Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease
Abstract
1. Introduction
2. Noncoding RNA Regulatory Network in DA-Neurons Development
2.1. miRNA Regulation of DA Neurons
2.2. Functional Roles of Long Noncoding RNAs in DA Neurons Development
3. Noncoding RNA Regulatory Network in DA Neuron Physiology
3.1. miRNA Regulation of DA Signaling
3.2. LncRNA Regulation of DA signaling
4. Noncoding RNAs Regulatory Network in Neurological Diseases
4.1. miRNA Regulation in Parkinson’s Disease
4.2. Long Noncoding RNA Regulation in Parkinson’s Disease
4.3. Circular RNAs as Parkinson’s Disease Biomarkers
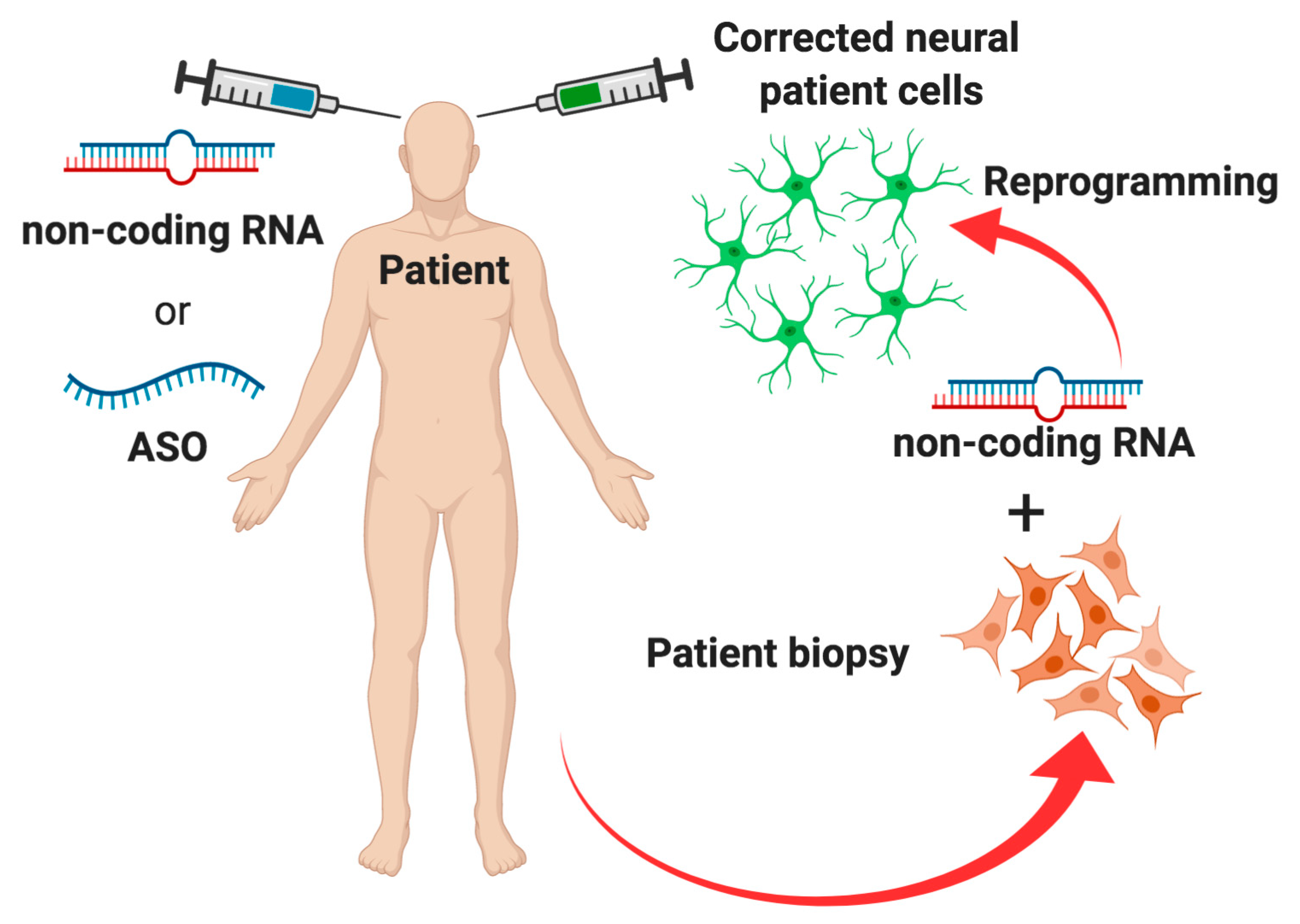
5. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Beaulieu, M.; Espinoza, S.; Gainetdinov, R.R. Dopamine receptors—IUPHAR Review 13. Br. J. Pharmacol. 2014, 172, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Barajas, C.; Coronel, I.; Florán, B. Dopamine Receptors and Neurodegeneration. Aging Dis. 2015, 6, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Dauer, W.; Przedborski, S. Parkinson’s Disease. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef]
- McNeill, E.M.; Van Vactor, D. MicroRNAs shape the neuronal landscape. Neuron 2012, 75, 363–379. [Google Scholar] [CrossRef]
- Cao, D.-D.; Li, L.; Chan, W.-Y. MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases. Int. J. Mol. Sci. 2016, 17, 842. [Google Scholar] [CrossRef]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 2007, 317, 1220–1224. [Google Scholar] [CrossRef]
- Aprea, J.; Lesche, M.; Massalini, S.; Prenninger, S.; Alexopoulou, D.; Dahl, A.; Hiller, M.; Calegari, F. Identification and expression patterns of novel long non-coding RNAs in neural progenitors of the developing mammalian cortex. Neurogenesis 2015, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martín, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.-F.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef]
- Liu, C.; Teng, Z.-Q.; Santistevan, N.J.; Szulwach, K.E.; Guo, W.; Jin, P.; Zhao, X. Epigenetic Regulation of miR-184 by MBD1 Governs Neural Stem Cell Proliferation and Differentiation. Cell Stem Cell 2010, 6, 433–444. [Google Scholar] [CrossRef]
- Cheng, L.-C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Boil. 2009, 16, 365–371. [Google Scholar] [CrossRef]
- Stappert, L.; Borghese, L.; Roese-Koerner, B.; Weinhold, S.; Koch, P.; Terstegge, S.; Uhrberg, M.; Wernet, P.; Brüstle, O. MicroRNA-Based Promotion of Human Neuronal Differentiation and Subtype Specification. PLoS ONE 2013, 8, e59011. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature 2008, 10, 987–993. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, R.; Pulcrano, S.; De Sanctis, C.; Volpicelli, F.; Guatteo, E.; Von Oerthel, L.; Latagliata, E.C.; Esposito, R.; Piscitelli, R.M.; Perrone-Capano, C.; et al. miR-34b/c Regulates Wnt1 and Enhances Mesencephalic Dopaminergic Neuron Differentiation. Stem Cell Rep. 2018, 10, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Anderegg, A.; Lin, H.P.; Chen, J.A.; Caronia-Brown, G.; Cherepanova, N.; Yun, B.; Joksimovic, M.; Rock, J.; Harfe, B.D.; Johnson, R.; et al. An Lmx1b-miR135a2 Regulatory Circuit Modulates Wnt1/Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool. PLoS Genet. 2013, 9, e1003973. [Google Scholar] [CrossRef] [PubMed]
- Hébert, S.S.; De Strooper, B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009, 32, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J.; Chen, S.; Le, W. The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog. Neurobiol. 2005, 77, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Cardenas, O.; Quintana-Hau, J.D.; Le, W.-D.; Smidt, M.P.; Cox, J.J.; De Mayo, F.; Burbach, J.P.H.; Conneely, O.M. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. USA 1998, 95, 4013–4018. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Boil. 2010, 189, 127–141. [Google Scholar] [CrossRef]
- Sun, G.; Ye, P.; Murai, K.; Lang, M.-F.; Li, S.; Zhang, H.; Li, W.; Fu, C.; Yin, J.; Wang, A.; et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef]
