In Search of High-Yielding and Single-Compound-Yielding Plants: New Sources of Pharmaceutically Important Saponins from the Primulaceae Family
Abstract
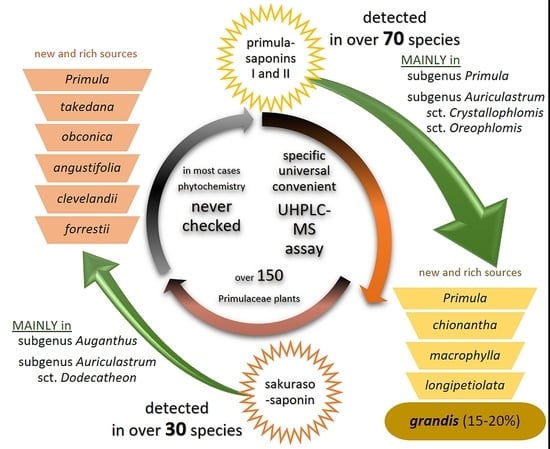
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Material
2.3. Preparation of Samples
2.4. Preparation of Standards
2.5. General NMR and HRMS Experimental Procedures
2.6. Purity of Standards
2.7. LC-MS Analytical Conditions
2.8. Validation of the Analytical Method
2.8.1. Specificity
2.8.2. Linearity, Range and Limits of Analysis
2.8.3. Precision, Accuracy and Stability
2.9. Clustering of Saponin Concentrations
3. Results
3.1. Method Development
3.2. Distribution of Saponins
3.2.1. Distribution of Primulasaponins
3.2.2. Distribution of Sakurasosaponin
3.2.3. Results of Clustering Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
List of Abbreviations
| [M–H]− | Pseudomolecular Ion (deprotonated molecule) |
| 2D | Two Dimensional (experiment in NMR) |
| A. | Androsace |
| APCI | Atmospheric-Pressure Chemical Ionization |
| COSY | 1H-1H Correlated Spectroscopy (experiment in NMR) |
| ELSD | Evaporative Light Scattering Detector |
| ESI | Electrospray Ionization |
| EU | European Union |
| H. | Hottonia |
| HCOOH | Formic Acid |
| HMBC | Heteronuclear Multiple Bond Correlation (experiment in NMR) |
| HRMS | High-Resolution Mass Spectrometry |
| HSQC | Heteronuclear Single Quantum Correlation (experiment in NMR) |
| LOD | Limit of Detection |
| LOQ | Limit of Quantification |
| LC | Liquid Chromatography |
| LC-MS | Liquid Chromatography coupled with Mass Spectrometry Detector |
| LC-UV | Liquid Chromatography coupled with Ultraviolet Absorbance Detector |
| MeOH | Methanol |
| MS/MS | Tandem Mass Spectrometry |
| NMR | Nuclear Magnetic Resonance (Spectroscopy) |
| P. | Primula |
| PSI | Primulasaponin I |
| PSII | Primulasaponin II |
| qHNMR | Quantitative Proton Nuclear Magnetic Resonance (Spectroscopy) |
| r | Coefficient of Correlation |
| r2 | Coefficient of Determination |
| RE | Relative Error |
| RSD | Relative Standard Deviation |
| sct. | Section |
| SD | Standard Deviation |
| sg. | Subgenus |
| SPE | Solid-Phase Extraction |
| ssct. | Subsection |
| ssp. | Subspecies |
| SSI | Sakurasosaponin |
| TLC | Thin-Layer Chromatography |
| UAE | Ultrasound-Assisted Extraction |
| UHPLC | Ultra-High-Performance Liquid Chromatography |
| var. | variety |
Appendix A
| Taxon (a) | Acronym | Taxon (b) | Acronym |
| genus Primula L. | sg. Aleuritia, sct. Minutissimae | ||
| sg. Aleuritia, sct. Aleuritia, ssct. Aleuritia | Primula primulina | PPRI_P_2014 | |
| Primula halleri | PHAL_P_2013, PHAL_K_2016 | sg. Aleuritia, sct. Muscarioides | |
| Primula scandinavica | PSCA_B_2013 | Primula concholoba | PCON_K_2014 |
| Primula scotica | PSCO_K_2016 | Primula muscarioides | PMUS_P_2013 |
| sg. Aleuritia, sct. Aleuritia, ssct. Algida | Primula vialii | PVIA_B_2013 | |
| Primula algida | PALG_K_2014 | sg. Aleuritia, sct. Oreophlomis | |
| Primula darialica | PDAR_K_2014 | Primula auriculata | PAUA_K_2015 |
| sg. Aleuritia, sct. Armerina | Primula luteola | PLUT_K_2015 | |
| Primula fasciculata | PFAS_K_2014 | Primula rosea ‘Gigas’ | PROG_B_2013 |
| Primula involucrata | PINV_P_2015 | Primula warschenewskiana | PWAR_K_2014 |
| Primula munroi ssp. yargongensis (->P. involucrata in [1]) | PMUY_P_2015 | sg. Aleuritia, sct. Petiolares, ssct. Edgeworthii | |
| Primula yargongensis (->P. involucrata in [1]) | PYAR_P_2015 | Primula moupinensis | PMOU_GP_2014 |
| Primula zambalensis | PZAM_K_2014 | Primula moupinensis ssp. barkamensis | PMOB_K_2015 |
