Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Force Field
3. Results
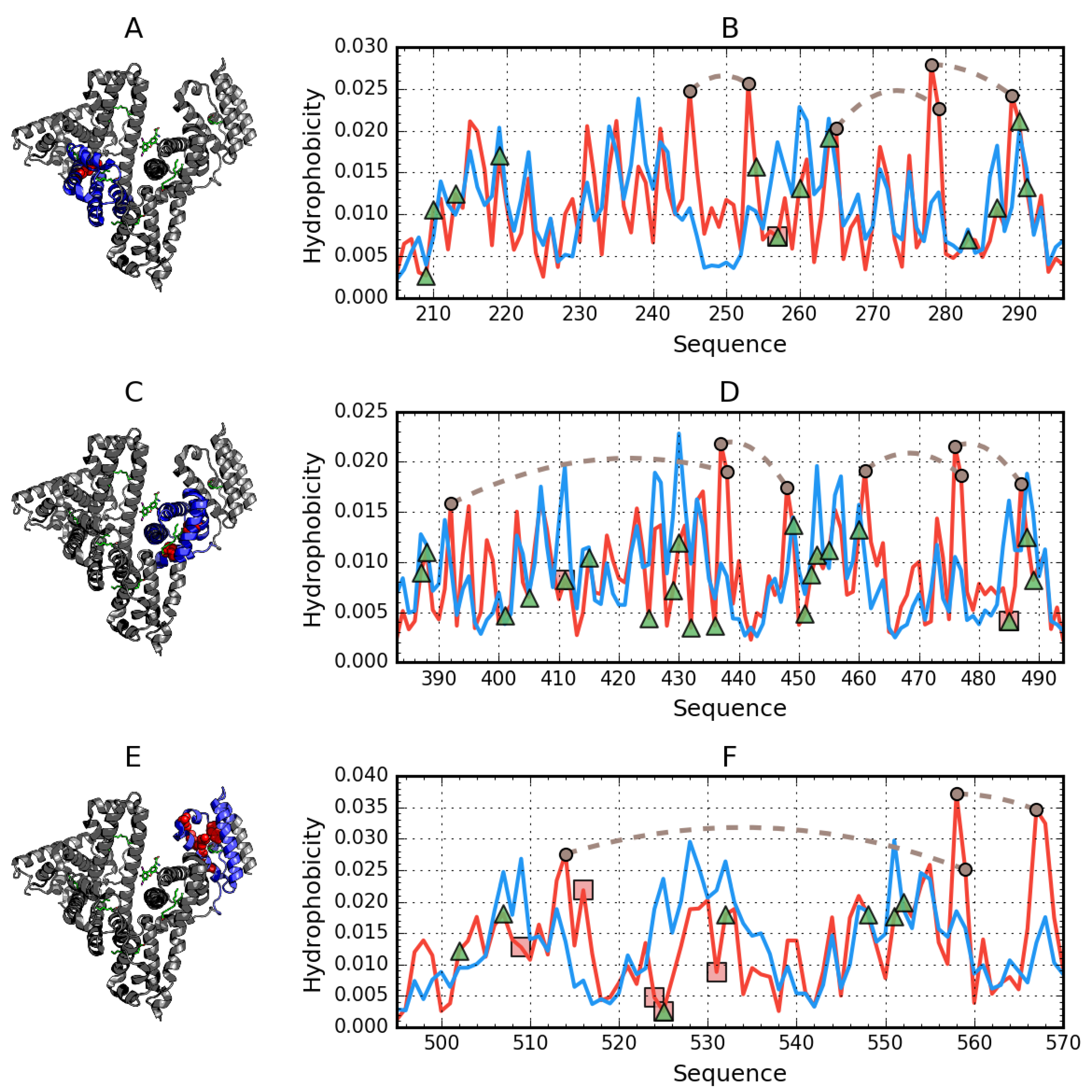
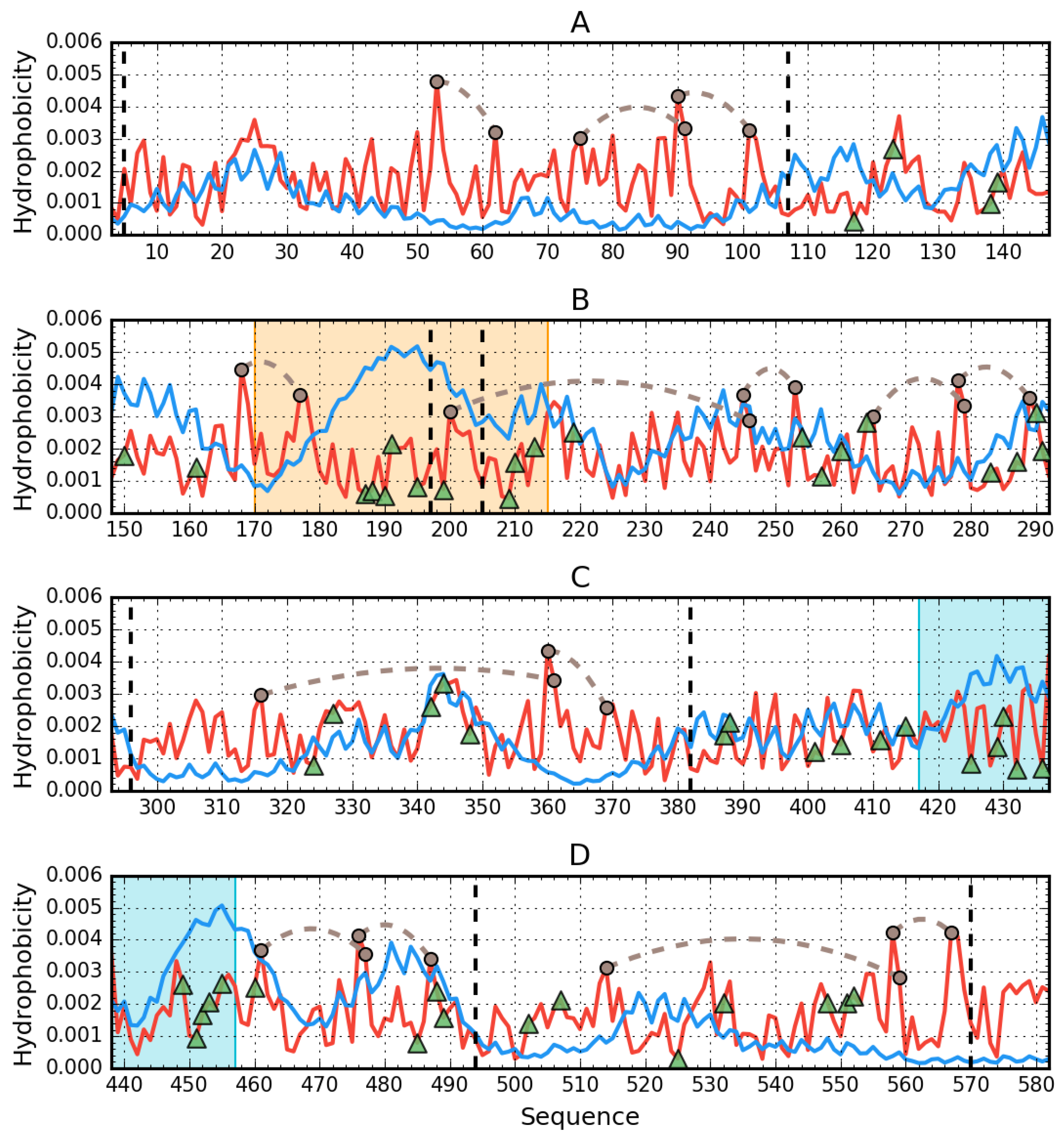
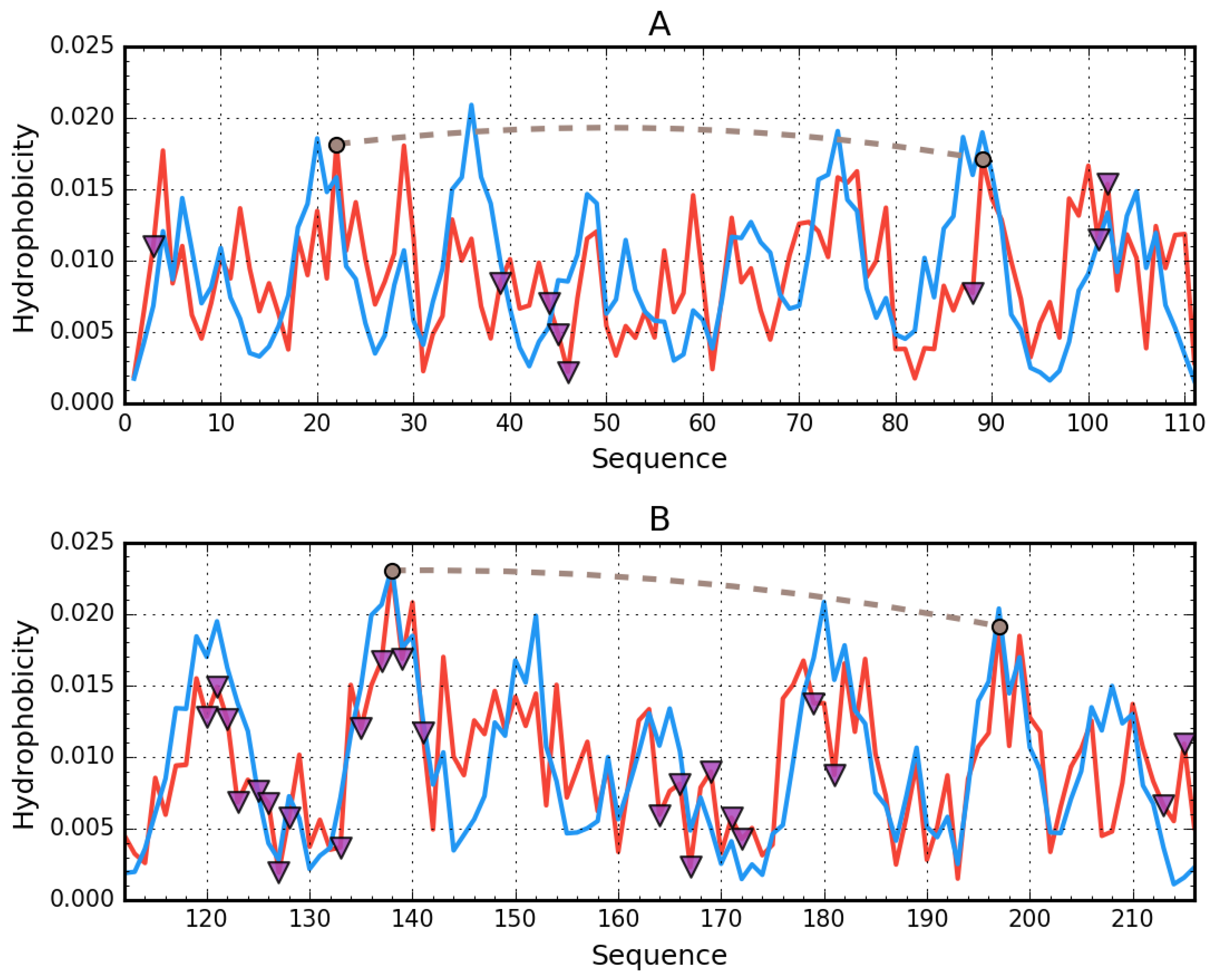
3.1. Assessment of the Status of Domains Comprising Human Albumin in the Context of Their Capacity for Binding Congo Red in Complex with Dox Based on the Fuzzy Oil Drop Model
3.2. Ligand Binding by Albumin
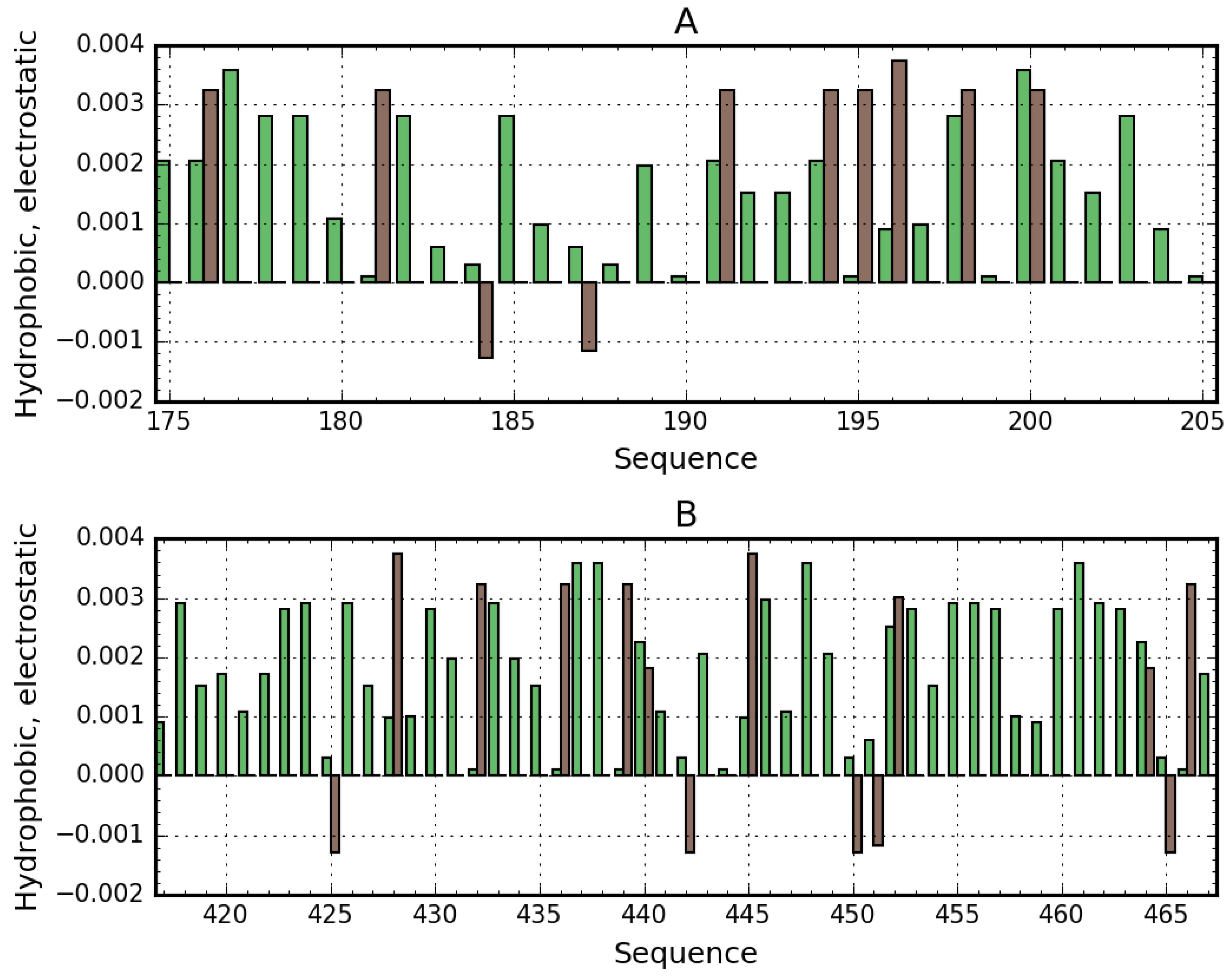
3.3. Albumin Molecule Analyzed as a Whole by Applying the Fuzzy Oil Drop Model
Interpretation of Results Obtained for the Complete Structure of Albumin
- its capacity for conformational rearrangements and therefore for accommodating a supramolecular ligand in its form of co-micelle (Congo red + Dox),
- markedly increased hydrophobicity,
- high positive electrostatic charge density (which promotes the binding of a negatively charged ligand—Figure 7).
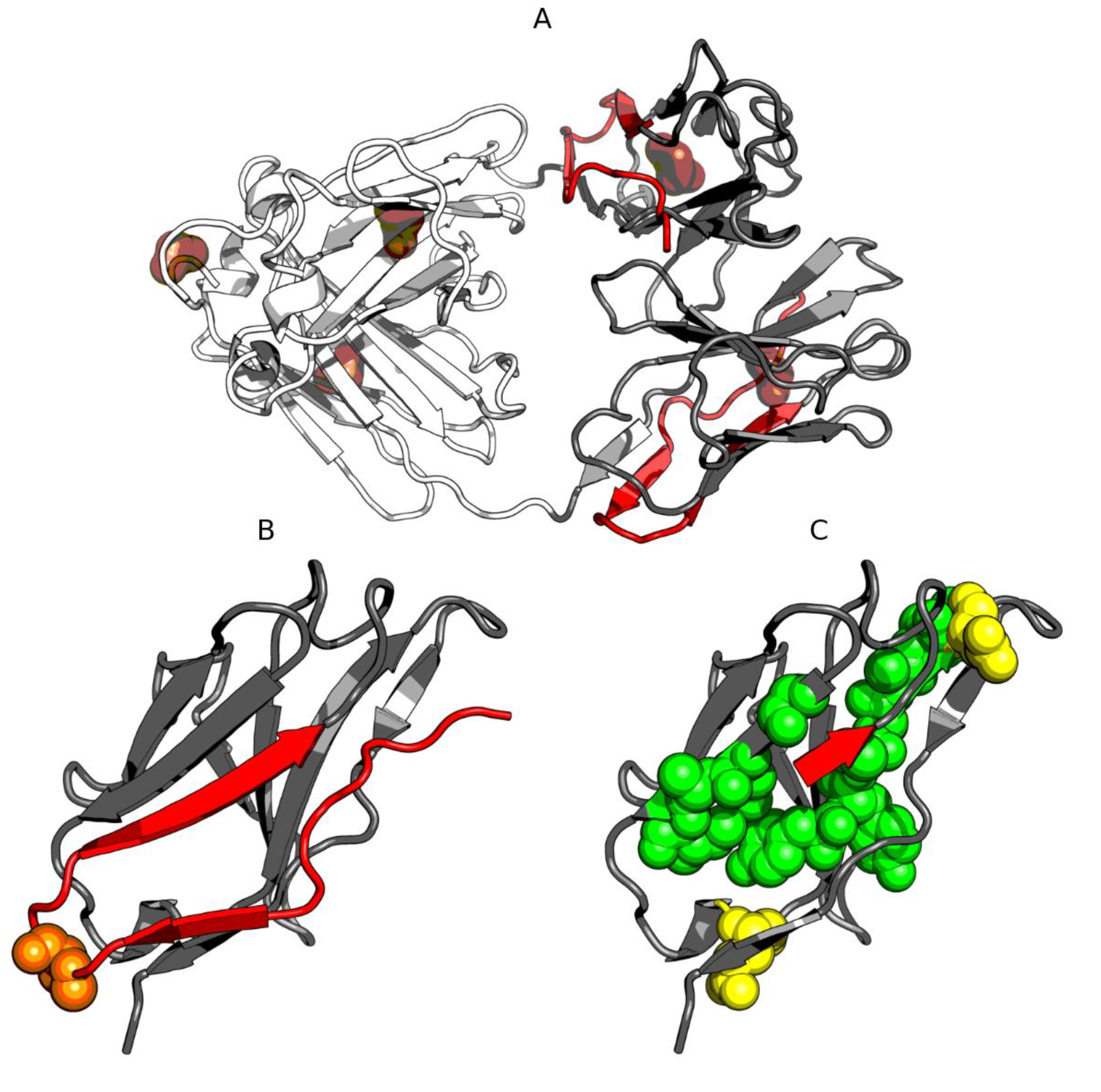
3.4. Light Chain (Lambda) of IgG in the Fuzzy Oil Drop Classification
3.5. Analysis of Pores, Clefts and Tunnels in the IgG Light Chain Dimer and in Albumin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rybarska, J.; Piekarska, B.; Stopa, B.; Zemanek, G.; Konieczny, L.; Roterman, I. Supramolecular Systems as Protein Ligands. In Self-Assembled Molecules—New Kind of Protein Ligands; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–20. [Google Scholar]