- Magill, S.T.; Cambronne, X.A.; Luikart, B.W.; Lioy, D.T.; Leighton, B.H.; Westbrook, G.L.; Mandel, G.; Goodman, R.H. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 20382–20387. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef]
- Tobón, K.E.; Chang, D.; Kuzhikandathil, E.V. MicroRNA 142-3p Mediates Post-Transcriptional Regulation of D1 Dopamine Receptor Expression. PLoS ONE 2012, 7, e49288. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, F.; Xia, X.; Wang, B.; Yao, J. MicroRNA-15a, microRNA-15b and microRNA-16 inhibit the human dopamine D1 receptor expression in four cell lines by targeting 3′UTR −12 bp to +154 bp. Artif. Cells Nanomed. Biotechnol. 2020, 48, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Leites, C.; He, D.; Schwartz, D.; Moy, W.; Shi, J.; Duan, J. MicroRNA-9 and MicroRNA-326 Regulate Human Dopamine D2 Receptor Expression, and the MicroRNA-mediated Expression Regulation Is Altered by a Genetic Variant. J. Boil. Chem. 2014, 289, 13434–13444. [Google Scholar] [CrossRef] [PubMed]
- Doxakis, E. Post-transcriptional Regulation of α-Synuclein Expression by mir-7 and mir-153. J. Boil. Chem. 2010, 285, 12726–12734. [Google Scholar] [CrossRef]
- Fragkouli, A.; Doxakis, E. miR-7 and miR-153 protect neurons against MPP+-induced cell death via upregulation of mTOR pathway. Front. Cell. Neurosci. 2014, 8, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A. Gene Co-Expression Network Analysis Implicates microRNA Processing in Parkinson’s Disease Pathogenesis. Neurodegener. Dis. 2018, 18, 191–199. [Google Scholar] [CrossRef]
- McMillan, K.J.; Murray, T.K.; Bengoa-Vergniory, N.; Cordero-Llana, O.; Cooper, J.; Buckley, A.; Wade-Martins, R.; Uney, J.B.; O’Neill, M.J.; Wong, L.F.; et al. Loss of MicroRNA-7 Regulation Leads to α-Synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo. Mol. Ther. 2017, 25, 2404–2414. [Google Scholar] [CrossRef]
- Wang, G.; Van Der Walt, J.M.; Mayhew, G.; Li, Y.-J.; Züchner, S.; Scott, W.K.; Martin, E.R.; Vance, J. Variation in the miRNA-433 Binding Site of FGF20 Confers Risk for Parkinson Disease by Overexpression of α-Synuclein. Am. J. Hum. Genet. 2008, 82, 283–289. [Google Scholar] [CrossRef]
- Cardo, L.F.; Coto, E.; Ribacoba, R.; Menéndez, M.; Morís, G.; Suárez, E.; Alvarez, V. MiRNA Profile in the Substantia Nigra of Parkinson’s Disease and Healthy Subjects. J. Mol. Neurosci. 2014, 54, 830–836. [Google Scholar] [CrossRef]
- Soreq, L.; Salomonis, N.; Bronstein, M.; Greenberg, D.S.; Israel, Z.; Bergman, H.; Soreq, H. Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front. Mol. Neurosci. 2013, 6, 10–30. [Google Scholar] [CrossRef]
- Gong, X.; Wang, H.; Ye, Y.; Shu, Y.; Deng, Y.; He, X.; Lu, G.; Zhang, S. miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am. J. Transl. Res. 2016, 8, 2127–2137. [Google Scholar] [PubMed]
- Gehrke, S.; Imai, Y.; Sokol, N.; Lu, B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 2010, 466, 637–641. [Google Scholar] [CrossRef]
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2012, 22, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The Long Noncoding RNA RMST Interacts with SOX2 to Regulate Neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.D.; Andersen, R.E.; Liu, S.J.; Nowakowski, T.J.; Hong, S.J.; Gertz, C.C.; Salinas, R.D.; Zarabi, H.; Kriegstein, A.R.; Lim, D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015, 16, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Chang, K.-Y.; Li, Z.; Gates, K.; Rana, Z.A.; Dang, J.; Zhang, D.; Han, T.; Yang, C.-S.; Cunningham, T.J.; et al. An Evolutionarily Conserved Long Noncoding RNA TUNA Controls Pluripotency and Neural Lineage Commitment. Mol. Cell 2014, 53, 1067. [Google Scholar] [CrossRef]
- Tochitani, S.; Hayashizaki, Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 2008, 372, 691–696. [Google Scholar] [CrossRef]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.E.; Dinger, M.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef]
