Inhibition of Rice Serotonin N-Acetyltransferases by MG149 Decreased Melatonin Synthesis in Rice Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Inhibitor Treatments
2.2. Cadmium Treatment
2.3. Chemical Compounds
2.4. Rice Recombinant SNAT1 and SNAT2 Enzymes
2.5. Measuring SNAT Enzyme Activity
2.6. Melatonin Quantification
2.7. Quantification of Tryptophan, Tryptamine, Serotonin, and N-Acetylserotonin
2.8. Laminar Joint Inclination Assay
2.9. Statistical Analyses
3. Results
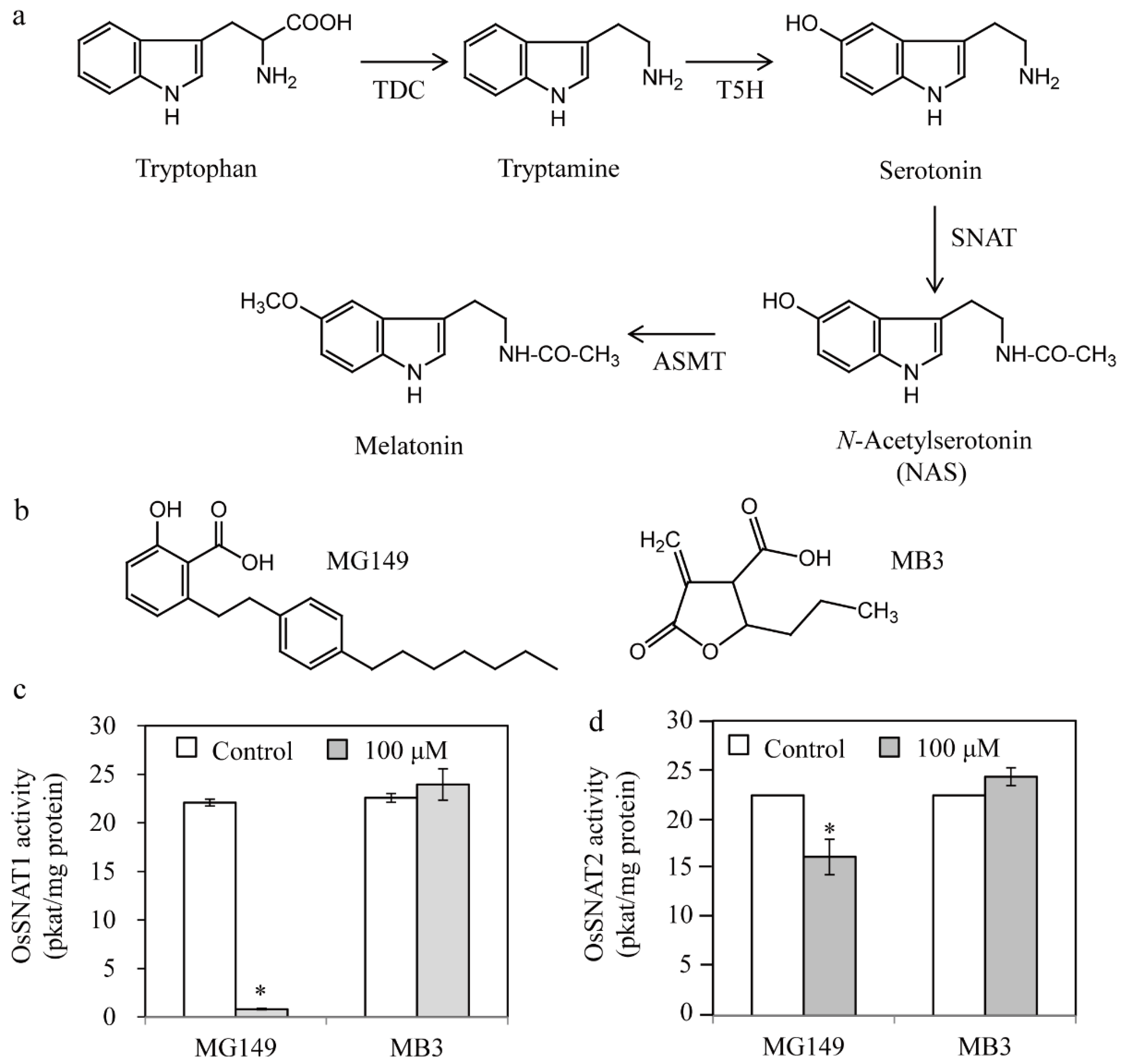
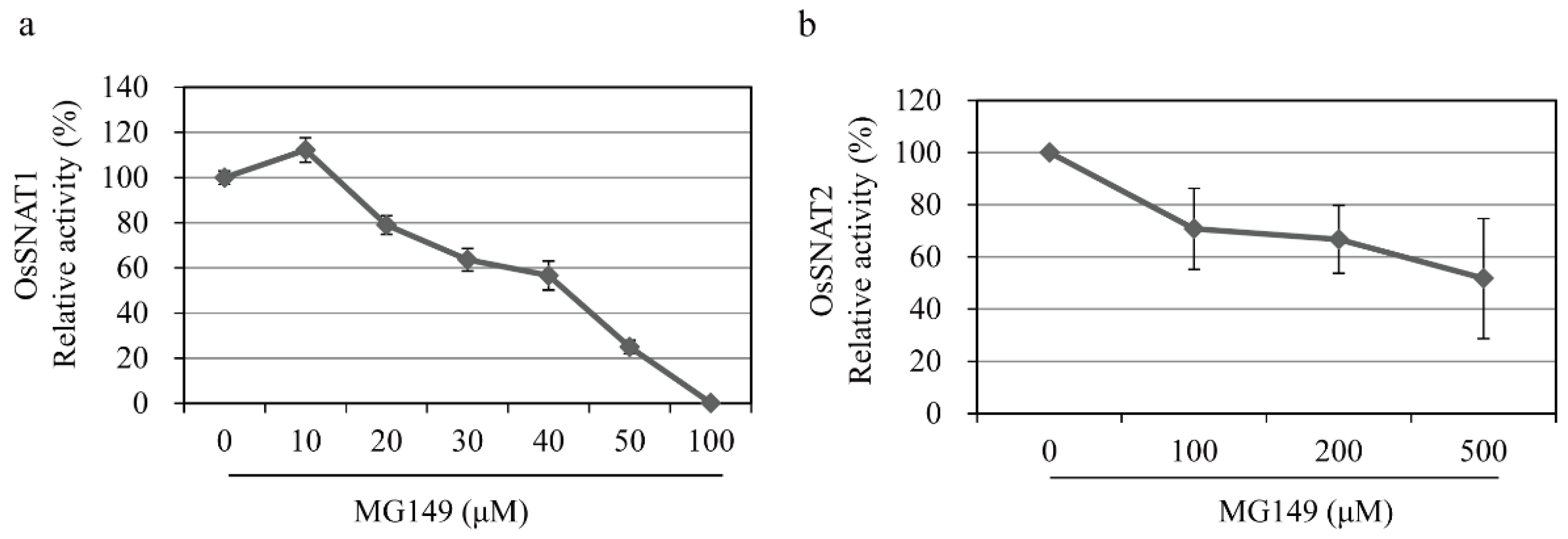
3.1. In Vitro Inhibition of Rice SNAT Enzymes by HAT Inhibitors
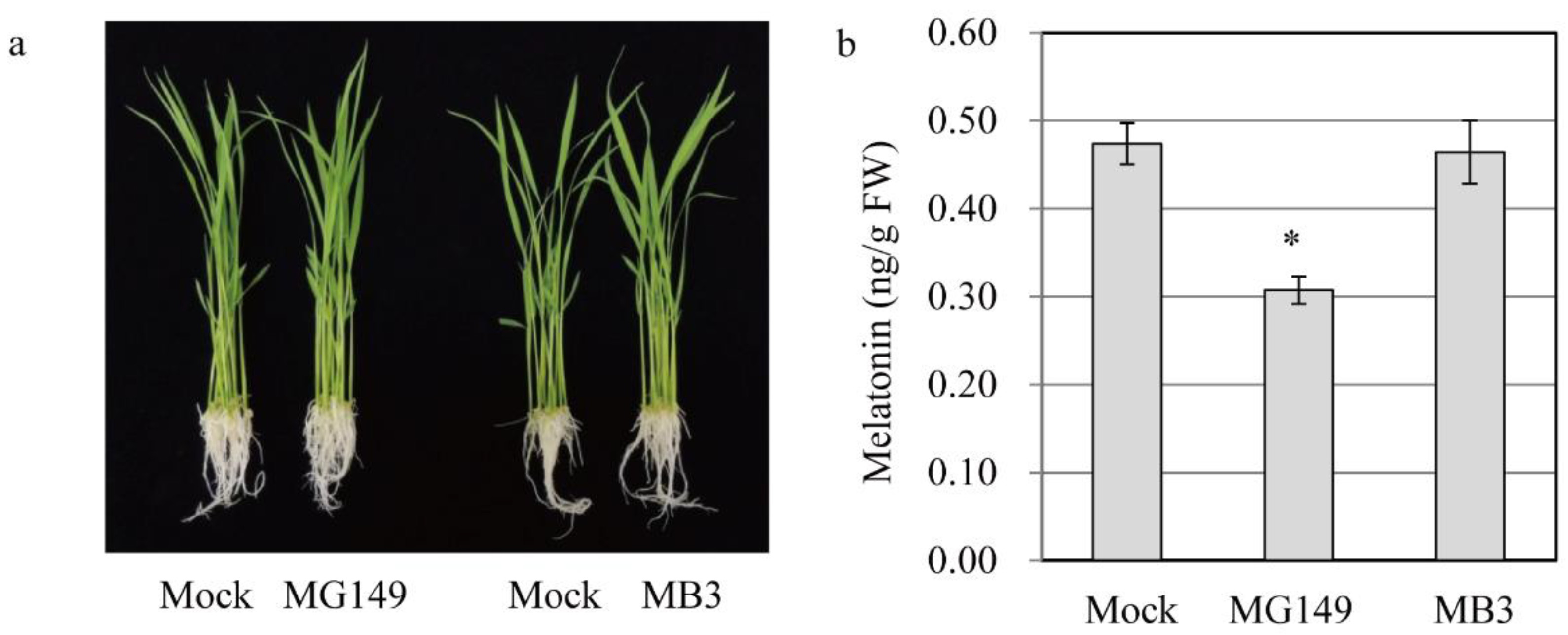
3.2. In Vivo Inhibition of Melatonin Synthesis by HAT Inhibitors
3.3. Quantification of Melatonin and Its Precursors with Cadmium Treatment
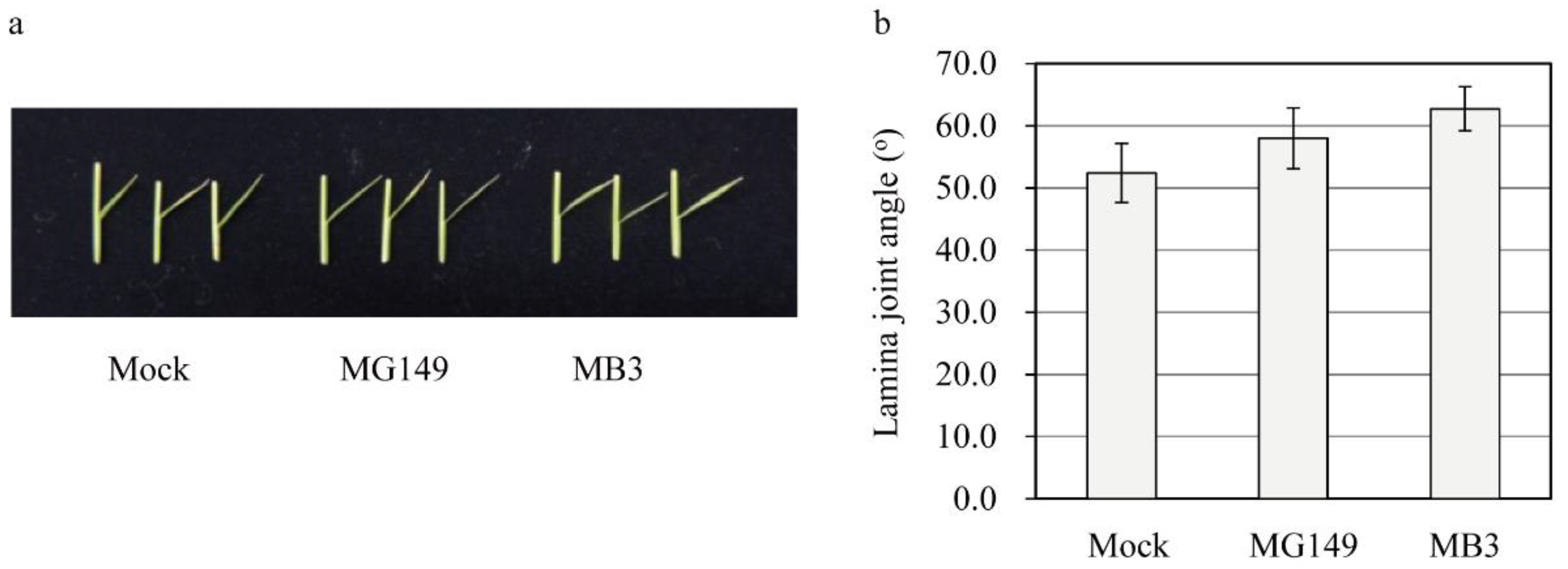
3.4. Measuring the Response of the Second Leaf Lamina Joint Angle to MG149
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Dubbels, R.; Reiter, R.J.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.W.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar] [PubMed]
- Tan, D.X.; Reiter, R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plant. J. Exp. Bot. 2020, 71, 4677–4689. [Google Scholar] [CrossRef]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Simultaneous suppression of two distinct serotonin N-acetyltransferase isogenes by RNA interference leads to severe decreases in melatonin and accelerated seed deterioration in rice. Biomolecules 2020, 10, 141. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Liu, L.; Wang, H.; Li, D.; Bai, Z.; Zhang, Y.; Sun, H.; Zhang, K.; Li, C. Exogenous melatonin accelerates seed germination in cotton (Gossypium hirsutum L.). PLoS ONE 2019, 14, e0216575. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Back, K. Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 2019, 66, e12537. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin in flowering, fruit set and fruit ripening. Plant Reprod. 2020, 33, 77–87. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019, 2, 10–36. [Google Scholar] [CrossRef]
- Ozgur, R.; Uzilday, B.; Turkan, I.; Sekmen, A.H. The effects of melatonin on transcriptional profile of unfolded protein response genes under endoplasmic reticulum stress in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2017, 35, 188–202. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin plays a pivotal role in conferring tolerance against endoplasmic reticulum stress via mitogen-activated protein kinases and bZIP60 in Arabidopsis thaliana. Melatonin Res. 2018, 1, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; He, Q.; Zhang, F.; Yu, J.; Li, C.; Zhao, T.; Zhang, Y.; Xie, Q.; Su, B.; Mei, L.; et al. Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis. Plant J. 2019, 100, 784–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Guo, Y.; Zhang, D.; He, S.; Gong, J.; Ma, H.; Gao, X.; Wang, Z.; Jiang, L.; Dun, X.; et al. Melatonin represses oil and anthocyanin accumulation in seeds. Plant Physiol. 2020, 183, 898–914. [Google Scholar] [CrossRef] [PubMed]
- Moustafa-Farag, M.; Elkelish, A.; Dafea, M.; Khan, M.; Arnao, M.B.; Abdelhamid, M.T.; El-Ezz, A.A.; Almoneafy, A.; Mahmoud, A.; Awad, M.; et al. Role of melatonin in plant tolerance to soil stressors: Salinity, pH and heavy metals. Molecules 2020, 25, 5359. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Laborda, P.; Liu, F. Exogenous melatonin enhances rice plant resistance against Xanthomonas oryzae pv. oryzae. Plant Dis. 2020, 104, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.R.; Bajwa, V.S.; Freixas-Coutin, J.A.; Saxena, P. Salt stress in Arabidopsis