- Jagusiak, A.; Rybarska, J.; Piekarska, B.; Stopa, B.; Konieczny, L. Supramolecular Congo Red as Specific Ligand of Antibodies Engaged in Immune Complex. In Self-Assembled Molecules—New Kind of Protein Ligands; Springer International Publishing: Cham, Switzerland, 2017; pp. 21–41. [Google Scholar]
- Peters, T., Jr. All about Albumin-Biochemistry, Genetics and Medical Applications; Academic Press: Cambridge, MA, USA, 1996. [Google Scholar]
- Petitpas, I.; Petersen, C.E.; Ha, C.-E.; Bhattacharya, A.A.; Zunszain, P.A.; Ghuman, J.; Bhagavan, N.V.; Curry, S. Structural basis of albumin-thyroxine interactions and familial dysalbuminemic hyperthyroxinemia. Proc. Natl. Acad. Sci. USA 2003, 100, 6440–6445. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.B.; Ainsworth, C.F.; Stevens, F.J.; Schiffer, M. Three quaternary structures for a single protein. Proc. Natl. Acad. Sci. USA 1996, 93, 7017–7021. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska, B.; Banach, M.; Konieczny, L.; Roterman, I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17, 1477–1507. [Google Scholar] [CrossRef]
- Fabian, P.; Stapor, K.; Banach, M.; Ptak-Kaczor, M.; Konieczny, L.; Roterman, I. Alternative Hydrophobic Core in Proteins—The Effect of Specific Synergy. Symmetry 2020, 12, 273. [Google Scholar] [CrossRef]
