In Silico Screening of Natural Products Isolated from Mexican Herbal Medicines against COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Descriptors and Computational Screening
2.2. Docking
2.3. Pharmacokinetic Assessment (PBPK Model Building and Evaluation)
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jahanshahlu, L.; Rezaei, N. Monoclonal antibody as a potential anti-COVID-19. Biomed. Pharmacother. 2020, 129, 110337. [Google Scholar] [CrossRef]
- Chilamakuri, R.; Agarwal, S. COVID-19: Characteristics and Therapeutics. Cells 2021, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Twomey, J.D.; Luo, S.; Dean, A.Q.; Bozza, W.P.; Nalli, A.; Zhang, B. COVID-19 update: The race to therapeutic development. Drug Resist. Updates 2020, 53, 100733. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. Chem. 2011, 46, 4769–4807. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Calixto, J.B. The role of natural products in modern drug discovery. An. Acad. Bras. Cienc. 2019, 91 (Suppl. 3), e20190105. [Google Scholar] [CrossRef] [PubMed]
- Chiang, L.-C.; Ng, L.-T.; Cheng, P.-W.; Chiang, W.; Lin, C.-C. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin. Exp. Pharmacol. Physiol. 2005, 32, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.-W.; Ng, L.-T.; Chiang, L.-C.; Lin, C.-C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006, 33, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Chen, C.; Zhang, H.-Q.; Guo, H.-Y.; Wang, H.; Wang, L.; Zhang, X.; Hua, S.-N.; Yu, J.; Xiao, P.-G.; et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005, 67, 18–23. [Google Scholar] [CrossRef]
