Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
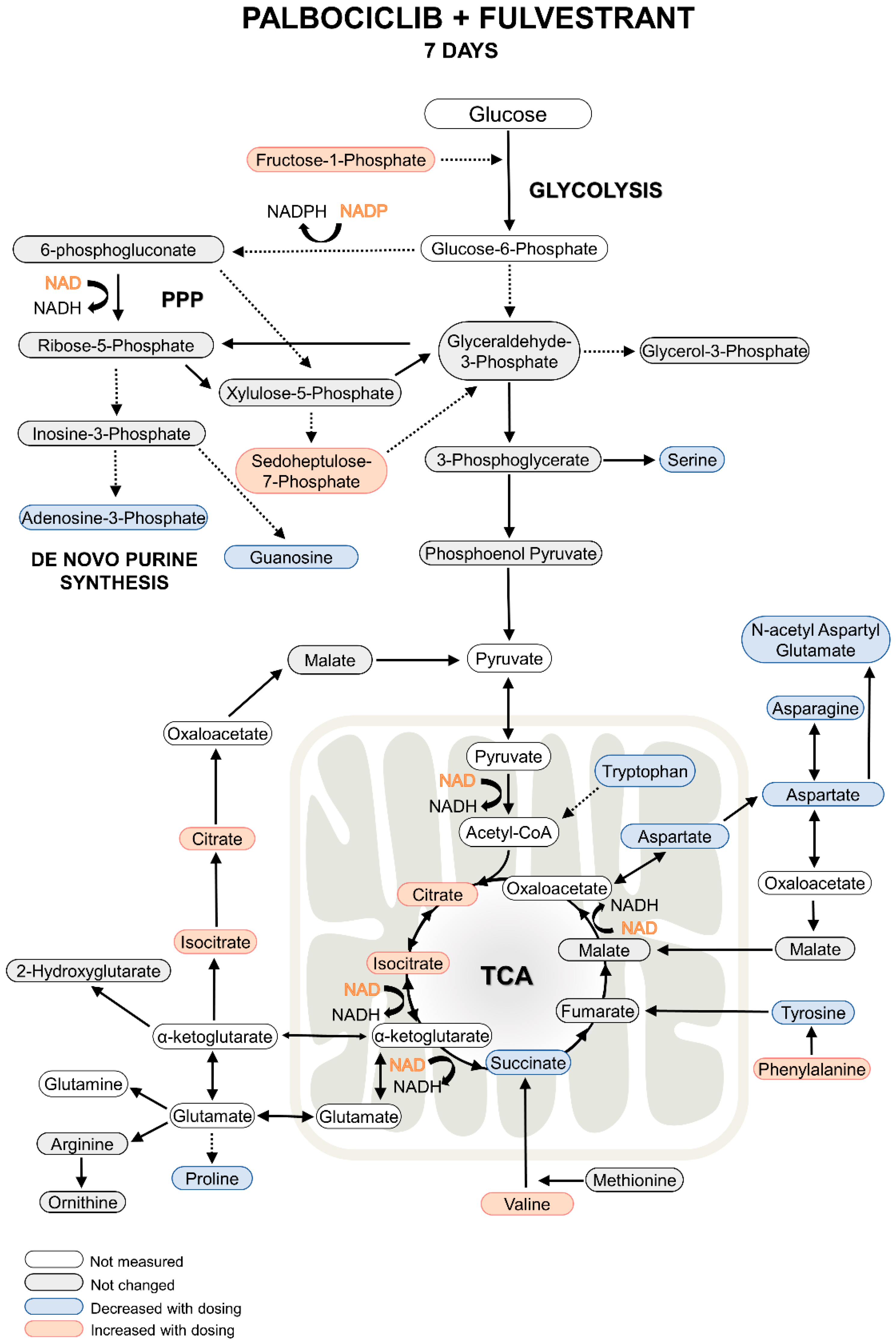
2.1. Metabolomics Analysis
2.2. Transcriptomics Analysis
2.3. Comparison with Previous Results
3. Materials and Methods
3.1. Cell Culture
3.2. Sample Preparation for Metabolomics Experiments
3.3. Untargeted Metabolomics Analysis
3.4. Targeted Metabolomics Analysis
3.5. Transcriptome Analysis
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cadoo, K.A.; Gucalp, A.; Traina, T.A. Palbociclib: An evidence-based review of its potential in the treatment of breast cancer. Breast Cancer Targ. Ther. 2014, 6, 123–133. [Google Scholar]
- Mayer, E. Targeting breast cancer with cdk inhibitors. Curr. Oncol. Rep. 2015, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Thijssen, B.; MscDermott, U.; Garnett, M.; Wessels, L.F.; Bernards, R. Targeting the rb-e2f pathway in breast cancer. Oncogene 2016, 35, 4829. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Palbociclib: First global approval. Drugs 2015, 75, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Mechcatie, E. Fda approves palbociclib with letrozole for advanced postmenopausal breast cancer. Oncol. Rep. 2015, 11, 7. [Google Scholar]
- Finn, R.S.; Crown, J.P.; Lang, I.; Boer, K.; Bondarenko, I.M.; Kulyk, S.O.; Ettl, J.; Patel, R.; Pinter, T.; Schmidt, M.; et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, her2-negative, advanced breast cancer (paloma-1/trio-18): A randomised phase 2 study. Lancet Oncol. 2015, 16, 25–35. [Google Scholar] [CrossRef]
- Turner, N.C.; Ro, J.; André, F.; Loi, S.; Verma, S.; Iwata, H.; Harbeck, N.; Loibl, S.; Huang Bartlett, C.; Zhang, K.; et al. Palbociclib in hormone-receptor–positive advanced breast cancer. N. Engl. J. Med. 2015, 373, 209–219. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Turner, N.C.; Bondarenko, I.; Ro, J.; Im, S.-A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, her2-negative metastatic breast cancer that progressed on previous endocrine therapy (paloma-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016, 17, 425–439. [Google Scholar]
- Finn, R.; Dering, J.; Conklin, D.; Kalous, O.; Cohen, D.; Desai, A.; Ginther, C.; Atefi, M.; Chen, I.; Fowst, C.; et al. Pd 0332991, a selective cyclin d kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009, 11, R77. [Google Scholar] [CrossRef]
- Zanuy, M.; Ramos-Montoya, A.; Villacanas, O.; Canela, N.; Miranda, A.; Aguilar, E.; Agell, N.; Bachs, O.; Rubio-Martinez, J.; Pujol, M.D.; et al. Cyclin-dependent kinases 4 and 6 control tumor progression and direct glucose oxidation in the pentose cycle. Metabolomics 2012, 8, 454–464. [Google Scholar] [CrossRef]