- Levitt, M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104, 59–107. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On Information and Sufficiency. Ann. Math. Stat. 2007, 22, 79–86. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, I. Protein-protein interaction encoded as an exposure of hydrophobic residues on the surface. In From Globular Proteins to Amyloids; Elsevier BV: Amsterdam, The Netherlands; Oxford, UK; Cambridge, MA, USA, 2020; pp. 79–89. [Google Scholar]
- Banach, M.; Konieczny, L.; Roterman, I. Ligand binding cavity encoded as a local hydrophobicity deficiency. In From Globular Proteins to Amyloids; Elsevier BV: Amsterdam, The Netherlands; Oxford, UK; Cambridge, MA, USA, 2020; pp. 91–93. [Google Scholar]
- Yamasaki, K.; Anraku, M. Stability of Albumin and Stabilization of Albumin Preparations; Otagiri, M., Chuang, V., Eds.; Albumin in Medicine; Springer: Singapore, 2016; pp. 1–24. [Google Scholar] [CrossRef]
- Kwiecińska, K.; Stachowicz-Kuśnierz, A.; Jagusiak, A.; Roterman, I.; Korchowiec, J. Impact of Doxorubicin on Self-Organization of Congo Red: Quantum Chemical Calculations and Molecular Dynamics Simulations. ACS Omega 2020, 5, 19377–19384. [Google Scholar] [CrossRef] [PubMed]
- Banach, M.; Kalinowska, B.; Konieczny, L.; Roterman, I. Role of Disulfide Bonds in Stabilizing the Conformation of Selected Enzymes—An Approach Based on Divergence Entropy Applied to the Structure of Hydrophobic Core in Proteins. Entropy 2016, 18, 67. [Google Scholar] [CrossRef]