- Lin, C.-W.; Tsai, F.-J.; Tsai, C.-H.; Lai, C.-C.; Wan, L.; Ho, T.-Y.; Hsieh, C.-C.; Chao, P.-D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005, 68, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, J.-Y.; Kim, D.; Nguyen, T.T.H.; Park, S.-J.; Chang, J.S.; Park, K.H.; et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010, 18, 7940–7947. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.-S.; Lee, J.; Lee, J.M.; Kim, Y.; Chin, Y.-W.; Jee, J.-G.; Keum, Y.-S.; Jeong, Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012, 22, 4049–4054. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Su, D.; Feng, Y.; Liu, K.; Song, Y. Antiviral herbs—Present and future. Infect. Disord. Drug Targets 2014, 14, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Suwannarach, N.; Kumla, J.; Sujarit, K.; Pattananandecha, T.; Saenjum, C.; Lumyong, S. Natural Bioactive Compounds from Fungi as Potential Candidates for Protease Inhibitors and Immunomodulators to Apply for Coronaviruses. Molecules 2020, 25, 1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.-F.; Xu, L.; Wei, M.-Y.; Wang, C.-Y.; Gu, Y.-C.; Shao, C.-L. Recent progresses in marine microbial-derived antiviral natural products. Arch. Pharm. Res. 2020, 43, 1215–1229. [Google Scholar] [CrossRef]
- Mata, R.; Figueroa, M.; Navarrete, A.; Rivero-Cruz, I. Chemistry and Biology of Selected Mexican Medicinal Plants. Prog. Chem. Org. Nat. Prod. 2019, 108, 1–142. [Google Scholar]
- Rodriguez-Fragoso, L.; Reyes-Esparza, J.; Burchiel, S.W.; Herrera-Ruiz, D.; Torres, E. Risks and benefits of commonly used herbal medicines in Mexico. Toxicol. Appl. Pharmacol. 2008, 227, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdivia-Correa, B.; Gómez-Gutiérrez, C.; Uribe, M.; Méndez-Sánchez, N. Herbal Medicine in Mexico: A Cause of Hepatotoxicity. A Critical Review. Int. J. Mol. Sci. 2016, 17, 235. [Google Scholar] [CrossRef] [Green Version]
