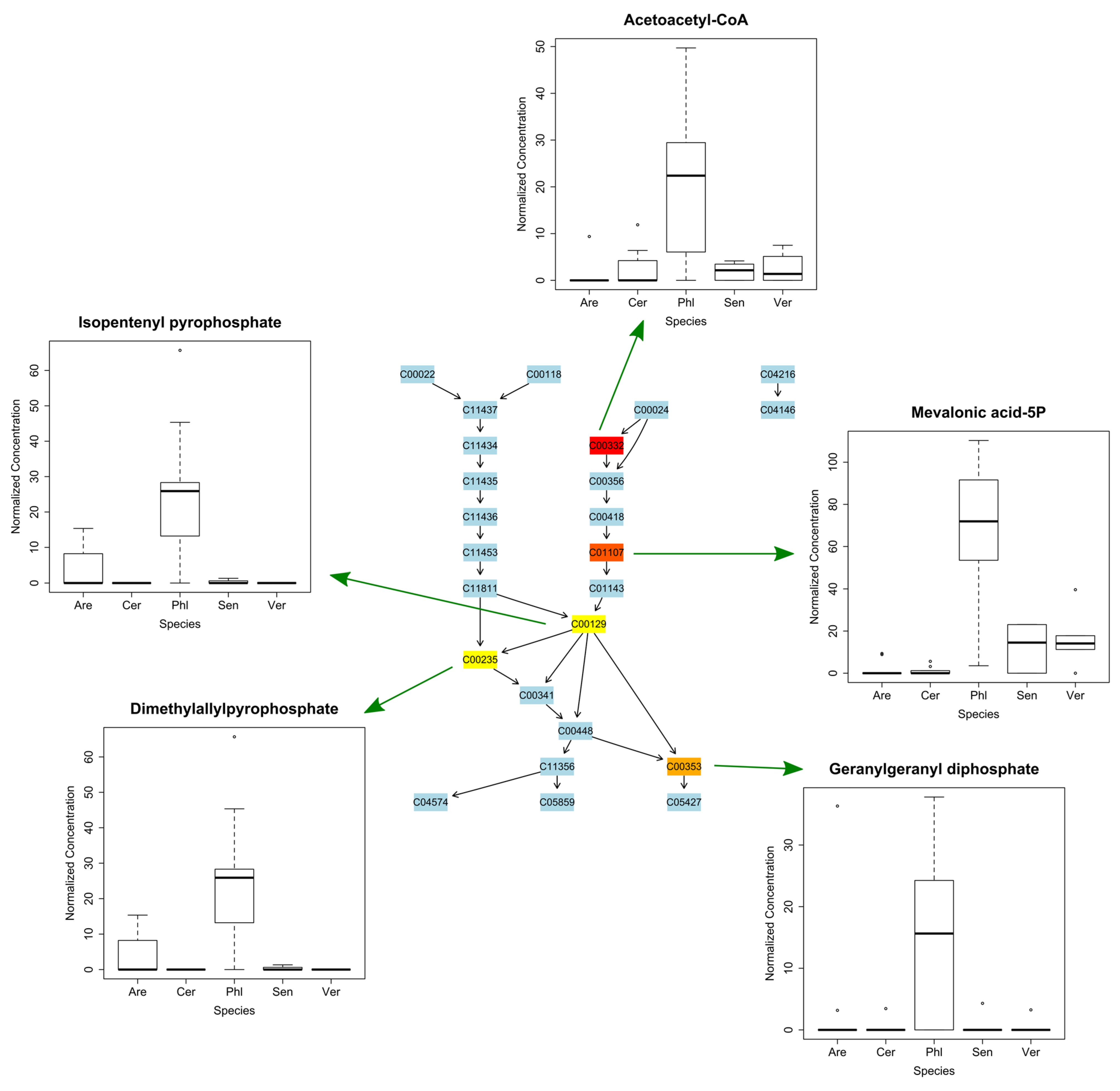
2.6. Putative Identification and Metabolite Mapping
3. Results
3.1. Inter-Specific Variation in Metabolic Phenotypes of Sand Dune Species in Common Garden
3.2. Intra-Specific Variation in Metabolic Profiles of Sand Dune Species in Common Garden
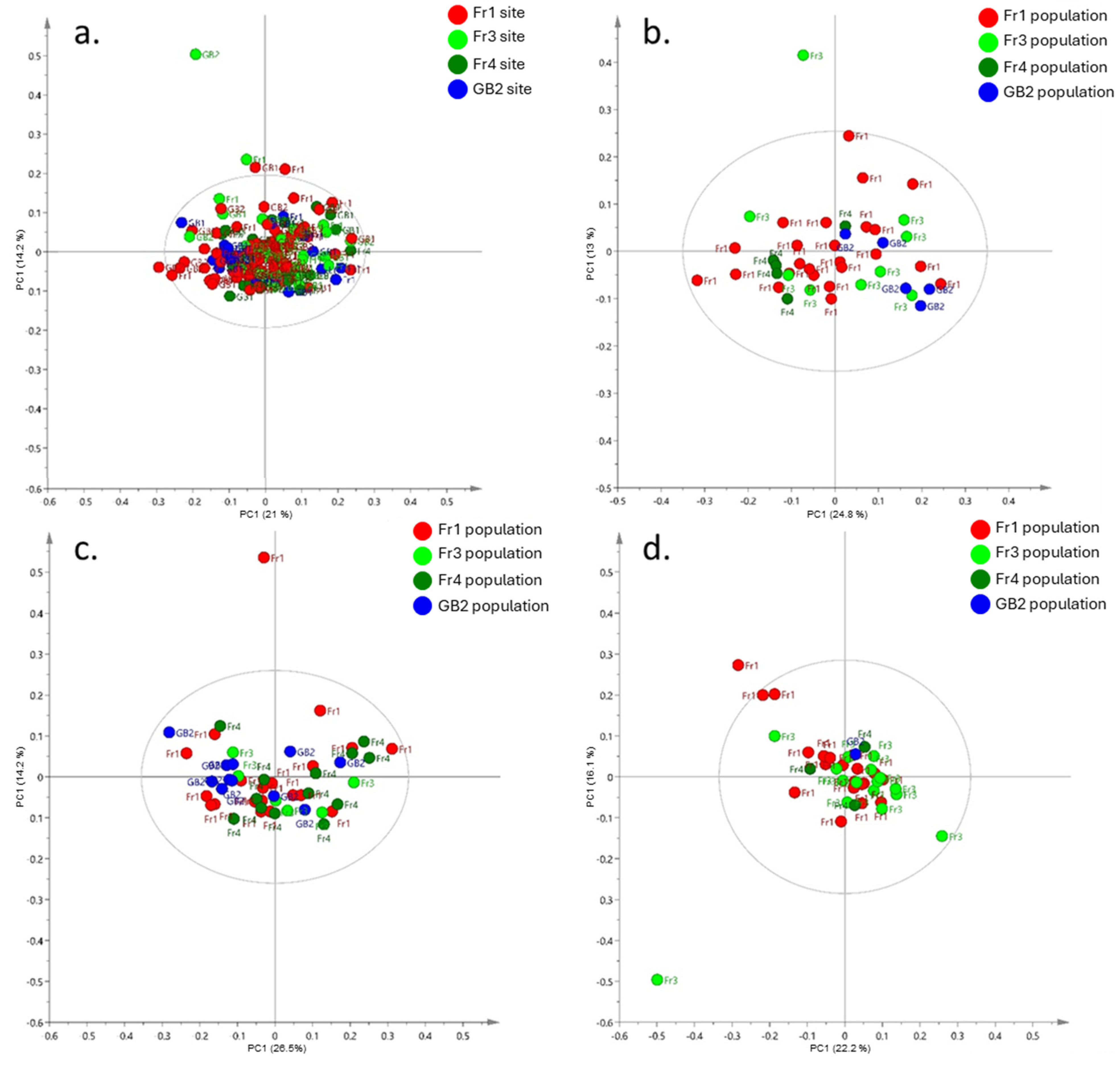
3.3. Metabolic Plasticity in Field Reciprocal Transplants
4. Discussion
4.1. Interspecific Variation in Metabolic Phenotypes
4.2. Intraspecific Variation in Metabolic Phenotypes
4.3. Reciprocal Transplants—Local Adaptation and Metabolic Plasticity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mitchell-Olds, T.; Willis, J.H.; Goldstein, D.B. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 2007, 8, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.B.; Linder, C.R.; Lynch, M.; Purugganan, M.; Somerville, S.; Thayer, S.S. Linking molecular insight and ecological research. Trends Ecol. Evol. 2002, 17, 409–414. [Google Scholar] [CrossRef]
- Macdonald, S.E.; Chinnappa, C.C.; Macdonald, E. Population differentiation for phenotypic plasticity in the Stellaria longipes. Am. J. Bot. 2011, 76, 1627–1637. [Google Scholar] [CrossRef]
- Schlichting, C.D.; Smith, H. Phenotypic plasticity: Linking molecular mechanisms with revolutionary outcomes. Evol. Ecol. 2002, 16, 189–211. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Paszkowski, J. Heat-induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genet. 2014, 10, e1004806. [Google Scholar] [CrossRef]
- Becklin, K.M.; Anderson, J.T.; Gerhart, L.M.; Wadgymar, S.M.; Wessinger, C.A.; Ward, J.K. Examining Plant Physiological Responses to Climate Change through an Evolutionary Lens. Plant Physiol. 2016, 172, 635–649. [Google Scholar] [CrossRef]
- Zuppinger-Dingley, D.; Flynn, D.F.B.; Brandl, H.; Schmid, B. Selection in monoculture vs. mixture alters plant metabolic fingerprints. J. Plant Ecol. 2015, 8, 549–557. [Google Scholar] [CrossRef]
- Morley, S.A.; Berman, J.; Barnes, D.K.A.; Carbonell, C.J.; Downey, R.V.; Peck, L.S. Extreme Phenotypic Plasticity in Metabolic Physiology of Antarctic Demosponges. Front. Ecol. Evol. 2016, 3, 157. [Google Scholar] [CrossRef]
- Sun, C.X.; Zhang, J.; Zhang, H.; Ni, Y.; Zhang, Q.; Chen, J.; Guan, Y. Plastic responses in the metabolome and functional traits of maize plants to temperature variations. Plant Biol. 2016, 18, 249–261. [Google Scholar] [CrossRef]