- Roterman, I.; Rybarska, J.; Konieczny, L.; Skowronek, M.; Stopa, B.; Piekarska, B.; Bakalarski, G. Congo Red bound to α-1-proteinase inhibitor as a model of supramolecular ligand and protein complex. Comput. Chem. 1998, 22, 61–70. [Google Scholar] [CrossRef]
- Król, M.; Roterman, I.; Piekarska, B.; Konieczny, L.; Rybarska, J.; Stopa, B. Local and Long-Range Structural Effects Caused by the Removal of the N-terminal Polypeptide Fragment from Immunoglobulin L Chain Lambda. Biopolymers 2003, 69, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Prot. Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Rybarska, J.; Konieczny, L.; Roterman, I.; Piekarska, B. The effect of azo dyes on the formation of immune complexes. Arch. Immunol. Ther. Exp. 1991, 39, 317–327. [Google Scholar]
- Kaszuba, J.; Konieczny, L.; Piekarska, B.; Rotterman, I.; Rybarska, J. Bis-azo dyes interference with effector activation of antibodies. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 1993, 44, 233–242. [Google Scholar]
- Roterman, I.; No, K.-T.; Piekarska, B.; Kaszuba, J.; Pawlicki, R.; Rybarska, J.; Konieczny, L. Bis-azo dyes—Studies on the mechanism of complex formation with IgG modulated be heating or antygen binding. J. Physiol Pharm. 1993, 44, 213–232. [Google Scholar]