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009–2018. JAMA 2020, 323, 844. [Google Scholar] [CrossRef]
- Ou-Yang, S.-S.; Lu, J.-Y.; Kong, X.-Q.; Liang, Z.-J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pech-Puch, D.; Pérez-Povedano, M.; Lenis-Rojas, O.A.; Rodríguez, J.; Jiménez, C. Marine Natural Products from the Yucatan Peninsula. Mar. Drugs 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Cansino, R.; Espitia-Pinzón, C.I.; Campos-Lara, M.G.; Guzmán-Gutiérrez, S.L.; Segura-Salinas, E.; Echeverría-Valencia, G.; Torras-Claveria, L.; Cuevas-Figueroa, X.M.; Reyes-Chilpa, R. Antimycobacterial and HIV-1 Reverse Transcriptase Activity of Julianaceae and Clusiaceae Plant Species from Mexico. Evid. Based. Complement. Alternat. Med. 2015, 2015, 183036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Mares, D.; Rivas-Galindo, V.M.; Salazar-Aranda, R.; Pérez-Lopez, L.A.; Waksman De Torres, N.; Pérez-Meseguer, J.; Torres-Lopez, E. Screening of north-east Mexico medicinal plants with activities against herpes simplex virus and human cancer cell line. Nat. Prod. Res. 2019, 33, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterisation of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Teague, S.J.; Davis, A.M.; Leeson, P.D.; Oprea, T. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed Engl. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Kong, R.; Yang, G.; Xue, R.; Liu, M.; Wang, F.; Hu, J.; Guo, X.; Chang, S. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020, 36, 5109–5111. [Google Scholar] [CrossRef]
- Miura, T.; Kamiya, Y.; Hina, S.; Kobayashi, Y.; Murayama, N.; Shimizu, M.; Yamazaki, H. Metabolic profiles of coumarin in human plasma extrapolated from a rat data set with a simplified physiologically based pharmacokinetic model. J. Toxicol. Sci. 2020, 45, 695–700. [Google Scholar] [CrossRef]