- Ip, J.Y.; Sone, M.; Nashiki, C.; Pan, Q.; Kitaichi, K.; Yanaka, K.; Abe, T.; Takao, K.; Miyakawa, T.; Blencowe, B.J.; et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016, 6, 27204–27218. [Google Scholar] [CrossRef]
- Zhong, J.; Chuang, S.-C.; Bianchi, R.; Zhao, W.; Lee, H.; Fenton, A.A.; Wong, R.K.S.; Tiedge, H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009, 29, 9977–9986. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, X.; Niu, W.; Yao, G.; Kong, L.; He, M.; Chen, C.; Lu, Z.; Cui, X.; Zhang, L. Overexpression Regulatory Role of lncRNA NONHSAT089447 in the Dopamine Signaling Pathway in Schizophrenic Patients. Med. Sci. Monit. 2019, 25, 4322–4332. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yu, J.; Zhao, J.; Zhou, Y.; Xiong, N.; Xu, J.; Wang, T.; Bell, R.L.; Qing, H.; Lin, Z. AZI23′UTR Is a New SLC6A3 Downregulator Associated with an Epistatic Protection Against Substance Use Disorders. Mol. Neurobiol. 2017, 55, 5611–5622. [Google Scholar] [CrossRef] [PubMed]
- Kraus, T.F.J.; Haider, M.; Spanner, J.; Steinmaurer, M.; Dietinger, V.; Kretzschmar, H.A. Altered Long Noncoding RNA Expression Precedes the Course of Parkinson’s Disease—A Preliminary Report. Mol. Neurobiol. 2016, 54, 2869–2877. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Carrieri, C.; Forrest, A.R.; Santoro, C.; Persichetti, F.; Carninci, P.; Zucchelli, S.; Gustincich, S. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 2015, 9, 114–125. [Google Scholar] [CrossRef]
- Zucchelli, S.; Cotella, D.; Takahashi, H.; Carrieri, C.; Cimatti, L.; Fasolo, F.; Jones, M.H.; Sblattero, D.; Sanges, R.; Santoro, C.; et al. SINEUPs: A new class of natural and synthetic antisense long non-coding RNAs that activate translation. RNA Boil. 2015, 12, 771–779. [Google Scholar] [CrossRef]
- Cai, L.; Tu, L.; Li, T.; Yang, X.; Ren, Y.; Gu, R.; Zhang, Q.; Yao, H.; Qu, X.; Wang, Q.; et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int. Immunopharmacol. 2019, 75, 105734–105745. [Google Scholar] [CrossRef]
- Lin, Q.; Hou, S.; Dai, Y.; Jiang, N.; Lin, Y. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Boil. Chem. 2019, 400, 1217–1228. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Lu, Y.; Gong, Z.; Jin, X.; Zhao, P.; Zhang, Y.; Wang, Z. LncRNA MALAT1 targeting miR-124-3p regulates DAPK1 expression contributes to cell apoptosis in Parkinson’s Disease. J. Cell. Biochem. 2020, 1–11. [Google Scholar] [CrossRef]
- Saglam, A.S.Y.; Alp, E.; Onen, H.I. Circular RNAs and Its Biological Functions in Health and Disease. Gene Expr. Phenotypic Trait. 2020, 1–37. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Guo, S.; Li, W.; Yu, P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015, 5, 12453–12465. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Liu, X.; Wang, L.; Qi, L.; Sun, W.; Wang, W.; Sun, Y.; Zhang, H. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 2018, 10, 1281–1293. [Google Scholar] [CrossRef]
- Arenas, E.; Denham, M.; Villaescusa, J.C. How to make a midbrain dopaminergic neuron. Development 2015, 142, 1918–1936. [Google Scholar] [CrossRef]
- Choe, Y.; Pleasure, S.J.; Mira, H. Control of Adult Neurogenesis by Short-Range Morphogenic-Signaling Molecules. Cold Spring Harb. Perspect. Boil. 2015, 8, 1–18. [Google Scholar] [CrossRef]
- Seiradake, E.; Jones, E.Y.; Klein, R. Structural Perspectives on Axon Guidance. Annu. Rev. Cell Dev. Boil. 2016, 32, 577–608. [Google Scholar] [CrossRef]
- Alavian, K.N.; Scholz, C.; Simon, H.H. Transcriptional regulation of mesencephalic dopaminergic neurons: The full circle of life and death. Mov. Disord. 2008, 23, 319–328. [Google Scholar] [CrossRef]
- Chakrabarty, K.; Von Oerthel, L.; Hellemons, A.; Clotman, F.; Espana, A.; Koerkamp, M.G.; Holstege, F.C.P.; Pasterkamp, R.J.; Smidt, M.P. Genome wide expression profiling of the mesodiencephalic region identifies novel factors involved in early and late dopaminergic development. Boil. Open 2012, 1, 693–704. [Google Scholar] [CrossRef][Green Version]