- Krol, M.; Roterman, I.; Drozd, A.; Konieczny, L.; Piekarska, B.; Rybarska, J.; Spolnik, P.; Stopa, B. The increased flexibility of CDR Loos generated in anibodies by Congo red complexation favors antygen binding. J. Biomol. Strucy Dyn. 2006, 23, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Jagusiak, A.; Konieczny, L.; Krol, M.; Marszalek, P.; Piekarska, B.; Roterman, I.; Rybarska, J.; Stopa, B.; Zemanek, G. Intramolecual immunological signal—Hypothesis revieved—Structural background of signalling revealed by using Congo red as a specific tool. Mini. Rev. Med. Chem. 2015, 14, 1104–1113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Konieczny, L.; Piekarska, B.; Rybarska, J.; Skowronek, M.; Stopa, B.; Tabor, B.; Daboros, W.; Pawlicki, R.; Roterman, I. The use of Congo red as a lyotropic liquid crystal to carry stains in a model immunotargeting system—Microscopic studiem. Folia Histochem. Cytobiol. 1997, 35, 203–210. [Google Scholar] [PubMed]
- Rybarska, J.; Piekarska, B.; Stopa, B.; Spolnik, P.; Zemanek, G.; Konieczny, L.; Roterman, I. In vivo accummulation of self-assembling dye Congo red in an area marked by specific immune complexes: Possible relevance to chemotherapy. Folia Histochem. Cytobiol. 2004, 42, 101–110. [Google Scholar] [PubMed]