thaliana seedlings: Role of indoleamines in stress alleviation. Melatonin Res. 2021, 4, 70–83. [Google Scholar] [CrossRef]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. The phytomelatonin receptor (PMRT1) Arabidopsis Cand2 is not a bona fide G protein-coupled melatonin receptor. Melatonin Res. 2020, 3, 177–186. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and reactive oxygen and nitrogen species: A model for the plant redox network. Melatonin Res. 2019, 2, 152–168. [Google Scholar] [CrossRef]
- Khan, A.; Numan, M.; Khan, A.L.; Lee, I.J.; Imran, M.; Asaf, S.; Al-Harrasi, A. Melatonin: Awakening the defense mechanisms during plant oxidative stress. Plants 2020, 9, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Byeon, Y.; Back, K. Transcriptional suppression of tryptamine 5-hydroxylase, a terminal serotonin biosynthetic gene, induces melatonin biosynthesis in rice (Oryza sativa L.). J. Pineal Res. 2013, 55, 131–137. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef] [PubMed]
- Dekker, F.J.; van den Bosch, T.; Martin, N.I. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov. Today 2014, 19, 654–660. [Google Scholar] [CrossRef] [Green Version]
- Aquea, F.; Timmermann, T.; Herrera-Vásquez, A. Chemical inhibition of the histone acetyltransferase activity in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2017, 483, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, D.; Lin, Z.; Guan, B.; Liu, D.; Yang, L.; Deng, X.; Mei, F.; Zhou, Z. Histone acetylation modification affects cell wall degradation and aerenchyma formation in wheat seminal roots under waterlogging. Plant Growth Reg. 2018, 87, 149–163. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Xie, Q.; Liu, Y.; Lv, H.; Bai, R.; Ma, R.; Li, X.; Zhang, X.; Guo, Y.D.; et al. SISNAT interacts with HSP40, a molecular chaperone, to regulate melatonin biosynthesis and promote thermotolerance in tomato. Plant Cell Physiol. 