- Warth, B.; Raffeiner, P.; Granados, A.; Huan, T.; Fang, M.; Forsberg, E.M.; Benton, H.P.; Goetz, L.; Johnson, C.H.; Siuzdak, G. Metabolomics reveals that dietary xenoestrogens alter cellular metabolism induced by palbociclib/letrozole combination cancer therapy. Cell Chem. Biol. 2018, 25, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Huan, T.; Forsberg, E.M.; Rinehart, D.; Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Fang, M.; Aisporna, A.; Hilmers, B.; Poole, F.L.; et al. Systems biology guided by xcms online metabolomics. Nat. Methods 2017, 14, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive xcms online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef]
- Forsberg, E.M.; Huan, T.; Rinehart, D.; Benton, H.P.; Warth, B.; Hilmers, B.; Siuzdak, G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using xcms online. Nat. Protoc. 2018, 13, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Benton, H.P.; Wong, D.M.; Trauger, S.A.; Siuzdak, G. Xcms2: Processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal. Chem. 2008, 80, 6382–6389. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Hobert, J.A.; Mester, J.L.; Moline, J.; Eng, C. Elevated plasma succinate in pten, sdhb, and sdhd mutation-positive individuals. Genet. Med. 2012, 14, 616–619. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Na, U.; Winge, D.R.; Rutter, J. Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Mu, X.; You, Q. Succinate: An initiator in tumorigenesis and progression. Oncotarget 2017, 8, 53819–53828. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, J.; Huang, Z.; Wei, P.; Liu, Y.; Hao, J.; Zhao, L.; Zhang, F.; Tu, Y.; Wei, T. Aberrantly upregulated trap1 is required for tumorigenesis of breast cancer. Oncotarget 2015, 6, 44495–44508. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzola, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, 10. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.A.; Winnike, J.H.; McRitchie, S.L.; Clark, R.F.; Pathmasiri, W.W.; Sumner, S.J. Metabolomics analysis of hormone-responsive and triple-negative breast cancer cell responses to paclitaxel identify key metabolic differences. J. Proteome Res. 2016, 15, 3225–3240. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. Xcms online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]





| Metabolite Name | Palbociclib | Fulvestrant | Combination | |||
|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value | |
| Succinate | 1.6 | 0.0025 | 1.8 | 0.0441 | 1.9 | 0.0021 |
| Malate | −4.0 | 0.0170 | −3.5 | 0.0381 | −34.0 | 0.0002 |
| Fumarate | −2.1 | 0.0189 | −2.1 | 0.0438 | −2.0 | 0.0003 |
| Phosphoenolpyruvate | 1.1 | 0.0450 | N.S | N.S | 1.2 | 0.0014 |
| 6-phosphogluconate | N.S | N.S | −3.7 | 0.0255 | −2.5 | 0.0496 |
| Sedoheptulose-7-phosphate | N.S | N.S | −3.6 | 0.0335 | −3.7 | 0.0003 |
| Serine | N.S | N.S | 1.7 | <0.0001 | N.S | N.S |
| Cholesterol sulfate | N.S | N.S | −5.2 | 0.0332 | N.S | N.S |
| Taurine | N.S | N.S | −2.1 | 0.0380 | N.S | N.S |
| Inosine monophosphate | N.S | N.S | N.S | N.S | −16.1 | 0.0052 |
| Fructose-1-phosphate | N.S | N.S | N.S | N.S | −2.8 | 0.0172 |