| sg. Aleuritia, sct. Capitatae | sg. Aleuritia, sct. Petiolares, ssct. Griffithii | ||
| Primula capitata ssp. mooreana | PCAM_B_2013 | Primula calderiana ssp. calderiana | PCAL_K_2014 |
| Primula glomerata | PGLO_K_2014 | Primula tanneri ssp. nepalensis | PTNN_K_2014 |
| sg. Aleuritia, sct. Crystalophlomis | sg. Aleuritia, sct. Petiolares, ssct. Petiolares | ||
| Primula purdomii | PPUR_P_2014 | Primula boothi var. repens | PBOR_K_2014 |
| sg. Aleuritia, sct. Crystalophlomis, ssct. Crystalophlomis | sg. Aleuritia, sct. Petiolares, ssct. Sonchifolia | ||
| Primula chionantha | PCHI_K_2014 | Primula sonchifolia ssp. sonchifolia | PSON_K_2014 |
| Primula chionantha ssp. chionantha | PCHC_K_2015 | sg. Aleuritia, sct. Proliferae | |
| Primula chionantha ssp. sinopurpurea | PCHS_K_2015 | Primula beesiana | PBES_B_2013 |
| Primula graminifolia (->P. chionantha in [1]) | PGRM_P_2015 | Primula bulleyana | PBUL_B_2013 |
| Primula longipetiolata | PLON_K_2014 | Primula japonica | PJAP_B_2013 |
| Primula macrophylla | PMAC_K_2015 | Primula prolifera | PPRO_K_2015 |
| Primula orbicularis | PORB_K_2015, PORB_P_2015 | Primula wilsonii var. anisodora | PWIA_K_2015 |
| sg. Aleuritia, sct. Crystalophlomis, ssct. Maximowiczii | sg. Aleuritia, sct. Pulchella | ||
| Primula maximowiczii var. maximowiczii | PMAX_K_2014 | Primula pulchella | PPUL_K_2015, PPUL_P_2015 |
| Primula tangutica | PTAN_P_2013 | Primula sharmae | PSHA_K_2016 |
| Primula woodwardii | PWOD_P_2013, PWOD_P_2014 | Primula stenocalyx | PSTE_P_2013, PSTE_P_2014 |
| sg. Aleuritia, sct. Davidii | sg. Aleuritia, sct. Sikkimensis | ||
| Primula bergenioides | PBER_K_2015 | Primula firmipes | PFIR_K_2014 |
| Primula ovaliifolia | POVA_K_2015 | Primula florindae | PFLO_B_2013 |
| sg. Aleuritia, sct. Denticulata | Primula florindae, red flowered | PFLR_K_2015 | |
| Primula denticulata ‘Alba’ | PDEA_B_2013 | Primula ioessa | PIOE_P_2013 |
| Primula denticulata | PDEN_K_2016 | Primula sikkimensis | PSIK_K_2014 |
| Primula monticola | PMON_K_2014, PMON_K_2015 | Primula waltonii | PWAL_K_2014 |
| Taxon (c) | Acronym | Taxon (d) | Acronym |
| sg. Aleuritia, sct. Soldanelloides | sg. Auriculastrum, sct. Auricula, ssct. Erythrodosum | ||
| Primula reidii | PREI_K_2016 | Primula daoensis | PDAO_K_2014 |
| Primula reidii var. williamsii ‘Alba’ | PREW_F_2015 | Primula hirsuta | PHIR_P_2013 |
| sg. Aleuritia, sct. Yunnanensis | sg. Auriculastrum, sct. Auricula, ssct. Rhopsidium | ||
| Primula florida (P. blinii) | PBLI_P_2013, PBLI_P_2014 | Primula allionii | PALL_F_2015 |
| Primula rupicola | PRUP_K_2016 | Primula integrifolia | PINT_P_2013 |
| sg. Auganthus, sct. Bullatae | Primula tyrolensis | PTYR_TK_2015 | |
| Primula bullata var. rufa | PBUR_K_2014 | sg. Auriculastrum, sct. Auricula, hybrids | |
| Primula forrestii | PFOR_K_2014 | Primula venusta | PVEN_B_2013 |
| sg. Auganthus, sct. Cortusoides, ssct. Cortusoides | Primula allioni × Primula villosa | PXAV_P_2013 | |
| Primula cortusoides | PCOR_B_2013 | sg. Auriculastrum, sct. Cuneifolia | |
| Primula polyneura | PPOL_K_2015 | Primula cuneifolia | PCUN_K_2016 |
| Primula sieboldii | PSIE_K_2014 | Primula cuneifolia ssp. heterodonta | PCUH_K_2016 |
| sg. Auganthus, sct. Cortusoides, ssct. Geraniifolia | sg. Auriculastrum, sct. Dodecatheon | ||
| Primula heucheriifolia | PHEU_K_2015 | Primula austrofrigida | PAUS_K_2014 |
| Primula jesoana | PJES_K_2014 | Primula clevelandii | PCLE_K_2016 |
| Primula kisoana | PKIS_K_2015 | Primula conjugens | PCJG_K_2014 |
| Primula palmata | PPAL_K_2014 | Primula jeffreyi | PJEF_K_2015, PJEF_K_2016 |