- Vernay, B.; Koch, M.; Vaccarino, F.M.; Briscoe, J.; Simeone, A.; Kageyama, R.; Ang, S.-L. Otx2 Regulates Subtype Specification and Neurogenesis in the Midbrain. J. Neurosci. 2005, 25, 4856–4867. [Google Scholar] [CrossRef] [PubMed]
- Abeliovich, A.; Hammond, R. Midbrain dopamine neuron differentiation: Factors and fates. Dev. Boil. 2007, 304, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. MicroRNAs as Regulators of Differentiation and Cell Fate Decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Liu, Y.; Huang, M.; Zhao, X.; Cheng, L. Wnt1-cre-mediated Conditional Loss of Dicer Results in Malformation of the Midbrain and Cerebellum and Failure of Neural Crest and Dopaminergic Differentiation in Mice. J. Mol. Cell Boil. 2010, 2, 152–163. [Google Scholar] [CrossRef]
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, T.P.; Erfle, H.; Sonntag, K.C.; Schütz, G.; Vinnikov, I.A.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Tonelli, D.D.P.; Calegari, F.; Fei, J.-F.; Nomura, T.; Osumi, N.; Heisenberg, C.-P.; Huttner, W.B. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotechnology 2006, 41, 727–732. [Google Scholar] [CrossRef]
- Tonelli, D.D.P.; Pulvers, J.N.; Haffner, C.; Murchison, E.P.; Hannon, G.J.; Huttner, W.B. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 2008, 135, 3911–3921. [Google Scholar] [CrossRef]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef]
- Andersson, E.; Tryggvason, U.; Deng, Q.; Friling, S.; Alekseenko, Z.; Robert, B.; Perlmann, T.; Ericson, J. Identification of Intrinsic Determinants of Midbrain Dopamine Neurons. Cell 2006, 124, 393–405. [Google Scholar] [CrossRef]
- Smidt, M.P.; Asbreuk, C.H.J.; Cox, J.J.; Chen, H.; Johnson, R.L.; Burbach, J.P.H. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat. Neurosci. 2000, 3, 337–341. [Google Scholar] [CrossRef]
- Ferri, A.L.M.; Lin, W.; Mavromatakis, Y.E.; Wang, J.C.; Sasaki, H.; Whitsett, J.A.; Ang, S.-L. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 2007, 134, 2761–2769. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.H.; Levesque, M.; Claxton, S.; Johnson, R.L.; Ang, S.-L. Lmx1a and Lmx1b Function Cooperatively to Regulate Proliferation, Specification, and Differentiation of Midbrain Dopaminergic Progenitors. J. Neurosci. 2011, 31, 12413–12425. [Google Scholar] [CrossRef] [PubMed]
- Caronia-Brown, G.; Anderegg, A.; Awatramani, R. Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 2016, 11, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, T.; Wang, Y.; Cui, H.; Tang, Y.; Zhang, X.; Chen, D.; Shen, N.; Le, W. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J. Cell Sci. 2012, 125, 1673–1682. [Google Scholar] [CrossRef]
- Baek, S.; Choi, H.; Kim, J. Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 2014, 1587, 23–32. [Google Scholar] [CrossRef]
- Hébert, S.S.; De Strooper, B. MOLECULAR BIOLOGY: miRNAs in Neurodegeneration. Science 2007, 317, 1179–1180. [Google Scholar] [CrossRef]
- Yoshimura, A.; Numakawa, T.; Odaka, H.; Adachi, N.; Tamai, Y.; Kunugi, H. Negative regulation of microRNA-132 in expression of synaptic proteins in neuronal differentiation of embryonic neural stem cells. Neurochem. Int. 2016, 97, 26–33. [Google Scholar] [CrossRef]
- Qian, Y.; Song, J.; Ouyang, Y.; Han, Q.; Chen, W.; Zhao, X.; Xie, Y.; Chen, Y.; Yuan, W.; Fan, C. Advances in Roles of miR-132 in the Nervous System. Front. Pharmacol. 2017, 8, 770–779. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Boil. 2004, 5, 13–24. [Google Scholar] [CrossRef]
- Ferretti, E.; De Smaele, E.; Miele, E.; Laneve, P.; Pò, A.; Pelloni, M.; Paganelli, A.; Di Marcotullio, L.; Caffarelli, E.; Screpanti, I.; et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008, 27, 2616–2627. [Google Scholar] [CrossRef]