| Metabolite Name | Palbociclib | Fulvestrant | Combination | |||
|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value | |
| Sedoheptulose-7-phosphate | 3.5 | 0.0076 | 13.2 | <0.0001 | 6.4 | 0.0012 |
| NADP | N.S | N.S | −2.6 | 0.0168 | −3.7 | 0.0030 |
| Citrate/Isocitrate | 9.3 | <0.0001 | 5.3 | <0.0001 | 3.8 | 0.0017 |
| Succinate | −1.9 | 0.0047 | −1.7 | <0.0001 | −1.8 | 0.0356 |
| Malate | 2.4 | 0.0130 | N.S | N.S | 1.7 | 0.0323 |
| Fructose-1-phosphate | 5.8 | 0.0036 | 19.5 | 0.0002 | 11.2 | 0.0006 |
| N-acetylaspartylglutamate | N.S | N.S | −3.4 | 0.0030 | −2.6 | 0.0051 |
| Aspartate | N.S | N.S | −2.6 | 0.0021 | −2.3 | 0.0041 |
| Serine | N.S | N.S | −2.7 | 0.0370 | −3.2 | 0.0245 |
| Proline | N.S | N.S | N.S | N.S | −2.5 | 0.0014 |
| Asparagine | N.S | N.S | −5.5 | 0.0021 | −8.4 | 0.0003 |
| Tryptophan | N.S | N.S | N.S | N.S | −2.9 | 0.0134 |
| Tyrosine | −10.0 | 0.0138 | −1008.1 | <0.0001 | −696.5 | <0.0001 |
| Phenylalanine | N.S | N.S | 4.2 | 0.0007 | 3.5 | 0.0008 |
| Valine | 1.8 | 0.0050 | 1.5 | 0.0003 | 1.7 | <0.0001 |
| Arginine | N.S | N.S | N.S | N.S | −3.5 | 0.0174 |
| Guanosine | −1.3 | 0.0496 | N.S | N.S | −4.5 | <0.0001 |
| Inosine monophosphate | −33.3 | <0.0001 | −37.3 | 0.0032 | −29.2 | 0.0021 |
| Adenosine monophosphate | −8.5 | 0.0008 | −11.1 | 0.0017 | −10.9 | 0.0019 |
| Pathway | Genes | Metabolites | ||
|---|---|---|---|---|
| Number/All | %Overlap | Number/All | %Overlap | |
| Adenosine nucleotides degradation II | 1/5 | 20 | 3/10 | 30.0 |
| Guanosine nucleotides degradation III | 1/3 | 33.3 | 2/9 | 22.2 |
| Urate biosynthesis/inosine 5’-phosphate degradation | 1/2 | 50.0 | 1/8 | 8.3 |
| Inosine 5’-phosphate biosynthesis II | 1/5 | 20.0 | 1/12 | N/A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Warth, B.; Palermo, A.; Rattray, N.J.W.; Lee, N.V.; Zhu, Z.; Hoang, L.T.; Cai, Y.; Mazurek, A.; Dann, S.; VanArsdale, T.; et al. Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells. Metabolites 2019, 9, 7. https://doi.org/10.3390/metabo9010007
Warth B, Palermo A, Rattray NJW, Lee NV, Zhu Z, Hoang LT, Cai Y, Mazurek A, Dann S, VanArsdale T, et al. Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells. Metabolites. 2019; 9(1):7. https://doi.org/10.3390/metabo9010007
Chicago/Turabian StyleWarth, Benedikt, Amelia Palermo, Nicholas J.W. Rattray, Nathan V. Lee, Zhou Zhu, Linh T. Hoang, Yuping Cai, Anthony Mazurek, Stephen Dann, Todd VanArsdale, and et al. 2019. "Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells" Metabolites 9, no. 1: 7. https://doi.org/10.3390/metabo9010007
APA StyleWarth, B., Palermo, A., Rattray, N. J. W., Lee, N. V., Zhu, Z., Hoang, L. T., Cai, Y., Mazurek, A., Dann, S., VanArsdale, T., Fantin, V. R., Shields, D., Siuzdak, G., & Johnson, C. H. (2019). Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells. Metabolites, 9(1), 7. https://doi.org/10.3390/metabo9010007