| sg. Auganthus, sct. Obconicolisteri | Primula latiloba | PLLB_K_2014 | |
| Primula obconica | POBC_K_2014 | Primula meadia | PMEA_K_2014 |
| sg. Auganthus, sct. Reinii | Primula pauciflora | PPAU_K_2014 | |
| Primula takedana | PTAK_E_2014 | Primula tetrandra | PTET_K_2014 |
| sg. Auriculastrum, sct. Amethystina | sg. Auriculastrum, sct. Parryi | ||
| Primula amethystina var. brevifolia | PAMB_K_2015 | Primula angustifolia | PANG_K_2016 |
| sg. Auriculastrum, sct. Auricula, ssct. Arthritica | Primula parryi | PPAR_K_2014 | |
| Primula glaucescens ssp. longobarda | PGLL_P_2013, PGLL_P_2014 | Primula rusbyi | PRUS_K_2014 |
| Primula spectabilis | PSPE_P_2013 | sg. Primula, sct. Primula | |
| sg. Auriculastrum, sct. Auricula, ssct. Auricula | Primula elatior | PELA_K_2014 | |
| Primula auricula | PAUR_B_2013 | Primula elatior var. amoena | PAMO_P_2014 |
| sg. Auriculastrum, sct. Auricula, ssct. Brevibracteatum | Primula juliae | PJUL_B_2013 | |
| Primula carniolica | PCAR_K_2014, PCAR_K_2015 | Primula megaseifolia | PMEG_K_2015 |
| Primula latifolia ‘Alba’ | PLAA_P_2013 | Primula veris (syn. P. officinalis) | PVRS_K_2014 |
| Primula marginata | PMGN_P_2013 | Primula vulgaris | PVUL_K_2014 |
| sg. Auriculastrum, sct. Auricula, ssct. Chamaecallis | sg. Primula, sct. Sredinskya | ||
| Primula minima | PMIN_I_2014 | Primula grandis | PGRA_K_2014, PGRA_K_2016 |
| Primula minima f. niveum | PNIV_P_2014 | sg. Primula, hybrids | |
| sg. Auriculastrum, sct. Auricula, ssct. Cyanaster | Primula margotae ‘Garryarde Guinevere’ | PMAG_B_2013 | |
| Primula glutinosa | PGLU_P_2013 | sg. Sphondyllia | |
| Primula × kewensis | PXKE_K_2016 | ||
| Primula verticillata | PVRT_K_2014 | ||
| Taxon (e) | Acronym | Taxon (f) | Acronym |
| genus Androsace L. | sct. Aizodium | ||
| sct. Andraspis | Androsace bulleyana | ABUL_F_2015 | |
| Androsace albana | AALB_K_2019 | genus Cortusa L. | |
| sct. Aretia, ssct. Aretia | Cortusa matthioli | CMAT_P_2013 | |
| Androsace cylindrica | ACYL_K_2015 | Cortusa matthioli ssp. matthioli | CMAT_K_2015 |
| Androsace lehmannii | ALEH_P_2014 | Cortusa matthioli ssp. caucasica | CCAU_K_2015, CCAU_K_2016 |
| Androsace mathildae | AMTH_TK_2015 | Cortusa matthioli ssp. sachalinensis | CSAC_P_2013, CSAC_K_2015 |
| sct. Aretia, ssct. Dicranothrix | Cortusa matthioli ssp. turkestanica | CTUR_P_2013 | |
| Androsace lacteal | ALAC_TK_2015 | genus Dionysia Fenzl. | |
| Androsace laggeri (= A. carnea var. laggeri) | ACAL_P_2014 | Dionysia khatamii | DKHA_F_2015 |
| Androsace obtusifolia | AOBT_P_2014 | Dionysia zschummelii | DZSH_F_2015 |
| sct. Chamaejasme, ssct. Hookerianae | genus Hottonia L. | ||
| Androsace limprichtii | ALIM_P_2014 | Hottonia inflata hb | HOIN_MO_2014 |
| sct. Chamaejasme, ssct. Mucronifoliae | Hottonia palustris hb | HOPA_PG_2014 | |
| Androsace mariae var. tibetica | AMAT_P_2014 | genus Lysimachia L. | |
| Androsace mucronifolia | AMUC_TK_2015 | Lysimachia nummularia ‘Gold’ | LYNG_PG_2014 |
| Androsace sempervivoides | ASPV_B_2015 | Lysimachia thyrsiflora | LYTH_MO_2014 |
| sct. Chamaejasme, ssct. Strigillosae | genus Omphalogramma (Franch.) Franch. | ||
| Androsace spinulifera | ASPI_P_2015 | Omphalogramma delavayi | ODEL_PO_2019 |
| Androsace strigillosa | ASTR_P_2014 | Omphalogramma tibeticum | OTIB_K_2019 |
| sct. Chamaejasme, ssct. Sublanatae | genus Soldanella L. | ||
| Androsace adenocephala | AADE_F_2015 | sct. Crateriflorae | |
| Androsace nortonii | ANOR_P_2014 | Soldanella alpina | SALP_K_2016 |
| sct. Chamaejasme, ssct. Villosae, series Chamaejasmoidae | Soldanella carpatica | SCAR_K_2014 | |
| Androsace brachystegia | ABRA_P_2014 | Soldanella cyanaster | SCYA_K_2014 |