- Valladares, F.; Matesanz, S.; Guilhaumon, F.; Araújo, M.B.; Balaguer, L.; Benito-Garzón, M.; Cornwell, W.; Gianoli, E.; van Kleunen, M.; Naya, D.E.; et al. The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecol. Lett. 2014, 17, 1351–1364. [Google Scholar] [CrossRef]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2008, 5, 3–21. [Google Scholar] [CrossRef]
- Brunetti, C.; George, R.M.; Tattini, M.; Field, K.; Davey, M.P. Metabolomics in plant environmental physiology. J. Exp. Bot. 2013, 64, 4011–4020. [Google Scholar] [CrossRef]
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.; Lup, J. Metabolomics-centered mining of plant metabolic diversity and function: Past decade and future perspectives. Mol. Plant 2023, 16, 43–63. [Google Scholar] [CrossRef]
- Walker, T.W.N.; Alexander, J.M.; Allard, P.-M.; Baines, O.; Baldy, V.; Bardgett, R.D.; Capdevila, P.; Coley, P.D.; David, B.; Defossez, E.; et al. Functional Traits 2.0: The power of the metabolome for ecology. J. Ecol. 2022, 110, 4–20. [Google Scholar] [CrossRef]
- Davey, M.P.; Norman, L.; Sterk, P.; Huete-Ortega, M.; Bunbury, F.; Kin Wai Loh, B.; Stockton, S.; Peck, L.S.; Convey, P.; Newsham, K.K.; et al. Snow algae communities in Antarctica: Metabolic and taxonomic composition. New Phytol. 2019, 222, 1242–1255. [Google Scholar] [CrossRef]
- Brosché, M.; Vinocur, B.; Alatalo, E.R.; Lamminmäki, A.; Teichmann, T.; Ottow, E.A.; Djilianov, D.; Afif, D.; Bogeat-Triboulot, M.B.; Altman, A.; et al. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol. 2005, 6, R101. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.C.; Steinfath, M.; Lisec, J.; Becher, M.; Witucka-Wall, H.; Törjék, O.; Fiehn, O.; Eckardt, A.; Willmitzer, L.; Selbig, J.; et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 4759–4764. [Google Scholar] [CrossRef]
- Benfey, P.N.; Mitchell-Olds, T. From genotype to phenotype: Systems biology meets natural variation. Science 2008, 320, 495–497. [Google Scholar] [CrossRef]
- Bowne, J.B.; Erwin, T.; Juttner, J.; Schnurbusch, T.; Langridge, P.; Bacic, A.; Roessner, U. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant 2012, 5, 418–429. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.J.; Prasad, S.H. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Biotechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Gray, G.R.; Heath, D. A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolic fingerprinting. Physiol. Plant. 2005, 124, 236–248. [Google Scholar] [CrossRef]