- Azizah, M.; Pripdeevech, P.; Thongkongkaew, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. UHPLC-ESI-QTOF-MS/MS-Based Molecular Networking Guided Isolation and Dereplication of Antibacterial and Antifungal Constituents of. Antibiotics 2020, 9, 606. [Google Scholar] [CrossRef]
- Flores-Ocelotl, M.R.; Rosas-Murrieta, N.H.; Moreno, D.A.; Vallejo-Ruiz, V.; Reyes-Leyva, J.; Domínguez, F.; Santos-López, G. Taraxacum officinale and Urtica dioica extracts inhibit dengue virus serotype 2 replication in vitro. BMC Complement. Altern. Med. 2018, 18, 95. [Google Scholar] [CrossRef]
- Murali, K.S.; Sivasubramanian, S.; Vincent, S.; Murugan, S.B.; Giridaran, B.; Dinesh, S.; Gunasekaran, P.; Krishnasamy, K.; Sathishkumar, R. Anti-chikungunya activity of luteolin and apigenin rich fraction from Cynodon dactylon. Asian Pac. J. Trop. Med. 2015, 8, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Su, W.; Jin, J.; Chen, J.; Li, X.; Zhang, X.; Sun, M.; Sun, S.; Fan, P.; An, D.; et al. Identification of luteolin as enterovirus 71 and coxsackievirus A16 inhibitors through reporter viruses and cell viability-based screening. Viruses 2014, 6, 2778–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Care, C.; Sornjai, W.; Jaratsittisin, J.; Hitakarun, A.; Wikan, N.; Triwitayakorn, K.; Smith, D.R. Discordant Activity of Kaempferol Towards Dengue Virus and Japanese Encephalitis Virus. Molecules 2020, 25, 1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifi-Rad, M.; Roberts, T.H.; Matthews, K.R.; Bezerra, C.F.; Morais-Braga, M.F.B.; Coutinho, H.D.M.; Sharopov, F.; Salehi, B.; Yousaf, Z.; Sharifi-Rad, M.; et al. Ethnobotany of the genus Taraxacum-Phytochemicals and antimicrobial activity. Phytother. Res. 2018, 32, 2131–2145. [Google Scholar] [CrossRef]