- Jia, X.; Wang, F.; Han, Y.; Geng, X.; Li, M.; Shi, Y.; Lu, L.; Chen, Y. miR-137 and miR-491 Negatively Regulate Dopamine Transporter Expression and Function in Neural Cells. Neurosci. Bull. 2016, 32, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Smrt, R.D.; Szulwach, K.E.; Pfeiffer, R.L.; Li, X.; Guo, W.; Pathania, M.; Teng, Z.-Q.; Luo, Y.; Peng, J.; Bordey, A.; et al. MicroRNA miR-137 Regulates Neuronal Maturation by Targeting Ubiquitin Ligase Mind Bomb-1. Stem Cells 2010, 28, 1060–1070. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and Transcriptional Co-Localization of Protein-Coding and Long Non-Coding RNA Pairs in the Developing Brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Uhde, C.W.; Vives, J.; Jaeger, I.; Li, M. Rmst Is a Novel Marker for the Mouse Ventral Mesencephalic Floor Plate and the Anterior Dorsal Midline Cells. PLoS ONE 2010, 5, e8641. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef]
- Bannon, M.J.; Savonen, C.; Jia, H.; Dachet, F.; Halter, S.D.; Schmidt, C.J.; Lipovich, L.; Kapatos, G. Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. J. Neurochem. 2015, 135, 50–59. [Google Scholar] [CrossRef]
- Bian, S.; Sun, T. Functions of Noncoding RNAs in Neural Development and Neurological Diseases. Mol. Neurobiol. 2011, 44, 359–373. [Google Scholar] [CrossRef]
- Kaasinen, V.; Vilkman, H.; Hietala, J.; Någren, K.; Helenius, H.; Olsson, H.; Farde, L.; Rinne, J.O. Age-related dopamine D2/D3 receptor loss in extrastriatal regions of the human brain. Neurobiol. Aging 2000, 21, 683–688. [Google Scholar] [CrossRef]
- Gerfen, C.R.; Miyachi, S.; Paletzki, R.; Brown, P. D1 Dopamine Receptor Supersensitivity in the Dopamine-Depleted Striatum Results from a Switch in the Regulation of ERK1/2/MAP Kinase. J. Neurosci. 2002, 22, 5042–5054. [Google Scholar] [CrossRef]
- Hisahara, S.; Shimohama, S. Dopamine Receptors and Parkinson’s Disease. Int. J. Med. Chem. 2011, 1–6. [Google Scholar] [CrossRef]
- Berke, J.D.; Hyman, S.E. Addiction, Dopamine, and the Molecular Mechanisms of Memory. Neuron 2000, 25, 515–532. [Google Scholar] [CrossRef]
- Missale, C.; Nash, S.R.; Robinson, S.W.; Jaber, M.; Caron, M.G. Dopamine receptors: From structure to function. Physiol. Rev. 1998, 78, 189–225. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Liang, X.; Liu, L.; Zhang, D.; Wan, C.; Gan, Z.; Yuan, L. High Throughput Sequencing Identifies MicroRNAs Mediating α-Synuclein Toxicity by Targeting Neuroactive-Ligand Receptor Interaction Pathway in Early Stage of Drosophila Parkinson’s Disease Model. PLoS ONE 2015, 10, e0137432. [Google Scholar] [CrossRef] [PubMed]
- Howes, O.D.; Kambeitz, J.; Kim, E.; Stahl, D.; Slifstein, M.; Abi-Dargham, A.; Kapur, S. The Nature of Dopamine Dysfunction in Schizophrenia and What This Means for Treatment. Arch. Gen. Psychiatry 2012, 69, 776–786. [Google Scholar] [CrossRef]
- Gitaí, D.L.G.; Fachin, A.L.; Mello, S.S.; Elias, C.F.; Bittencourt, J.C.; Leite, J.P.; Passos, G.A.; Garcia-Cairasco, N.; Paçó-Larson, M.L. The non-coding RNA BC1 is down-regulated in the hippocampus of Wistar Audiogenic Rat (WAR) strain after audiogenic kindling. Brain Res. 2011, 1367, 114–121. [Google Scholar] [CrossRef]
- Chen, S.; Sun, X.; Niu, W.; Kong, L.; He, M.; Li, W.; Zhong, A.; Lu, J.; Zhang, L. Aberrant Expression of Long Non-Coding RNAs in Schizophrenia Patients. Med. Sci. Monit. 2016, 22, 3340–3351. [Google Scholar] [CrossRef]
- Eom, T.-Y.; Han, S.B.; Kim, J.; Blundon, J.A.; Wang, Y.-D.; Yu, J.; Anderson, K.; Kaminski, D.B.; Sakurada, S.M.; Pruett-Miller, S.M.; et al. Schizophrenia-related microdeletion causes defective ciliary motility and brain ventricle enlargement via microRNA-dependent mechanisms in mice. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Carrick, W.T.; Burks, B.; Cairns, M.J.; Kocerha, J. Noncoding RNA Regulation of Dopamine Signaling in Diseases of the Central Nervous System. Front. Mol. Biosci. 2016, 3, 69–77. [Google Scholar] [CrossRef]
- Doura, M.B.; Unterwald, E.M. MicroRNAs Modulate Interactions between Stress and Risk for Cocaine Addiction. Front. Cell. Neurosci. 2016, 10, 125–135. [Google Scholar] [CrossRef]
- Higuchi, Y.; Soga, T.; Parhar, I.S. Potential Roles of microRNAs in the Regulation of Monoamine Oxidase A in the Brain. Front. Mol. Neurosci. 2018, 11, 339–348. [Google Scholar] [CrossRef]