| Androsace chamaejasme ssp. carinata | ACHC_P_2014 | Soldanella dimoniei | SDIM_K_2014 |
| Androsace zambalensis | AZAM_P_2014 | Soldanella villosa | SVIL_K_2014 |
| sct. Chamaejasme, ssct. Villosae, series Euvillosae | sct. Tubiflorae | ||
| Androsace dasyphylla | ADAS_P_2014 | Soldanella minima | SMIN_F_2015, SMIN_TK_2015 |
| Androsace robusta ssp. purpurea | AROP_P_2014 | genus Vitaliana Sesl. | |
| Androsace sarmentosa | ASAR_K_2015 | Vitaliana primuliflora | VPRI_B_2015 |
| sct. Pseudoprimula | Vitaliana primuliflora ssp. assoana | VPAS_TK_2015 | |
| Androsace elatior | AELA_F_2015 | Vitaliana primuliflora ssp. praetutiana | VPPR_B_2015, VPPR_B_2017 |
| sct. Douglasia | |||
| Androsace montana (= Douglasia montana) | AMON_F_2015 |
| Taxon (a) | Average % of Dry Mass | Taxon (b) | Average % of Dry Mass | Taxon (c) | Average % of Dry Mass | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PSI | PSII | SSI | PSI | PSII | SSI | PSI | PSII | SSI | |||||
| genus Primula | sg. Auganthus, sct. Bullatae | sg. Auriculastrum, sct. Cuneifolia | |||||||||||
| sg. Aleuritia, sct. Armerina | P. bullata var. rufa | nd | nd | 1.07 | P. cuneifolia | Nd | nd | 2.31 | |||||
| P. zambalensis | t | nd | 2.97 | P. forrestii | t | nd | 8.00 | P. cuneifolia ssp. heterodonta | Nd | nd | 3.28 | ||
| sg. Aleuritia, sct. Crystallophlomis | sg. Auganthus, sct. Cortusoides | sg. Auriculastrum, sct. Dodecatheon | |||||||||||
| P. chionantha | 5.57 | nd | nd | P. cortusoides | nd | nd | 4.37 | P. austrofrigida | T | nd | 2.62 | ||
| P. chionantha ssp. chionantha | 6.35 | nd | nd | P. heucherifolia | t | nd | 2.24 | P. clevelandii | t | nd | 8.59 | ||
| P. chionantha ssp. sinopurpurea | 4.31 | nd | nd | P. kisoana | t | nd | 1.68 | P. conjugens | 3.86 | nd | 1.26 | ||
| P. graminifolia | 1.22 | nd | nd | P. palmata | t | nd | 2.13 | P. jeffreyi | 2.85 | nd | 1.00 | ||
| P. longipetiolata | 9.48 | nd | nd | P. polyneura | t | nd | 2.22 | P. latiloba | t | nd | 0.77 | ||
| P. macrophylla | 7.06 | nd | nd | P. sieboldii | t | nd | 4.20 | P. meadia | t | nd | 3.56 | ||
| P. orbicularis | 0.60 | nd | nd | sg. Auganthus, sct. Obconicolisteri | P. pauciflora | t | nd | 5.08 | |||||
| P. purdomii | t | nd | nd | P. obconica | t | nd | 8.59 | P. tetrandra | t | nd | 6.90 | ||
| sg. Aleuritia, sct. Davidii | sg. Auganthus, sct. Reinii | sg. Auriculastrum, sct. Parryi | |||||||||||
| P. bergenioides | 0.73 | nd | nd | P. takedana | t | nd | 9.09 | P. angustifolia | 0.35 | nd | 8.60 | ||
| P. ovaliifolia | t | nd | nd | sg. Auriculastrum, sct. Auricula | P. parryi | 1.13 | nd | 0.44 | |||||
| sg. Aleuritia, sct. Oreophlomis | P. allionii | t | nd | nd | P. rusbyi | t | nd | 0.85 | |||||
| P. auriculata | 3.24 | nd | nd | P. allionii × P. villosa | t | nd | nd | sg. Primula, sct. Primula | |||||
| P. luteola | 1.75 | nd | nd | P. auricula | t | nd | nd | P. elatior var. amoena | t | nd | nd | ||
| P. rosea ‘Gigas’ | 4.06 | nd | nd | P. carniolica | t | nd | nd | P. elatior | 3.30 | nd | nd | ||
| P. warschenewskiana | 5.21 | nd | nd | P. glaucescens ssp. longobarda | t | nd | 0.48 | P. elatior rhiz | 4.63 | nd | nd | ||
| sg. Aleuritia, sct. Petiolares | P. glutinosa | t | nd | nd | P. juliae | 0.79 | t | nd | |||||
| P. boothi var. repens | 1.04 | nd | nd | P. hirsuta | t | nd | nd | P. juliae rhiz | 0.21 | t | nd | ||
| P. moupinensis | t | nd | nd | P. integrifolia | t | nd | nd | P. megaseifolia | 3.24 | 2.65 | nd | ||
| P. moupinensis ssp. barkamensis | 1.17 | nd | nd | P. latifolia ‘Alba’ | t | nd | nd | P. veris (syn. P. officinalis) | 2.84 | t | nd | ||