- Davey, M.P.; Burrell, M.M.; Woodward, F.; Quick, W.P. Population-specific metabolic phenotypes of Arabidopsis lyrata ssp. petraea. New Phytol. 2008, 177, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.P.; Woodward, F.I.; Quick, W.P. Intraspecfic variation in cold-temperature metabolic phenotypes of Arabidopsis lyrata ssp. petraea. Metabolomics 2008, 5, 138–149. [Google Scholar] [CrossRef]
- Guy, C.; Kaplan, F.; Kopka, J.; Selbig, J.; Hincha, D.K. Metabolomics of temperature stress. Physiol. Plant. 2008, 132, 220–235. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Du, Y.; Chen, S.; Tang, H. Dynamic metabonomic responses of Tobacco (Nicotiana tabacum) plants to salt stress. J. Prot. Res. 2011, 10, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef]
- Bölling, C.; Fiehn, O. Metabolite profiling of Chlamydomonas reinhardti under nutrient deprivation. Plant Physiol. 2005, 139, 1995–2005. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, J.; Zhang, H.; Ni, Y.; Zhang, Q.; Chen, J.; Guan, Y. The responses of Arabidopsis thaliana to cadmium exposure explored via metabolite profiling. Chemosphere 2010, 78, 840–845. [Google Scholar] [CrossRef]
- Li, H.; Madden, J.L.; Potts, B.M. Variation in volatile leaf oils of the Tasmanian Eucalyptus species—1. Subgenus Monocalyptus. Biochem. System. Ecol. 1995, 23, 299–318. [Google Scholar] [CrossRef]
- Field, K.J.; Lake, J.A. Environmental metabolomics links genotype to phenotype and predicts genotype abundance in wild plant populations. Physiol. Plant. 2011, 142, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Kunin, W.E.; Vergeer, P.; Kenta, T.; Davey, M.P.; Burke, T.; Woodward, F.I.; Quick, P.; Mannarelli, M.E.; Watson-Haigh, N.S.; Butlin, R. Variation at range margins across multiple spatial scales: Environmental temperature, population genetics and metabolomic phenotype. Proc. R. Soc. B Biol. Sci. 2009, 276, 1495–1506. [Google Scholar] [CrossRef]
- Bustos-Segura, C.; Poelman, E.H.; Reichelt, M.; Gershenzon, J.; Gols, R. Intraspecific chemical diversity among neighbouring plants correlates positively with plant size and herbivore load but negatively with herbivore damage. Ecol. Lett. 2017, 20, 87–97. [Google Scholar] [CrossRef]