- Junn, E.; Lee, K.W.; Jeong, B.S.; Chan, T.W.; Im, J.Y.; Mouradian, M.M. Repression of α-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, L. α-Synuclein in Parkinson’s Disease. Cold Spring Harb. Perspect. Med. 2011, 2, 1–24. [Google Scholar] [CrossRef]
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R.; et al. A Parkinson’s disease Circ RNA s Resource reveals a link between circ SLC 8A1 and oxidative stress. EMBO Mol. Med. 2020, 1–19. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541. [Google Scholar] [CrossRef]
- Barbash, S.; Shifman, S.; Soreq, H. Global coevolution of human microRNAs and their target genes. Mol. Boil. Evol. 2014, 31, 1237–1247. [Google Scholar] [CrossRef]
- Simchovitz, A.; Soreq, L.; Soreq, H. Transcriptome profiling in Parkinson’s leukocytes: From early diagnostics to neuroimmune therapeutic prospects. Curr. Opin. Pharmacol. 2016, 26, 102–109. [Google Scholar] [CrossRef]
- Singh, A.; Sen, D. MicroRNAs in Parkinson’s disease. Exp. Brain Res. 2017, 235, 2359–2374. [Google Scholar] [CrossRef]
- Soreq, L.; Guffanti, A.; Salomonis, N.; Simchovitz, A.; Israel, Z.; Bergman, H.; Soreq, H. Long Non-Coding RNA and Alternative Splicing Modulations in Parkinson’s Leukocytes Identified by RNA Sequencing. PLoS Comput. Boil. 2014, 10, e1003517. [Google Scholar] [CrossRef]
- Leggio, L.; Vivarelli, S.; L’Episcopo, F.; Tirolo, C.; Caniglia, S.; Testa, N.; Marchetti, B.; Iraci, N. microRNAs in Parkinson’s Disease: From Pathogenesis to Novel Diagnostic and Therapeutic Approaches. Int. J. Mol. Sci. 2017, 18, 2698. [Google Scholar] [CrossRef]
- Cardo, L.F.; Coto, E.; De Mena, L.; Ribacoba, R.; Morís, G.; Menéndez, M.; Alvarez, V. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J. Neurol. 2013, 260, 1420–1422. [Google Scholar] [CrossRef]
- Bengoa-Vergniory, N.; Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Alpha-synuclein oligomers: A new hope. Acta Neuropathol. 2017, 134, 819–838. [Google Scholar] [CrossRef] [PubMed]
- Dehay, B.; Bourdenx, M.; Gorry, P.; Przedborski, S.; Vila, M.; Hunot, S.; Singleton, A.; Olanow, C.W.; Merchant, K.M.; Bezard, E.; et al. Targeting α-synuclein for treatment of Parkinson’s disease: Mechanistic and therapeutic considerations. Lancet Neurol. 2015, 14, 855–866. [Google Scholar] [CrossRef]
- Espinoza, S.; Scarpato, M.; Damiani, D.; Managò, F.; Mereu, M.; Contestabile, A.; Peruzzo, O.; Carninci, P.; Santoro, C.; Papaleo, F.; et al. SINEUP Non-coding RNA Targeting GDNF Rescues Motor Deficits and Neurodegeneration in a Mouse Model of Parkinson’s Disease. Mol. Ther. 2020, 28, 642–652. [Google Scholar] [CrossRef]
- Dang, T.T.P. Ubiquitin Carboxyl-Terminal Hydrolase L1 in Parkinson’s Disease. Ubiquitin Proteasome Syst. Curr. Insights Mech. Cell. Regul. Dis. 2019. [Google Scholar] [CrossRef]
- Lyu, Y.; Bai, L.; Qin, C. Long noncoding RNAs in neurodevelopment and Parkinson’s disease. Anim. Model. Exp. Med. 2019, 2, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.B.; Ordóñez, A.J.L.; Fdez, E.; Madero-Pérez, J.; Gonnelli, A.; Drouyer, M.; Chartier-Harlin, M.C.; Taymans, J.-M.; Bubacco, L.; Greggio, E.; et al. GTP binding regulates cellular localization of Parkinson’s disease-associated LRRK2. Hum. Mol. Genet. 2017, 26, 2747–2767. [Google Scholar] [CrossRef]
- Sanchez, Y.; Huarte, M. Long Non-Coding RNAs: Challenges for Diagnosis and Therapies. Nucleic Acid Ther. 2013, 23, 15–20. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Reference service: A thing of the past? Ref. Libr. 1961, 115–123. [Google Scholar] [CrossRef]
- Sharpless, W.R.J.; Norman, E. Legal Education in the Soviet Union and Eastern Europe. Nat. Biotechnol. 2015, 5, 738–749. [Google Scholar]
- Stoll, L.; Sobel, J.A.; Rodríguez-Trejo, A.; Guay, C.; Lee, K.; Venø, M.T.; Kjems, J.; Laybutt, D.R.; Regazzi, R. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol. Metab. 2018, 9, 69–83. [Google Scholar] [CrossRef]