| P. sonchifolia ssp. sonchifolia | 0.67 | nd | nd | P. marginata | t | nd | nd | P. vulgaris | 6.23 | 3.26 | nd | ||
| sg. Aleuritia, sct. Proliferae | P. minima | t | nd | nd | P. vulgaris rhiz | 3.30 | 2.27 | nd | |||||
| P. besiana | t | nd | nd | P. minima f. niveum | t | nd | nd | sg. Primula, sct. Sredinskya | |||||
| P. japonica ‘Alba’ | 1.05 | nd | nd | P. tyrolensis | t | nd | nd | P. grandis (PGRA_K_2014) | 23.88 | nd | nd | ||
| P. prolifera | t | nd | nd | P. venusta | t | nd | nd | P. grandis (PGRA_K_2016) | 15.72 | nd | nd | ||
| sg. Aleuritia, sct. Pulchella | P. grandis rhiz (PGRA_K_2016) | 14.57 | nd | nd | |||||||||
| P. pulchella | t | nd | t | ||||||||||
| Taxon (d) | Average % of Dry Mass | ||
|---|---|---|---|
| PSI | PSII | SSI | |
| sg. Primula, hybrids | |||
| P. margotae ‘GarryardeGuinevere’ | 4.80 | 0.32 | nd |
| P. margotae rhiz | 2.21 | 0.23 | nd |
| sg. Primula, trade samples | |||
| Primulae radix 1 (PR1) | 8.60 | 4.90 | nd |
| Primulae radix 2 (PR2) | 6.16 | t | nd |
| Primulae radix 3 (PR3) | 2.66 | t | nd |
| Primulae radix 4 (PR4) | 2.18 | t | nd |
| sg. Sphondylia, hybrids | |||
| P. × kewensis | 0.33 | nd | nd |
| genus Androsace | |||
| sct. Aretia | |||
| A. lehmanii | nd | nd | 1.12 |
| sct. Chamaejasme | |||
| A. adenocephala | nd | nd | t |
| A. brachystegia | nd | nd | t |
| A. chamaejasme ssp. carinata | nd | nd | t |
| A. limprichtii | nd | nd | 0.36 |
| A. robusta ssp. purpurea | nd | nd | 0.76 |
| A. sarmentosa | nd | nd | 1.28 |
| A. strigillosa | t | nd | t |
| genus Hottonia | |||
| H. inflata hb | 0.36 | nd | 2.27 |
| H. palustris hb | 0.31 | nd | 0.62 |
References
- Richards, J.A. Primula, 2nd ed.; Timber Press: Portland, OR, USA, 2003; ISBN 0-88192-580-2. [Google Scholar]
- Halda, J.J. The Genus Primula: In Cultivation and the Wild; Tethys Books: Englewood, CO, USA, 1992; ISBN 0-9632289-0-0. [Google Scholar]
- Smith, G.; Lowe, D. The Genus Androsace: A Monograph for Gardeners and Botanists; Alpine Garden Society: Pershore, UK, 1997; ISBN 0-900048-67-0. [Google Scholar]
- Grey-Wilson, C. The Genus Dionysia; Alpine Garden Society: Woking, UK, 1989; ISBN 0-900048-51-4. [Google Scholar]
- Hu, Q. On the geographical distribution of the Primulaceae. J. Trop. Subtrop. Bot. 1994, 2, 1–14. [Google Scholar]
- Valant-Vetschera, K.M.; Bhutia, T.D.; Wollenweber, E. Exudate flavonoids of Primula spp: Structural and biogenetic chemodiversity. Nat. Prod. Commun. 2009, 4, 365–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.; Ganzera, M.; Stuppner, H. Analysis of phenolic glycosides and saponins in Primula elatior and Primula veris (primula root) by liquid chromatography, evaporative light scattering detection and mass spectrometry. J. Chromatogr. A 2006, 1112, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Colombo, P.S.; Flamini, G.; Rodondi, G.; Giuliani, C.; Santagostini, L.; Fico, G. Phytochemistry of European Primula species. Phytochemistry 2017, 143, 132–144. [Google Scholar] [CrossRef] [PubMed]
- EMA Committee on Herbal Medicinal Products (HMPC). Assessment Report on Primula veris L. and/or Primula elatior (L.) Hill, radix. 2012. Available online: https://www.ema.europa.eu/en/documents/herbal-report/final-assessment-report-primula-veris-l/primula-elatior-l-hill-radix_en.pdf (accessed on 19 March 2012).
- Primrose root (Primulae radix) monograph. In European Pharmacopoeia, 3rd ed.; Council of Europe: Strasbourg, France, 1999; pp. 804–805.
- Primrose root (Primulae radix) monograph. In European Pharmacopoeia, 10th ed.; Council of Europe: Strasbourg, France, 2020; pp. 1588–1589.