2020, 61, 909–921. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y.; Uozu, S.; et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Lee, K.; Jang, K.; Seo, P.J. Circadian expression profiles of chromatin remodeling factor genes in Arabidopsis. J. Plant Res. 2015, 128, 187–199. [Google Scholar] [CrossRef]
- Ghizzoni, M.; Wu, J.; Gao, T.; Haisma, H.J.; Dekker, F.J. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur. J. Med. Chem. 2012, 47, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Choi, G.H.; Back, K. Cadmium-induced melatonin synthesis in rice requires light, hydrogen peroxide, and nitric oxide: Key regulatory roles for tryptophan decarboxylase and caffeic acid O-methyltransferase. J. Pineal Res. 2017, 63, e12441. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Hwang, O.J.; Lee, H.J.; Lee, K.; Back, K. Coordinated regulation of melatonin synthesis and degradation genes in rice leaves in response to cadmium treatment. J. Pineal Res. 2015, 58, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Hwang, O.J.; Back, K. Melatonin deficiency confers tolerance to multiple abiotic stresses in rice via decreased brassinosteroid levels. Int. J. Mol. Sci. 2019, 20, 5173. [Google Scholar] [CrossRef] [Green Version]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol. Cell Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Bourke, E.; Scobie, M.; Famme, M.A.; Koolmeister, T.; Helleday, T.; Eriksson, L.A.; Lowndes, N.F.; Brown, J.A.L. Rational design and validation of a Tip60 histone acetyltransferase inhibitor. Sci. Rep. 2014, 4, 5372. [Google Scholar] [CrossRef]
- Khalil, E.M.; Cole, P.A. A potent inhibitor of the melatonin rhythm enzyme. J. Am. Chem. Soc. 1998, 120, 6195–6196. [Google Scholar] [CrossRef]