- Piekarska, B.; Konieczny, L.; Rybarska, J.; Stopa, B.; Zemanek, G.; Szneler, E.; Krol, M.; Nowak, M.; Roterman, I. Heat-induced formation of a specific binding site for self-assembled Congo red in the V domain of immunoglobulin L lambda chain. Biopolymers 2001, 9, 446–456. [Google Scholar] [CrossRef]
- Zemanek, G.; Jagusiak, A.; Rybarska, J.; Piwowar, P.; Chłopaś, K.; Roterman, I. Protein conditioning for binding Congo red and other supramolecuar ligands. In Self-Assembled Molecules—New Kind of Protein Ligands; Springer Open: Cham, Switzerland, 2018; pp. 43–60. [Google Scholar]
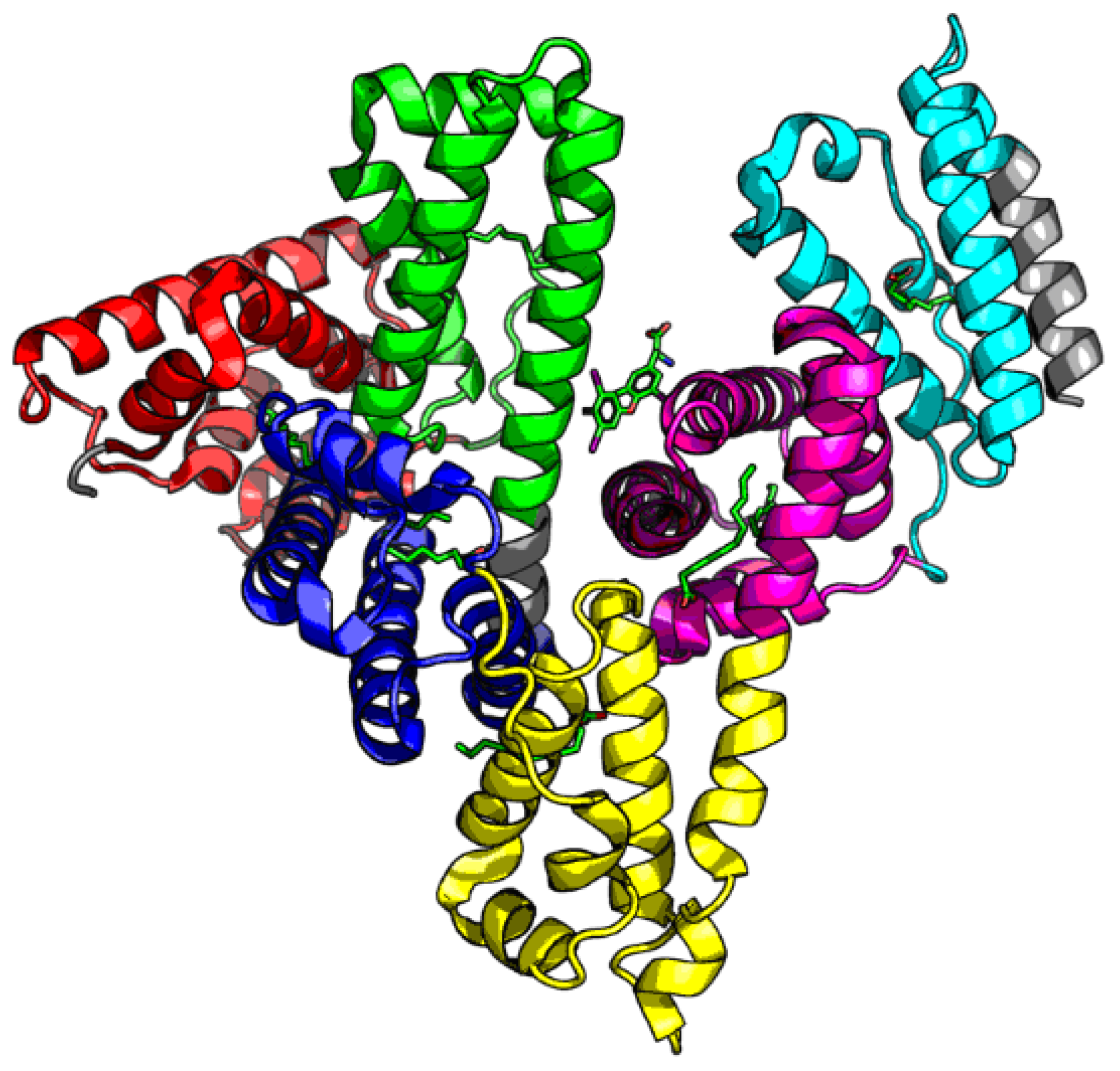





| PDB- ID | Name | Source | Reference | |
|---|---|---|---|---|
| 1HK4 | Albumin | HUMAN | [4] | |
| 4BJL | Light chain of IgG | HUMAN | Bence-Jones dimer | [5] |
| Protein | Fragment | RD | |||||
|---|---|---|---|---|---|---|---|
| H | vdW | Ele | |||||
| Albumin (1HK4) | 3–584 | 0.741 | 0.853 | 0.917 | |||
| Domain Status | |||||||
| Domain | Domains in Molecule | Individual Domains | |||||
| H | vdW | Ele | H | vdW | Ele | ||
| AI | 5–107 | 0.692 | 0.843 | 0.860 | 0.547 | 0.764 | 0.834 |
| AII | 108–197 | 0.678 | 0.759 | 0.853 | 0.558 | 0.738 | 0.875 |
| AIII | 205–296 | 0.503 | 0.745 | 0.752 | 0.434 | 0.746 | 0.795 |
| BI | 297–382 | 0.718 | 0.820 | 0.908 | 0.542 | 0.665 | 0.847 |
| BII | 383–494 | 0.504 | 0.730 | 0.906 | 0.461 | 0.786 | 0.936 |
| BIII | 495–570 | 0.723 | 0.864 | 0.958 | 0.465 | 0.788 | 0.942 |