- Kumar, L.; Haque, R.; Baghel, T.; Nazir, A. Circular RNAs: The Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol. Neurobiol. 2016, 54, 7224–7234. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jia, Y.-J.; Wang, Q.; Shao, X.-Q.; Lv, R. Circular RNA: A new star in neurological diseases. Int. J. Neurosci. 2016, 127, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Maloney, L.T.; Hempel de Ibarra, N. Blackawton bees: Commentary on Blackawton, PS et al. Biol. Lett. 2011, 7, 166–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Efe, J.A.; Zhu, S.; Talantova, M.; Yuan, X.; Wang, S.; Lipton, S.A.; Zhang, K.; Ding, S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA 2011, 108, 7838–7843. [Google Scholar] [CrossRef]
- Liu, X.; Li, F.; Stubblefield, E.A.; Blanchard, B.; Richards, T.L.; Larson, G.A.; He, Y.; Huang, Q.; Tan, A.C.; Zhang, D.; et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2011, 22, 321–332. [Google Scholar] [CrossRef]
- Dell’Anno, M.T.; Caiazzo, M.; Leo, D.; Dvoretskova, E.; Medrihan, L.; Colasante, G.; Giannelli, S.; Theka, I.; Russo, G.; Mus, L.; et al. Remote control of induced dopaminergic neurons in parkinsonian rats. J. Clin. Investig. 2014, 124, 3215–3229. [Google Scholar] [CrossRef]
- Cervo, P.R.D.V.; Romanov, R.A.; Spigolon, G.; Masini, D.; Martín-Montañez, E.; Toledo, E.M.; La Manno, G.; Feyder, M.; Pifl, C.; Ng, Y.-H.; et al. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 2017, 35, 444–452. [Google Scholar] [CrossRef]
- Yu, X.; Nagai, J.; Khakh, B.S. Improved tools to study astrocytes. Nat. Rev. Neurosci. 2020, 21, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Qian, H.; Hu, J.; Zhou, B.; Zhou, Y.; Hu, X.; Karakhanyan, A.; Pang, Z.P.; Fu, X.-D. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 2016, 19, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Kang, X.; Hu, J.; Zhang, D.; Liang, Z.; Meng, F.; Zhang, X.; Xue, Y.; Maimon, R.; Dowdy, S.F.; et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 2020, 582, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef]
- Zhou, H.; Su, J.; Hu, X.; Zhou, C.; Li, H.; Chen, Z.; Xiao, Q.; Wang, B.; Wu, W.; Sun, Y.; et al. Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell 2020, 181, 590–603. [Google Scholar] [CrossRef]
- Uehara, T.; Choong, C.-J.; Nakamori, M.; Hayakawa, H.; Nishiyama, K.; Kasahara, Y.; Baba, K.; Nagata, T.; Yokota, T.; Tsuda, H.; et al. Amido-bridged nucleic acid (AmNA)-modified antisense oligonucleotides targeting α-synuclein as a novel therapy for Parkinson’s disease. Sci. Rep. 2019, 9, 7567–7580. [Google Scholar] [CrossRef]
- Bennett, C.F.; Krainer, A.R.; Cleveland, D.W. Antisense Oligonucleotide Therapies for Neurodegenerative Diseases. Annu. Rev. Neurosci. 2019, 42, 385–406. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H.; et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef]
- Rodrigues, F.B.; Wild, E.J. Huntington’s Disease Clinical Trials Corner: February 2018. J. Huntingt. Dis. 2018, 7, 89–98. [Google Scholar] [CrossRef]
- Man, J.H.; Groenink, L.; Caiazzo, M. Cell reprogramming approaches in gene- and cell-based therapies for Parkinson’s disease. J. Control. Release 2018, 286, 114–124. [Google Scholar] [CrossRef]


| miRNA Subgroups | Name | Function | References |
|---|---|---|---|
| miRNAs in development | miR-let-7b | Regulates neural stem cell (NSC) proliferation and differentiation | [16] |
| miR-184 | Binds Numbl transcript | [17] | |
| miR-124 | Suppresses Sox9 expression, promotes differentiation of NPs | [18] | |
| miR-9 | Inhibits NSC proliferation, promotes differentiation | [19] | |
| miR-125 | Differentiation of neural progenitors | [20,21] | |
| miR-34b/c | Modulates Wnt1 signaling, promotes cell cycle exit, and induces dopaminergic differentiation | [22] | |
| miR135a2 | Modulates Wnt1/Wnt morphogen signaling | [23] | |
| miR-133b | Maturation and function of DA neuron development | [10,24] | |
| miR-132 | Differentiation of DA neurons. | [25,26] | |
| miR-181a | Promotes neuroepithelial-like stem cell switch from self-renewal to neuronal differentiation | [20] | |
| miR-137 | Negatively regulates neuronal maturation of adult NSC proliferation and cell fate determination | [27,28] | |