- Clancy, M.V.; Zytynska, S.E.; Moritz, F.; Witting, M.; Schmitt-Kopplin, P.; Weisser, W.W.; Schnitzler, J.-P. Metabotype variation in a field population of tansy plants influences aphid host selection. Plant Cell Environ. 2018, 41, 2791–2805. [Google Scholar] [CrossRef] [PubMed]
- Ziaja, D.; Muller, C. Intraspecific chemodiversity provides plant individual- and neighbourhood-mediated associational resistance towards aphids. Front. Plant Sci. 2023, 14, 1145918. [Google Scholar] [CrossRef]
- Clarke, A.; Gaston, K.J. Climate, energy and diversity. Proc. R. Soc. B Biol. Sci. 2006, 273, 2257–2266. [Google Scholar] [CrossRef]
- Okie, J.G.; Van Horn, D.J.; Storch, D.; Barrett, J.E.; Gooseff, M.N.; Kopsova, L.; Takacs-Vesbach, C.D. Niche and metabolic principles explain patterns of diversity and distribution: Theory and a case study with soil bacterial communities. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142630. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Junker, R.R. Chemical phenotype as important and dynamic niche dimension of plants. New Phytol. 2022, 234, 1168–1174. [Google Scholar] [CrossRef]
- Lande, R.; Arnold, S. The measurement of selection on correlated characters. Evolution 1983, 37, 1210–1226. [Google Scholar] [CrossRef]
- Doxford, S.W.; Ooi, M.K.J.; Freckleton, R.P. Spatial and temporal variability in positive and negative plant-bryophyte interactions along a latitudinal gradient. J. Ecol. 2013, 101, 465–474. [Google Scholar] [CrossRef]
- Vergnon, R.; Ooi, M.K.J.; Freckleton, R.P. Complex relationships between competing guilds along large-scale environmental gradients. Am. Nat. 2017, 189, 407–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vergeer, P.; Kunin, W.E. Adaptation at range margins: Common garden trials and the performance of Arabidopsis lyrata across its northwestern European range. New Phytol. 2013, 197, 989–1001. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Roessner, U.; Hansen, M.A.E.; Smedsgaard, J.; Nielsen, J. Metabolome Analysis: An Introduction; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef]
- Goodacre, R.; Vaidyanathan, S.; Bianchi, G.; Kell, D.B. Metabolic profiling using direct infusion electrospray ionisation mass spectrometry for the characterisation of olive oils. Analyst 2002, 127, 1457–1462. [Google Scholar] [CrossRef]
- Brown, M.; Dunn, W.B.; Dobson, P.; Patel, Y.; Winder, C.L.; Francis-McIntyre, S.; Begley, P.; Carroll, K.; Broadhurst, D.; Tseng, A.; et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst 2009, 134, 1322–1332. [Google Scholar] [CrossRef]