| Fragment | RD for Complex | |
|---|---|---|
| DIMER | TWO CHAINS | 0.729 |
| INDIVIDUAL | DOMAIN VL | 0.737/0.611 |
| INDIVIDUAL | DOMAIN CL | 0.768/0.711 |
| CHAIN A | 0.709 | |
| CHAIN B | 0.740 | |
| SS bonds | 22—89 | 0.731/0.816 |
| 138—197 | 0.607/0.736 | |
| P-P | 0.643 | |
| NO P-P | 0.724 |
| RD for Domains Treated as Individual Units | ||
|---|---|---|
| Fragment | RD | |
| VL | 1–111 | 0.573/0.582 |
| P-P | 0.515/0.722 | |
| NO P-P | 0.576/0.572 | |
| SS | 22–89 | 0.541/0.544 |
| β-sheet | 8–2 * | 0.611/0.585 |
| 18–22 * | 0.402/0.425 | |
| CL | 112–216 | 0.424/0.356 |
| P-P | 0.505/0.377 | |
| NO P-P | 0.391/0.338 | |
| SS | 138–197 | 0.376/0.343 |
| β-sheet | 118–122 * | 0.492/0.520 |
| 147–155 * | 0.470/0.502 | |
| RD Values for Dimers VL-VL CL-CL | ||
|---|---|---|
| Domains | Fragments | RD |
| VL-VL | (1 − 111) + (1 − 111) | 0.716 |
| CL-CL | (112 − 216) + (112 − 216) | 0.495 |
| P-P | 0.599/0.366 | |
| NO P-P | 0.695/0.438 | |
| SS | 22–89 | 0.713/0.702 |
| 138–197 | 0.493/0.485 | |
| β-sheet | 8–12 * | 0.778/0.789 |
| 18–22 * | 0.355/0.444 | |
| 118–122 * | 0.559/0.547 | |
| 147–155 * | 0.564/0.543 |
| Clefts Volume Å3 | Tunnels Radius Å/Free Radius Å | Pores Radius Å/Free Radius Å/Length Å | |||
|---|---|---|---|---|---|
| 4BJL | 1HK4 | 4BJL | 1HK4 | 4BJL | 1HK4 |