- Siems, K.; Jaensch, M.; Jakupović, J. Structures of the two saponins isolated from commercially available root extract of Primula sp. Planta Med. 1998, 64, 272–274. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, I.; Yoshikawa, M.; Kobayashi, K.; Imakura, Y.; Im, K.S.; Ikenishi, Y. Saponin and sapogenol. XXVIII. Reinvestigation of the branching positions in the glucuronide moieties of three glucuronide saponins: Desacyl-jegosaponin, deacyl-boninsaponin A and sakuraso-saponin. Chem. Pharm. Bull. (Tokyo) 1980, 28, 296–300. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T. Primula sieboldii E. Morren. In International Collation of Traditional and Folk Medicine; Guo, J.-X., Ed.; World Scientific: Singapore, 2001; Volume 4, p. 80. [Google Scholar]
- Koczurkiewicz, P.; Kowolik, E.; Podolak, I.; Wnuk, D.; Piska, K.; Łabędź-Masłowska, A.; Wójcik-Pszczoła, K.; Pękala, E.; Czyż, J.; Michalik, M. Synergistic cytotoxic and anti-invasive effects of mitoxantrone and triterpene saponins from Lysimachia ciliata on human prostate cancer cells. Planta Med. 2016, 82, 1546–1552. [Google Scholar] [CrossRef] [Green Version]
- Frenkel, N.; Makky, A.; Sudji, I.R.; Wink, M.; Tanaka, M. Mechanistic investigation of interactions between steroidal saponin digitonin and cell membrane models. J. Phys. Chem. B 2014, 118, 14632–14639. [Google Scholar] [CrossRef]
- Eid, S.Y.; El-Readi, M.Z.; Wink, M. Synergism of three-drug combinations of sanguinarine and other plant secondary metabolites with digitonin and doxorubicin in multi-drug resistant cancer cells. Phytomedicine 2012, 19, 1288–1297. [Google Scholar] [CrossRef]
- Sedaghat Doost, A.; van Camp, J.; Dewettinck, K.; van der Meeren, P. Production of thymol nanoemulsions stabilized using quillaja saponin as a biosurfactant: Antioxidant activity enhancement. Food Chem. 2019, 293, 134–143. [Google Scholar] [CrossRef]
- Tschiggerl, C.; Bucar, F. Influence of saponin plants on the volatile fraction of thyme in herbal teas. Fitoterapia 2011, 82, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Fleck, J.D.; Betti, A.H.; Da Silva, F.P.; Troian, E.A.; Olivaro, C.; Ferreira, F.; Verza, S.G. Saponins from Quillaja saponaria and Quillaja brasiliensis: Particular chemical characteristics and biological activities. Molecules 2019, 24, 171. [Google Scholar] [CrossRef] [Green Version]
- Hegde, V.D.; Silver, J.; Patel, M.G.; Bryant, R.; Pai, J.; Das, P.R.; Puar, M.S.; Cox, P.A. Phospholipase D inhibitors from a Myrsine species. J. Nat. Prod. 1995, 58, 1492–1497. [Google Scholar] [CrossRef] [PubMed]
- Vermeersch, M.; Foubert, K.; da Luz, R.I.; Van Puyvelde, L.; Pieters, L.; Cos, P.; Maes, L. Selective antileishmania activity of 13,28-epoxy-oleanane and related triterpene saponins from the plant families Myrsinaceae, Primulaceae, Aceraceae and Icacinaceae. Phyther. Res. 2009, 23, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Girardi, C.; Vásquez-Ocmin, P.G.; Castillo, D.; Sauvain, M.; Rojas, R.; Fabre, N.; Haddad, M. Biological activities of 13, 28-epoxyoleanane triterpene saponins from two peruvian Myrsinaceae. Rev. la Soc. Química del Perú 2012, 78, 188–197. [Google Scholar]
- Li, Q.; Li, W.; Hui, L.-P.; Zhao, C.-Y.; He, L.; Koike, K. 13,28-Epoxy triterpenoid saponins from Ardisia japonica selectively inhibit proliferation of liver cancer cells without affecting normal liver cells. Bioorg. Med. Chem. Lett. 2012, 22, 6120–6125. [Google Scholar] [CrossRef]
- Wang, R.; Xiao, X.; Wang, P.-Y.; Wang, L.; Guan, Q.; Du, C.; Wang, X.-J. Stimulation of autophagic activity in human glioma cells by anti-proliferative ardipusilloside I isolated from Ardisia pusilla. Life Sci. 2014, 110, 15–22. [Google Scholar] [CrossRef]
- Cao, W.-Y.; Wang, Y.-N.; Wang, P.-Y.; Lei, W.; Feng, B.; Wang, X.-J. Ardipusilloside-I metabolites from human intestinal bacteria and their antitumor activity. Molecules 2015, 20, 20569–20581. [Google Scholar] [CrossRef] [Green Version]
- Mbaveng, A.T.; Ndontsa, B.L.; Kuete, V.; Nguekeu, Y.M.M.; Çelik, İ.; Mbouangouere, R.; Tane, P.; Efferth, T. A naturally occuring triterpene saponin ardisiacrispin B displayed cytotoxic effects in multi-factorial drug resistant cancer cells via ferroptotic and apoptotic cell death. Phytomedicine 2018, 43, 78–85. [Google Scholar] [CrossRef]
- Patten, A.M.; Vassão, D.G.; Wolcott, M.P.; Davin, L.B.; Lewis, N.G. Trees: A remarkable biochemical bounty. In Comprehensive Natural Products II; Elsevier: Kidlington, UK, 2010; pp. 1173–1296. ISBN 978-0-08-045381-1. [Google Scholar]
- Ashihara, H.; Kato, M.; Chuang-Xing, Y. Biosynthesis and metabolism of purine alkaloids in leaves of cocoa tea (Camellia ptilophylla). J. Plant. Res. 1998, 111, 599–604. [Google Scholar] [CrossRef]
- Ma, L.; Li, W.; Wang, H.; Kuang, X.; Li, Q.; Wang, Y.; Xie, P.; Koike, K. A simple and rapid method to identify and quantitatively analyze triterpenoid saponins in Ardisia crenata using ultrafast liquid chromatography coupled with electrospray ionization quadrupole mass spectrometry. J. Pharm. Biomed. Anal. 2015, 102, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Foam index. In European Pharmacopoeia, 10th ed.; Council of Europe: Strasbourg, France, 2020; p. 317.