- Overy, S.A.; Walker, H.J.; Malone, S.; Howard, T.P.; Baxter, C.J.; Sweetlove, L.J.; Hill, S.A.; Quick, W.P. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J. Exp. Bot. 2005, 56, 287–296. [Google Scholar] [CrossRef]
- Broadhurst, D.I.; Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006, 2, 171–196. [Google Scholar] [CrossRef]
- Draper, J.; Enot, D.P.; Parker, D.; Beckmann, M.; Snowdon, S.; Lin, W.; Zubair, H. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour ‘rules’. BMC Bioinf. 2009, 10, 227. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Prot. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin biosynthesis in Arabidopsis. Phytochem. Rev. 2006, 5, 39–47. [Google Scholar] [CrossRef]
- Parsons, A.J.; Rasmussen, S.; Liu, Q.; Xue, H.; Ball, C.; Shaw, C. Plant growth—Resource or strategy limited: Insights from responses to gibberellin. Grass For. Sci. 2013, 68, 577–588. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, X.; Li, Y.; Tang, W.; Han, L. Differential expression of gibberellin 20 oxidase gene induced by abiotic stresses in Zoysiagrass (Zoysia japonica). Biologia 2012, 67, 681–688. [Google Scholar] [CrossRef]
- Wilson, R.N.; Heckman, J.W.; Somerville, C.R. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992, 100, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Langal, D.; Ober, D.; Pelse, P. The evolution of pyrrolizidine alkaloid biosynthesis and diversity in the Senecioneae. Phytochem. Rev. 2011, 10, 3–74. [Google Scholar] [CrossRef]
- Macel, M.; Klinkhamer, P.G.L. Chemotype of Senecio jacobaea affects damage by pathogens and insect herbivores in the field. Evol. Ecol. 2010, 24, 237–250. [Google Scholar] [CrossRef]
- Metlen, K.L.; Aschehoug, E.T.; Callaway, R.M. Plant behavioral ecology: Dynamic plasticity in secondary metabolites. Plant Cell Environ. 2009, 32, 641–653. [Google Scholar] [CrossRef]
- Greenway, H.; Munns, R. Mechanisms of Salt Tolerance in Non-Halophytes. Ann. Rev. Plant Physiol. 1980, 31, 149–190. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Schwabe, F.; Erban, A.; Udvardi, M.K.; Kopka, J. Comparative metabolomics of drought acclimation in model and forage legumes. Plant Cell Environ. 2012, 35, 136–149. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, P.; Jiang, Y.; Fu, J. Metabolomic analysis revealed differential adaptation to salinity and alkalinity stress in Kentucky Bluegrass Poa pratensis. Plant Mol. Biol. Report. 