| 11,137.50 1591.73 951.33 604.12 729.84 512.58 429.05 340.03 340.03 341.72 | 1713.78 * 2131.31 * 2035.55 * 1690.88 1125.98 1002.80 600.33 530.30 529.03 810.42 | 1.16/21.1 1.39/16.1 1.45/16.9 1.40/17.9 1.31/20.2 | 1.26/28.8 * 1.29/29.5 * 1.25/38.4 * 1.29/39.3 * 1.25/39.3 * 1.30/40.1 * 1.26/48.9 * 1.31/49.7 * | ABSENT | /3.58/27.8 1.16/1.17/37.0 1.57/3.20/42.9 1.97/4.70/48.3 1.96/4.72/55.2 * 1.88/2.88/58.6 * 1.17/1.21/98.6 * 2.09/2.10/98.0 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magdalena, P.-K.; Klaudia, K.; Jacek, K.; Katarzyna, C.; Mateusz, B.; Irena, R.; Anna, J. Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies. Biomolecules 2021, 11, 501. https://doi.org/10.3390/biom11040501
Magdalena P-K, Klaudia K, Jacek K, Katarzyna C, Mateusz B, Irena R, Anna J. Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies. Biomolecules. 2021; 11(4):501. https://doi.org/10.3390/biom11040501
Chicago/Turabian StyleMagdalena, Ptak-Kaczor, Kwiecińska Klaudia, Korchowiec Jacek, Chłopaś Katarzyna, Banach Mateusz, Roterman Irena, and Jagusiak Anna. 2021. "Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies" Biomolecules 11, no. 4: 501. https://doi.org/10.3390/biom11040501
APA StyleMagdalena, P.-K., Klaudia, K., Jacek, K., Katarzyna, C., Mateusz, B., Irena, R., & Anna, J. (2021). Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies. Biomolecules, 11(4), 501. https://doi.org/10.3390/biom11040501