- Shin, J.C.; Jung, H.Y.; Harikishore, A.; Kwon, O.D.; Yoon, H.S.; Kim, K.T.; Choi, B.H. The flavonoid myricetin reduces nocturnal melatonin levels in the blood through the inhibition of serotonin N-acetyltransferase. Biochem. Biophys. Res. Commun. 2013, 440, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Hwang, O.J.; Reiter, R.J.; Back, K. Flavonoids inhibit both rice and sheep serotonin N-acetyltransferases and reduce melatonin levels in plants. J. Pineal Res. 2018, 65, e12512. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. Melatonin regulates chloroplast protein quality control via a mitogen-activated protein kinase signaling pathway. Antioxidants 2021, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Hwang, O.J.; Back, K. Suppression of rice cryptochrome 1b decreases both melatonin and expression of brassinosteroid biosynthetic genes resulting in salt tolerance. Molecules 2021, 26, 1075. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Choi, G.-H.; Back, K. Inhibition of Rice Serotonin N-Acetyltransferases by MG149 Decreased Melatonin Synthesis in Rice Seedlings. Biomolecules 2021, 11, 658. https://doi.org/10.3390/biom11050658
Lee K, Choi G-H, Back K. Inhibition of Rice Serotonin N-Acetyltransferases by MG149 Decreased Melatonin Synthesis in Rice Seedlings. Biomolecules. 2021; 11(5):658. https://doi.org/10.3390/biom11050658
Chicago/Turabian StyleLee, Kyungjin, Geun-Hee Choi, and Kyoungwhan Back. 2021. "Inhibition of Rice Serotonin N-Acetyltransferases by MG149 Decreased Melatonin Synthesis in Rice Seedlings" Biomolecules 11, no. 5: 658. https://doi.org/10.3390/biom11050658
APA StyleLee, K., Choi, G.-H., & Back, K. (2021). Inhibition of Rice Serotonin N-Acetyltransferases by MG149 Decreased Melatonin Synthesis in Rice Seedlings. Biomolecules, 11(5), 658. https://doi.org/10.3390/biom11050658