- Włodarczyk, M.; Szumny, A.; Gleńsk, M. Lanostane-Type Saponins from Vitaliana primuliflora. Molecules 2019, 24, 1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kew Herbarium’s Digital Collection. Available online: http://apps.kew.org/herbcat/gotoHerbariumGrowthPage.do (accessed on 28 February 2020).
- Pauli, G.F.; Jaki, B.U.; Lankin, D.C. Quantitative 1H NMR: Development and potential of a method for natural products analysis. J. Nat. Prod. 2005, 68, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.-F.; He, C.-H.; Peng, C.-I.; Hu, C.-M.; Hao, G. Circumscription of Primula subgenus Auganthus (Primulaceae) based on chloroplast DNA sequences. J. Syst. Evol. 2010, 48, 123–132. [Google Scholar] [CrossRef]
- Mast, A.R.; Reveal, J.L. Transfer of Dodecatheon to Primula (Primulaceae). Brittonia 2007, 59, 79–82. [Google Scholar] [CrossRef]
- Bender, S.F.; van der Heijden, M.G.A. Soil biota enhance agricultural sustainability by improving crop yield, nutrient uptake and reducing nitrogen leaching losses. J. Appl. Ecol. 2015, 52, 228–239. [Google Scholar] [CrossRef] [Green Version]
- Thirkell, T.J.; Charters, M.D.; Elliott, A.J.; Sait, S.M.; Field, K.J. Are mycorrhizal fungi our sustainable saviours? Considerations for achieving food security. J. Ecol. 2017, 105, 921–929. [Google Scholar] [CrossRef] [Green Version]
- Schneeweiss, G.M.; Schönswetter, P.; Kelso, S.; Niklfeld, H. Complex biogeographic patterns in Androsace (Primulaceae) and related genera: Evidence from phylogenetic analyses of nuclear internal transcribed spacer and plastid trnL-F sequences. Syst. Biol. 2004, 53, 856–876. [Google Scholar] [CrossRef] [Green Version]
- Mast, A.R.; Kelso, S.; Richards, J.A.; Lang, D.J.; Feller, D.M.S.; Conti, E. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int. J. Plant Sci. 2001, 162, 1381–1400. [Google Scholar] [CrossRef] [Green Version]


| Standard | n ± SD | A ± SD | B ± SD | r | r2 | LOD [ng/mL] | LOQ [ng/mL] |
|---|---|---|---|---|---|---|---|
| PSI | 0.890 ± 0.072 | 4,574,451 ± 83,044 | 24.73 ± 1.26 | 0.9998 | 0.9996 | 6.7 | 20.3 |
| PSII | 0.776 ± 0.110 | 3,274,550 ± 100,794 | 23.41 ± 2.24 | 0.9994 | 0.9988 | 6.4 | 19.4 |
| SSI | 0.943 ± 0.110 | 3,951,182 ± 86,066 | 20.29 ± 1.62 | 0.9995 | 0.9990 | 7.4 | 22.5 |
| Standard | Repeatability (RSD)/(Intra-Day Precision) | Intermediate Precision (RSD)/(Inter-Day Precision) | ||||
| Level | 1 µg/mL | 10 µg/mL | 100 µg/mL | 1 µg/mL | 10 µg/mL | 100 µg/mL |
| PSI | 2.3 | 2.0 | 0.4 | 5.6 | 1.6 | 1.2 |
| PSII | 4.0 | 1.7 | 0.7 | 6.1 | 1.5 | 1.1 |
| SSI | 2.8 | 1.2 | 1.0 | 3.0 | 1.2 | 0.9 |
| Standard | Accuracy (%RE) | Stability (RSD)/(14 days) | ||||
| Level | 1 µg/mL | 10 µg/mL | 100 µg/mL | 1 µg/mL | 10 µg/mL | 100 µg/mL |
| PSI | +21.2 | +3.5 | +0.1 | 3.2 | 3.3 | 2.6 |
| PSII | +30.6 | +5.5 | −0.7 | 10.1 | 1.8 | 4.2 |
| SSI | +15.4 | +4.8 | +0.4 | 6.8 | 2.2 | 1.4 |
| Sample Acronym | Group | Sample Acronym | Group | Sample Acronym | Group | Sample Acronym | Group |
|---|---|---|---|---|---|---|---|
| PFOR_K_2014 | 1 | HOIN_MO_2014 | 2 | PCHS_K_2015 | 3 | PCHI_K_2014 | 6 |
| POBC_K_2014 | 1 | PZAM_K_2014 | 2 | PAUA_K_2015 | 3 | PCHC_K_2015 | 6 |
| PTAK_E_2014 | 1 | PCOR_B_2013 | 2 | PROG_B_2013 | 3 | PLON_K_2014 | 6 |
| PCLE_K_2016 | 1 | PPOL_K_2015 | 2 | PCJG_K_2014 | 3 | PMAC_K_2015 | 6 |
| PTET_K_2014 | 1 | PSIE_K_2014 | 2 | PJEF_K_2015 | 3 | PWAR_K_2014 | 6 |
| PANG_K_2016 | 1 | PHEU_K_2015 | 2 | PELA_K_2014 | 3 | PR2 | 6 |
| PPAL_K_2014 | 2 | PMEG_K_2015 | 3 | ||||
| PCUN_K_2016 | 2 | PVRS_K_2014 | 3 | PVUL_K_2014 | 7 | ||
| PCUH_K_2016 | 2 | PR3 | 3 | PR1 | 7 | ||
| PAUS_K_2014 | 2 | PR4 | 3 | ||||
| PMEA_K_2014 | 2 | PMAG_B_2013 | 3 | PGRA_K_2014 | 8 | ||
| PPAU_K_2014 | 2 | PGRA_K_2016 | 8 |