2015, 33, 56–68. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.-J. The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J. Exp. Bot. 2006, 57, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Samis, K.E.; Villalobos, A.L.; Eckert, C.G. Strong genetic differentiation but not local adaptation toward the range limit of a coastal dune plant. Evolution 2016, 70, 2520–2536. [Google Scholar] [CrossRef] [PubMed]
- Buckley, J.; Daly, R.; Cobbold, C.A.; Burgess, K.; Mable, B.K. Changing environments and genetic variation: Natural variation in inbreeding does not compromise short-term physiological responses. Proc. R. Soc. B Biol. Sci. 2019, 286, 1915. [Google Scholar] [CrossRef] [PubMed]





| Species | Common Garden * | Reciprocal Transplant |
|---|---|---|
| Arenaria serpyllifolia | GB1, GB2, Fr1, Fr2, Fr3, Fr4 (3) | None survived |
| Cerastium diffusum | Fr1, Fr2, Fr4 (3) | None survived |
| Phleum arenarium | GB2, Fr1, Fr2, Fr3, Fr4 (3) | Not selected for experiment |
| Senecio vulgaris | Fr2, Fr4 (3) | Not selected for experiment |
| Veronica arvensis | Fr3, Fr4 (3) | In site Fr1 = Fr1 (24), Fr3 (9), Fr4 (5), GB2 (5) In site GB1 = Fr1 (20), Fr3 (6), Fr4 (13), GB2 (11) In site GB2 = Fr1 (21), Fr3 (17), Fr4 (3), GB2 (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davey, M.P.; George, R.M.; Ooi, M.K.J.; Burrell, M.M.; Freckleton, R.P. Metabolic Niches and Plasticity of Sand-Dune Plant Communities Along a Trans-European Gradient. Metabolites 2025, 15, 217. https://doi.org/10.3390/metabo15040217
Davey MP, George RM, Ooi MKJ, Burrell MM, Freckleton RP. Metabolic Niches and Plasticity of Sand-Dune Plant Communities Along a Trans-European Gradient. Metabolites. 2025; 15(4):217. https://doi.org/10.3390/metabo15040217
Chicago/Turabian StyleDavey, Matthew P., Rachel M. George, Mark K. J. Ooi, Mike M. Burrell, and Robert P. Freckleton. 2025. "Metabolic Niches and Plasticity of Sand-Dune Plant Communities Along a Trans-European Gradient" Metabolites 15, no. 4: 217. https://doi.org/10.3390/metabo15040217
APA StyleDavey, M. P., George, R. M., Ooi, M. K. J., Burrell, M. M., & Freckleton, R. P. (2025). Metabolic Niches and Plasticity of Sand-Dune Plant Communities Along a Trans-European Gradient. Metabolites, 15(4), 217. https://doi.org/10.3390/metabo15040217