- Mokdad-Bzeouich, I.; Mustapha, N.; Chaabane, F.; Ghedira, Z.; Ghedira, K.; Ghoul, M.; Chebil, L.; Chekir-Ghedira, L. Oligomerization of esculin improves its antibacterial activity and modulates antibiotic resistance. J. Antibiot. 2015, 68, 148–152. [Google Scholar] [CrossRef]
- Silva-Mares, D.; Torres-López, E.; Rivas-Estilla, A.M.; Cordero-Pérez, P.; Waksman-Minsky, N.; Rivas-Galindo, V.M. Plants from northeast Mexico with anti-HSV activity. Nat. Prod. Commun. 2013, 8, 297–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeganegi, M.; Tabatabaei Yazdi, F.; Mortazavi, S.A.; Asili, J.; Alizadeh Behbahani, B.; Beigbabaei, A. Equisetum telmateia extracts: Chemical compositions, antioxidant activity and antimicrobial effect on the growth of some pathogenic strain causing poisoning and infection. Microb. Pathog. 2018, 116, 62–67. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Froese, N.T.; Van Acker, R.C. Distribution and interference of dandelion (Taraxacum officinale) in spring canola. Weed Sci. 2003, 51, 435–442. [Google Scholar] [CrossRef]
- Ríos, J.L.V.; Espinosa García, F.J. Catálogo de Malezas de México; Fondo De Cultura Economica: San Diego, CA, USA, 1998. [Google Scholar]
- Villaseñor, J.L.; Espinosa-Garcia, F.J. The alien flowering plants of Mexico. Divers. Distrib. 2004, 10, 113–123. [Google Scholar] [CrossRef]
- Edwards, S.E.; da Costa Rocha, I.; Williamson, E.M.; Heinrich, M. Phytopharmacy: An Evidence-Based Guide to Herbal Medicinal Products; John Wiley & Sons: Hoboken, NJ, USA, 2015; ISBN 9781118543450. [Google Scholar]