| Taxon | n | PSI | PSII | SSI | Taxon (Continuation) | n | PSI | PSII | SSI | Taxon (Continuation) | n | PSI | PSII | SSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| genus Primula L., sg. Aleuritia | genus Primula L., sg. Auganthus | genus Androsace L. | ||||||||||||
| sct. Aleuritia, ssct. Aleuritia | 3 | sct. Bullatae | 2 | sct. Aizodium | 1 | |||||||||
| sct. Aleuritia, ssct. Algida | 2 | sct. Cortusoides, ssct. Cortusoides | 3 | sct. Andraspis | 1 | |||||||||
| sct. Armerina | 5 | sct. Cortusoides, ssct. Geraniifolia | 4 | sct. Aretia, ssct. Aretia | 3 | |||||||||
| sct. Capitatae | 2 | sct. Obconicolisteri | 1 | sct. Aretia, ssct. Dicranothrix | 3 | |||||||||
| sct. Crystalophlomis | 1 | sct. Reinii | 1 | sct. Chamaejasme, ssct. Hookerianae | 1 | |||||||||
| sct. Crystalophlomis, ssct. Crystalophlomis | 7 | genus Primula L., sg. Auriculastrum | sct. Chamaejasme, ssct. Mucronifoliae | 3 | ||||||||||
| sct. Crystalophlomis, ssct. Maximowiczii | 3 | sct. Amethystina | 1 | sct. Chamaejasme, ssct. Strigillosae | 2 | |||||||||
| sct. Davidii | 2 | sct. Auricula, ssct. Arthritica | 2 | sct. Chamaejasme, ssct. Sublanatae | 2 | |||||||||
| sct. Denticulata | 3 | sct. Auricula, ssct. Auricula | 1 | sct. Chamaejasme, ssct. Villosae | ||||||||||
| sct. Minutissimae | 1 | sct. Auricula, ssct. Brevibracteatum | 3 | series Chamaejasmoidae | 3 | |||||||||
| sct. Muscarioides | 3 | sct. Auricula, ssct. Chamaecallis | 2 | series Euvillosae | 3 | |||||||||
| sct. Oreophlomis | 4 | sct. Auricula, ssct. Cyanaster | 1 | sct. Douglasia | 1 | |||||||||
| sct. Petiolares, ssct. Edgeworthii | 2 | sct. Auricula, ssct. Erythrodosum | 2 | sct. Pseudoprimula | 1 | |||||||||
| sct. Petiolares, ssct. Griffithii | 2 | sct. Auricula, ssct. Rhopsidium | 3 | |||||||||||
| sct. Petiolares, ssct. Petiolares | 1 | sct. Auricula, hybrids | 2 | genus Cortusa L. | 6 | |||||||||
| sct. Petiolares, ssct. Sonchifolia | 1 | sct. Cuneifolia | 2 | genus Dionysia Fenzl. | 2 | |||||||||
| sct. Proliferae | 5 | sct. Dodecatheon | 8 | genus Hottonia L. | 2 | |||||||||
| sct. Pulchella | 3 | sct. Parryi | 3 | genus Lysimachia L. | 2 | |||||||||
| sct. Sikkimensis | 6 | genus Primula L., sg. Primula | genus Omphalogramma (Franch.) Franch. | 2 | ||||||||||
| sct. Soldanelloides | 2 | sct. Primula | 6 | genus Soldanella L. | 6 | |||||||||
| sct. Yunnanensis | 2 | sct. Sredinskya | 1 | genus Vitaliana Sesl. | 3 | |||||||||
| genus Primula, sg. Sphondyllia | 2 | sct. Primula, hybrids | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Włodarczyk, M.; Pasikowski, P.; Osiewała, K.; Frankiewicz, A.; Dryś, A.; Gleńsk, M. In Search of High-Yielding and Single-Compound-Yielding Plants: New Sources of Pharmaceutically Important Saponins from the Primulaceae Family. Biomolecules 2020, 10, 376. https://doi.org/10.3390/biom10030376
Włodarczyk M, Pasikowski P, Osiewała K, Frankiewicz A, Dryś A, Gleńsk M. In Search of High-Yielding and Single-Compound-Yielding Plants: New Sources of Pharmaceutically Important Saponins from the Primulaceae Family. Biomolecules. 2020; 10(3):376. https://doi.org/10.3390/biom10030376
Chicago/Turabian StyleWłodarczyk, Maciej, Paweł Pasikowski, Kinga Osiewała, Aleksandra Frankiewicz, Andrzej Dryś, and Michał Gleńsk. 2020. "In Search of High-Yielding and Single-Compound-Yielding Plants: New Sources of Pharmaceutically Important Saponins from the Primulaceae Family" Biomolecules 10, no. 3: 376. https://doi.org/10.3390/biom10030376
APA StyleWłodarczyk, M., Pasikowski, P., Osiewała, K., Frankiewicz, A., Dryś, A., & Gleńsk, M. (2020). In Search of High-Yielding and Single-Compound-Yielding Plants: New Sources of Pharmaceutically Important Saponins from the Primulaceae Family. Biomolecules, 10(3), 376. https://doi.org/10.3390/biom10030376