| miRNAs in physiology (DA signaling network) | miR-132/ miR-212 cluster | Mediates dendritic growth and spine formation | [29] |
| miR-134 | Negatively regulates the size of dendritic spines | [30] | |
| miR-142-3p | Modulates the D1 signaling | [31] | |
| miRNA-15a, miRNA-15b, and miRNA 16 | Inhibit the DRD1 gene expression | [32] | |
| miR-137 | Enhances D2 receptor expression | [33] | |
| miR-326 and miR-9 | Post-transcriptional regulation of DRD2 by both microRNAs | [33] | |
| miRNAs in neurological diseases | miR-7 and miR-153 | Regulate post-transcriptionally α-synuclein | [34,35] |
| miR34b/c, and miR-214 | Bind directly the 3′ UTR of alpha-synuclein. | [36] | |
| miR-7 | Its depletion is related with alpha-synuclein accumulation and with neuron loss | [37] | |
| miR-433 | Causes the overexpression of alpha-synuclein (SNCA) | [38] | |
| miR-331-5p | Upregulated miRNA in Parkinson’s disease (PD) patients | [39] | |
| miR-20a, miR-16, and miR-320 | Specifically altered in PD patients | [40] | |
| miR- 133b | Controlling midbrain DA (mDA) neuron differentiation | [10] | |
| miR-124 | Increases neuronal autophagy and apoptosis | [41] | |
| let-7 and mir-184 | Linked to defects in cell division and cell death. | [42] | |
| miR-205 | Upregulation of LRRK2 protein expression | [43] |
| Long Noncoding RNA Subgroups | Name | Function | References |
|---|---|---|---|
| Long noncoding RNAs in development | RMST | Neuronal differentiation | [44] |
| Pnky | Controls neurogenesis of ventricular–subventricular zone stem cells | [45] | |
| TUNA | Promotes the differentiation of NSCs into glial cells | [46] | |
| NEAT1 | Regulates the NSCs differentiation into oligodendrocytes | [47,48] | |
| Gomafu (known also as MIAT) | Modulates dopaminergic transmission and neurobehavioral phenotypes | [49] | |
| Long noncoding RNAs in physiology (DA signaling network) | BC1 | Regulates the postsynaptic signaling | [50] |
| NONHSAT089447, NONHSAT021545, and NONHSAT041499 | Regulatory role on the DA receptors signaling pathway, upregulated in schizophrenic patients | [51] | |
| AZI23′UTR | Transcriptional regulation of human SLC6A3 (DAT) and a crucial risk factor for substance abuse disorders | [52] | |
| Long noncoding RNAs in neurological diseases | H19, MALAT1, SNHG1, and TncRNA | Are upregulated in PD patients | [53] |
| Uchl1 | Is responsible to remove DNA damage and prevents cell apoptosis | [54] | |
| UCHL1-AS (Antisense transcript of UCHL1) | Promotes translation and expression of UCHL1 which is strongly down regulated in neurochemical models of PD in vitro and in vivo | [55,56] | |
| NEAT1 | Overexpressed in the substantia nigra of PD. Neuroprotective role against drug-induced oxidative stress. | [57] | |
| UCA1 | Inhibits the PI3K/Akt signaling pathway | [57] | |
| HOTAIR | Affects the progression of PD | [58] | |
| MALAT | In PD mice induces apoptosis of DA neurons. | [59,60] | |
| ciRS-7 (CDR1as) | Negatively regulates miR-7 activities. | [61,62,63,64] | |
| circSNCA | Act as a sponge for miR-7 regulating alpha-synuclein expression. | [65] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascale, E.; Divisato, G.; Palladino, R.; Auriemma, M.; Ngalya, E.F.; Caiazzo, M. Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease. Biomolecules 2020, 10, 1269. https://doi.org/10.3390/biom10091269
Pascale E, Divisato G, Palladino R, Auriemma M, Ngalya EF, Caiazzo M. Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease. Biomolecules. 2020; 10(9):1269. https://doi.org/10.3390/biom10091269
Chicago/Turabian StylePascale, Emilia, Giuseppina Divisato, Renata Palladino, Margherita Auriemma, Edward Faustine Ngalya, and Massimiliano Caiazzo. 2020. "Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease" Biomolecules 10, no. 9: 1269. https://doi.org/10.3390/biom10091269
APA StylePascale, E., Divisato, G., Palladino, R., Auriemma, M., Ngalya, E. F., & Caiazzo, M. (2020). Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease. Biomolecules, 10(9), 1269. https://doi.org/10.3390/biom10091269