- González-Castejón, M.; Visioli, F.; Rodriguez-Casado, A. Diverse biological activities of dandelion. Nutr. Rev. 2012, 70, 534–547. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-N.; Yao, Z.-L.; Yang, D.; Ke, J.; Wu, Q.-L.; Li, J.-K.; Zhou, X.-D. Chemical Constituents from and Their Antifungal and Herbicidal Activities. Biomolecules 2020, 10, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seregheti, T.M.Q.; Pinto, A.P.R.; Gonçalves, M.d.C.; Antunes, A.D.S.; Almeida, W.A.d.S.; Machado, R.S.; Silva, J.N.; Ferreira, P.M.P.; Pessoa, C.; Santos, V.M.R.D.; et al. Antiproliferative and photoprotective activities of the extracts and compounds from Calea fruticosa. Braz. J. Med. Biol. Res. 2020, 53, e9375. [Google Scholar] [CrossRef]
- Abdallah, H.; Farag, M.; Osman, S.; Kim, D.H.; Kang, K.; Pan, C.-H.; Abdel-Sattar, E. Isolation of major phenolics from Launaea spinosa and their protective effect on HepG2 cells damaged with t-BHP. Pharm. Biol. 2016, 54, 536–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
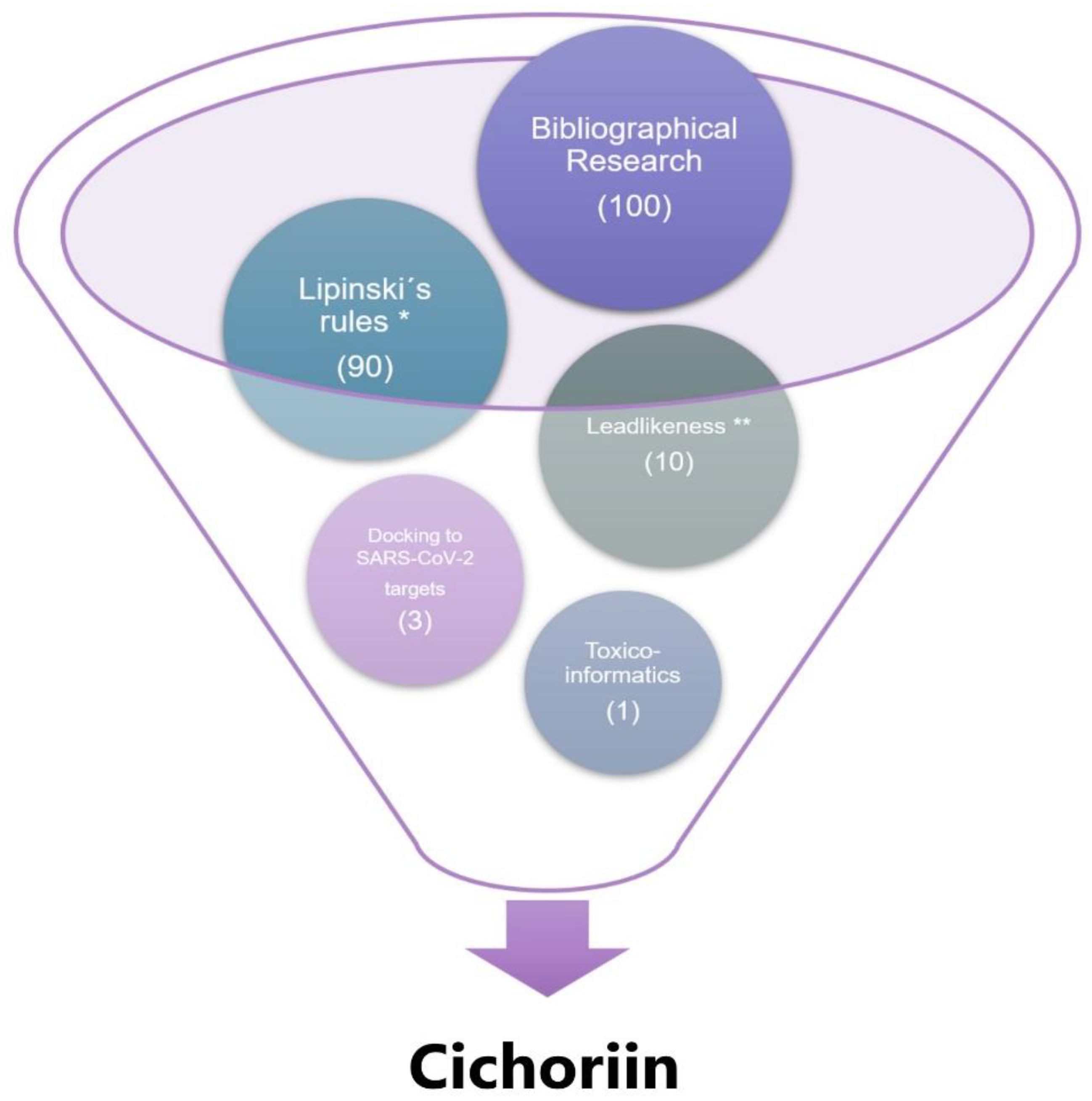
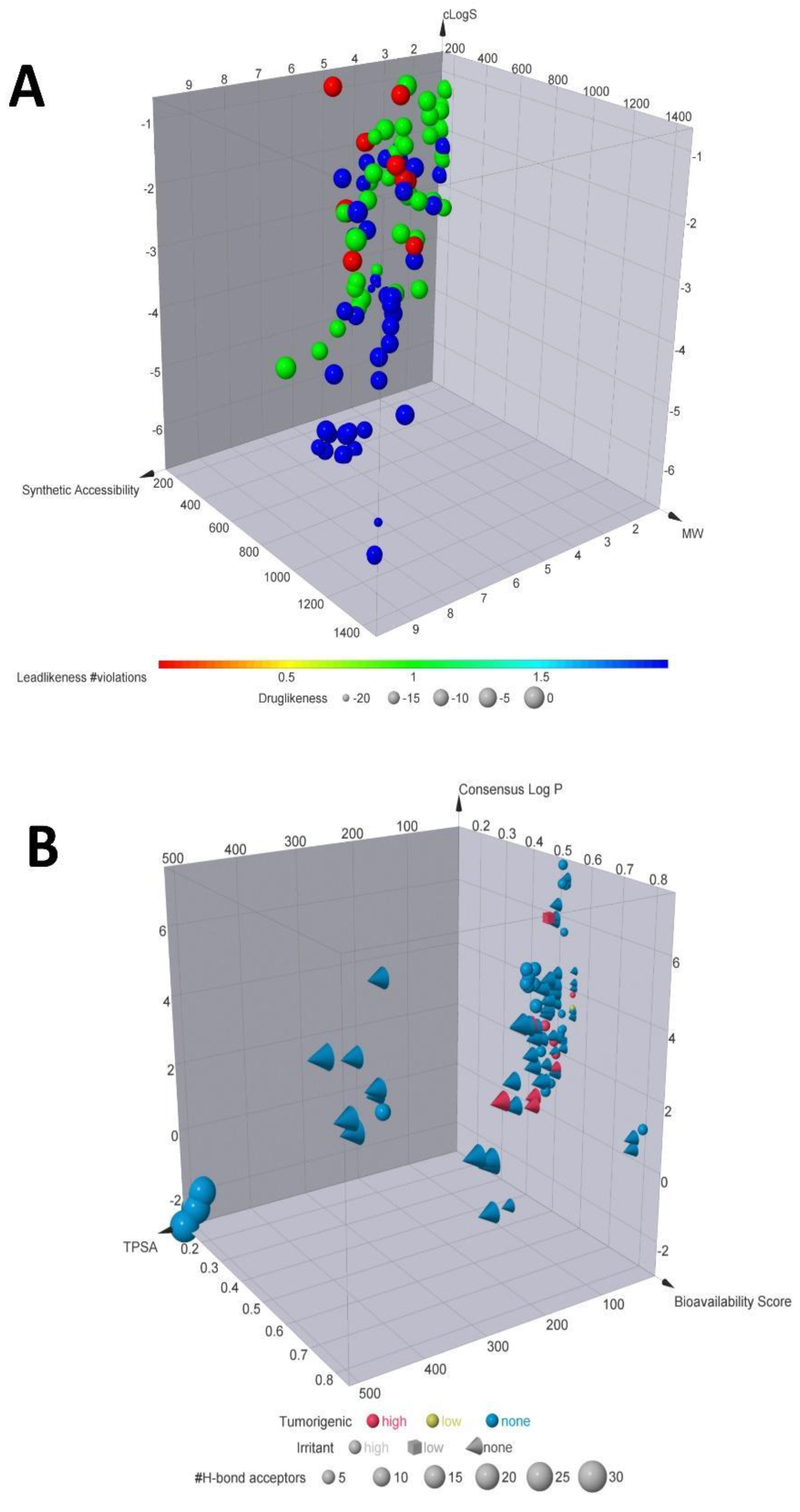
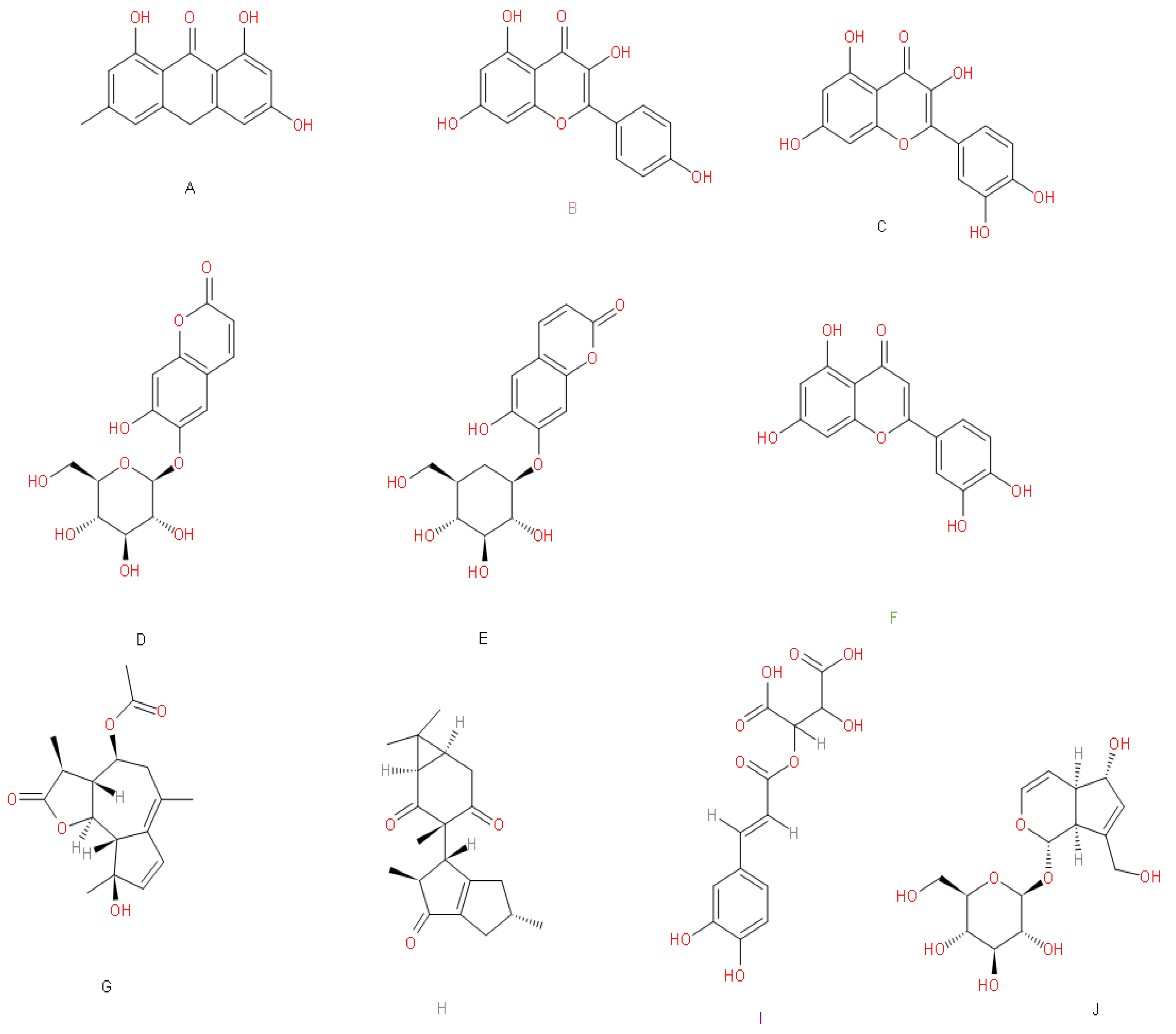

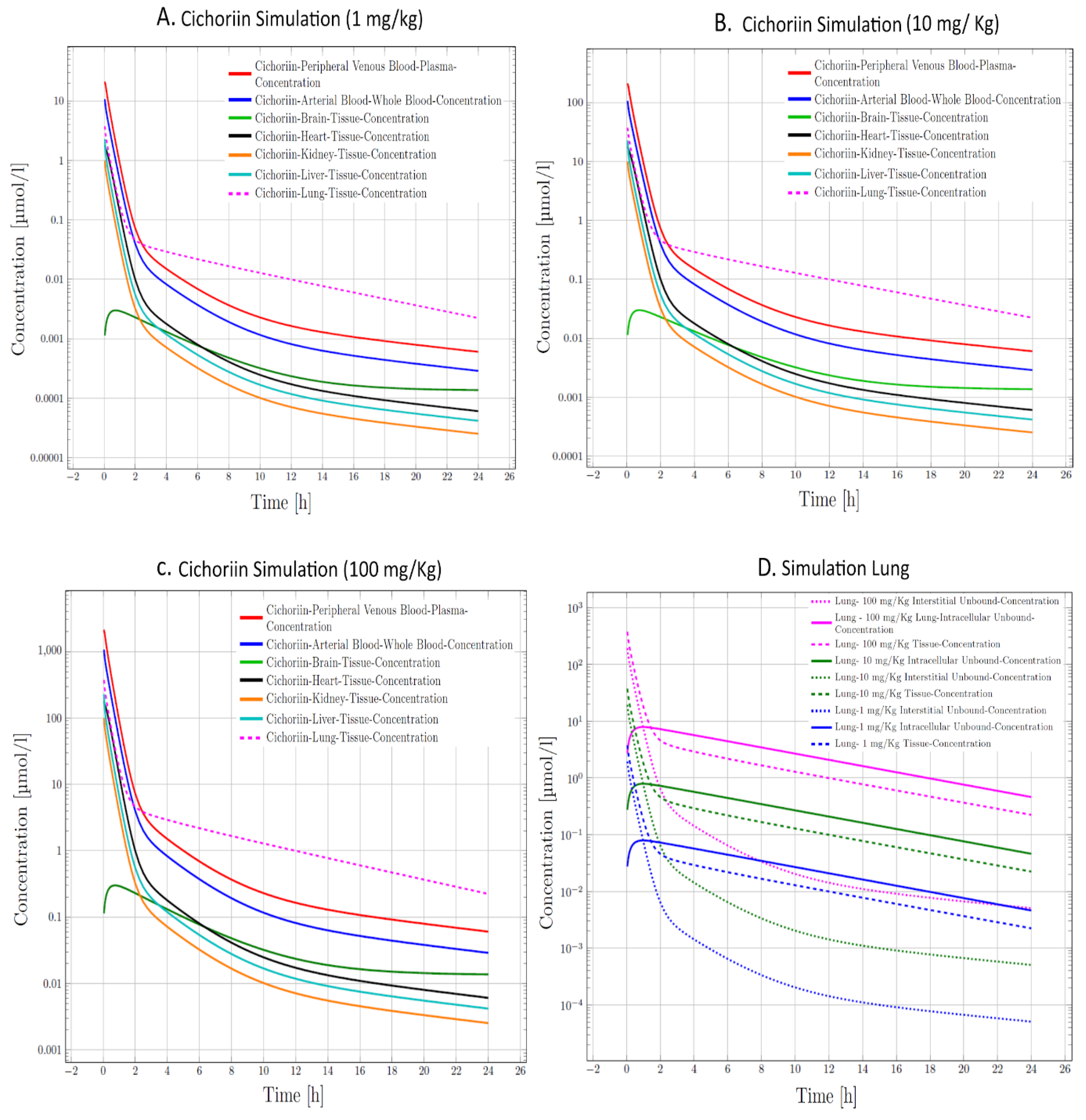
| A | C | EA | E | K | L | M | MTA | Q | R | |
|---|---|---|---|---|---|---|---|---|---|---|
| Protease | −6.5 | −7.4 | −7.1 | −7.7 | −7.4 | −7.3 | −7.1 | −6.7 | −7.3 | −6.4 |
| Papain-like protease | −7.7 | −8.3 | −7.9 | −8.7 | −8.2 | −8.3 | −8.5 | −7.2 | −8.4 | −9.4 |
| Nsp3 (207-379, AMP site) | −7.7 | −7.1 | −7.5 | −7.5 | −7.8 | −8.1 | −7.1 | −6.8 | −8.3 | −8.3 |
| Nsp3 (207-379, MES site) | −6.8 | −8.4 | −8.1 | −7.8 | −8.6 | −7.9 | −7.6 | −7.7 | −8.6 | −8.1 |
| RdRp (RTP site) | −9.4 | −9.5 | −9.2 | −9.4 | −10.2 | −8.4 | −7.5 | −9 | −10.5 | −8 |
| RdRp (RNA site) | −6.6 | −7.5 | -6.7 | −7.7 | −7.4 | −7.5 | −7.2 | −7 | −7.5 | −7.5 |
| Helicase (ADP site) | −6.3 | −6.7 | −6.3 | −6.6 | −6.3 | −6.6 | −6.6 | −6.3 | −6.5 | −6 |
| Helicase (NCB site) | −7 | −7.4 | −7 | −7.2 | −7.2 | −7 | −7.1 | −7.1 | −7.2 | −7.7 |
| Nsp14 (ExoN) | −7.8 | −8.8 | −8.6 | −8.3 | −8.7 | −8.7 | −8.1 | −7.3 | −8.7 | −8.9 |
| Nsp14 (N7-MTase) | −6.6 | −7.5 | −6.8 | −7.2 | −7.3 | −7 | −6.1 | −6 | −7.4 | −6.5 |
| Nsp15 (endoribonuclease) | −6.5 | −7.2 | −7.3 | −6.9 | −7 | −7.2 | −6.6 | −6.4 | −6.8 | −7.3 |
| Nsp16 (GTA site) | −7.3 | −8.3 | −7.9 | −7.9 | −8.6 | −8.8 | −7.5 | −7 | -8.7 | −7.5 |
| Nsp16 (MGP site) | −6 | −7.2 | −7.1 | −7.2 | −7 | −6.7 | −6.3 | −6.2 | −6.9 | −7.4 |
| Nsp16 (SAM site) | −7.5 | −8.1 | −7.5 | −8 | −8.7 | −8.9 | −6.8 | −7.2 | −8.7 | −7.4 |
| N protein (NCB site) | −6.6 | −7.7 | −8.7 | −7.8 | −8.1 | −7.9 | −6.5 | −6.7 | −8.4 | −9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivero-Segura, N.A.; Gomez-Verjan, J.C. In Silico Screening of Natural Products Isolated from Mexican Herbal Medicines against COVID-19. Biomolecules 2021, 11, 216. https://doi.org/10.3390/biom11020216
Rivero-Segura NA, Gomez-Verjan JC. In Silico Screening of Natural Products Isolated from Mexican Herbal Medicines against COVID-19. Biomolecules. 2021; 11(2):216. https://doi.org/10.3390/biom11020216
Chicago/Turabian StyleRivero-Segura, Nadia A., and Juan C. Gomez-Verjan. 2021. "In Silico Screening of Natural Products Isolated from Mexican Herbal Medicines against COVID-19" Biomolecules 11, no. 2: 216. https://doi.org/10.3390/biom11020216
APA StyleRivero-Segura, N. A., & Gomez-Verjan, J. C. (2021). In Silico Screening of Natural Products Isolated from Mexican Herbal Medicines against COVID-19. Biomolecules, 11(2), 216. https://doi.org/10.3390/biom11020216







