Highly Conserved C-Terminal Region of Indian Hedgehog N-Fragment Contributes to Its Auto-Processing and Multimer Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of HH Mutants
2.2. Construction of Stable Cell Line and Gel-Filtration Chromatography
2.3. IHH Activity Assay in C3H10T1/2 Cells
2.4. Transfection and Analysis of HH Mutants
2.5. Prediction of IHH Protein Structure
2.6. Cell Immunofluorescence of IHH Mutants
3. Results
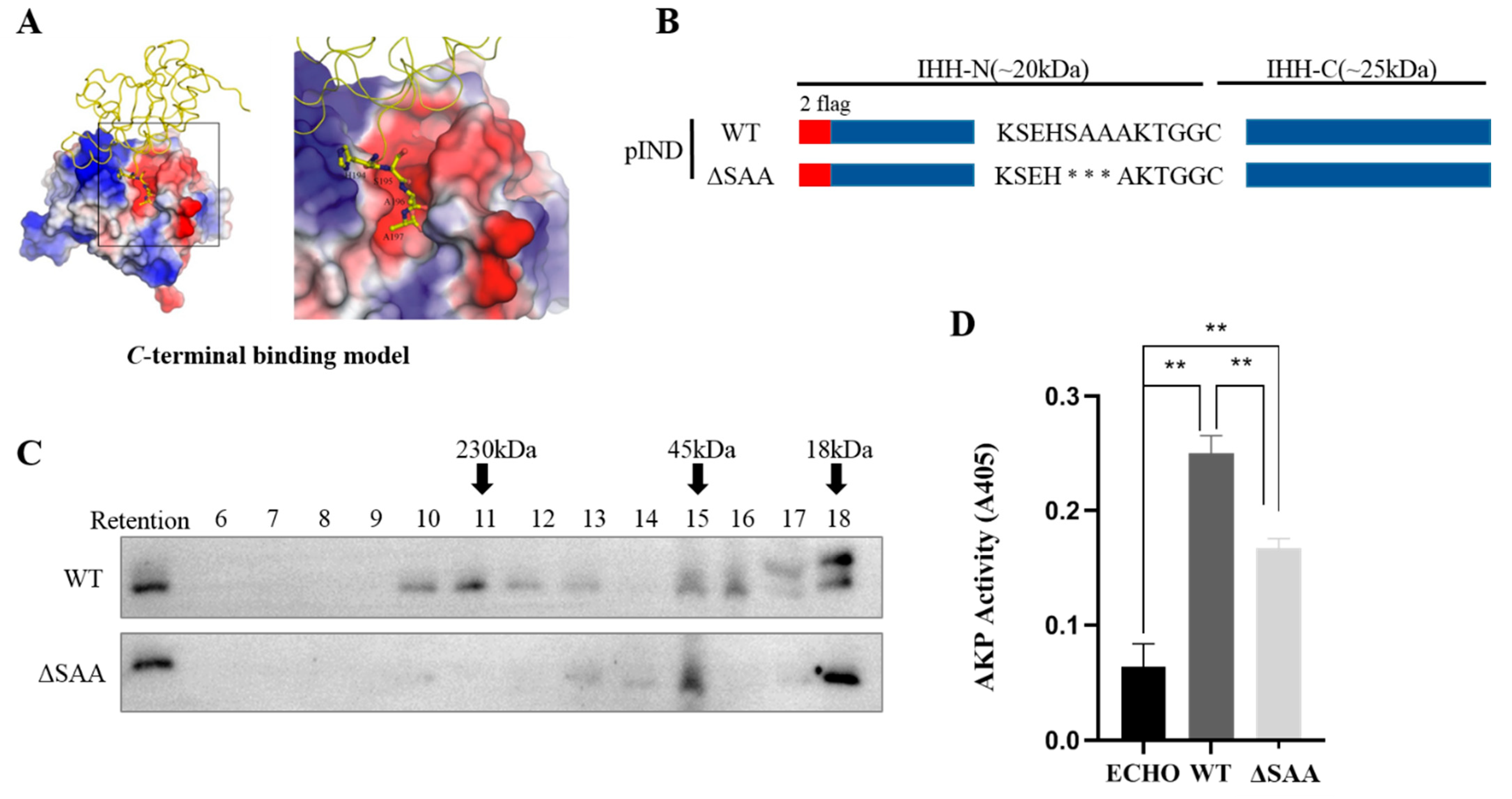
3.1. S195, A196, and A197 Are Involved in Multimerization of IHH-N Protein by Maintaining Intermolecular Interactions
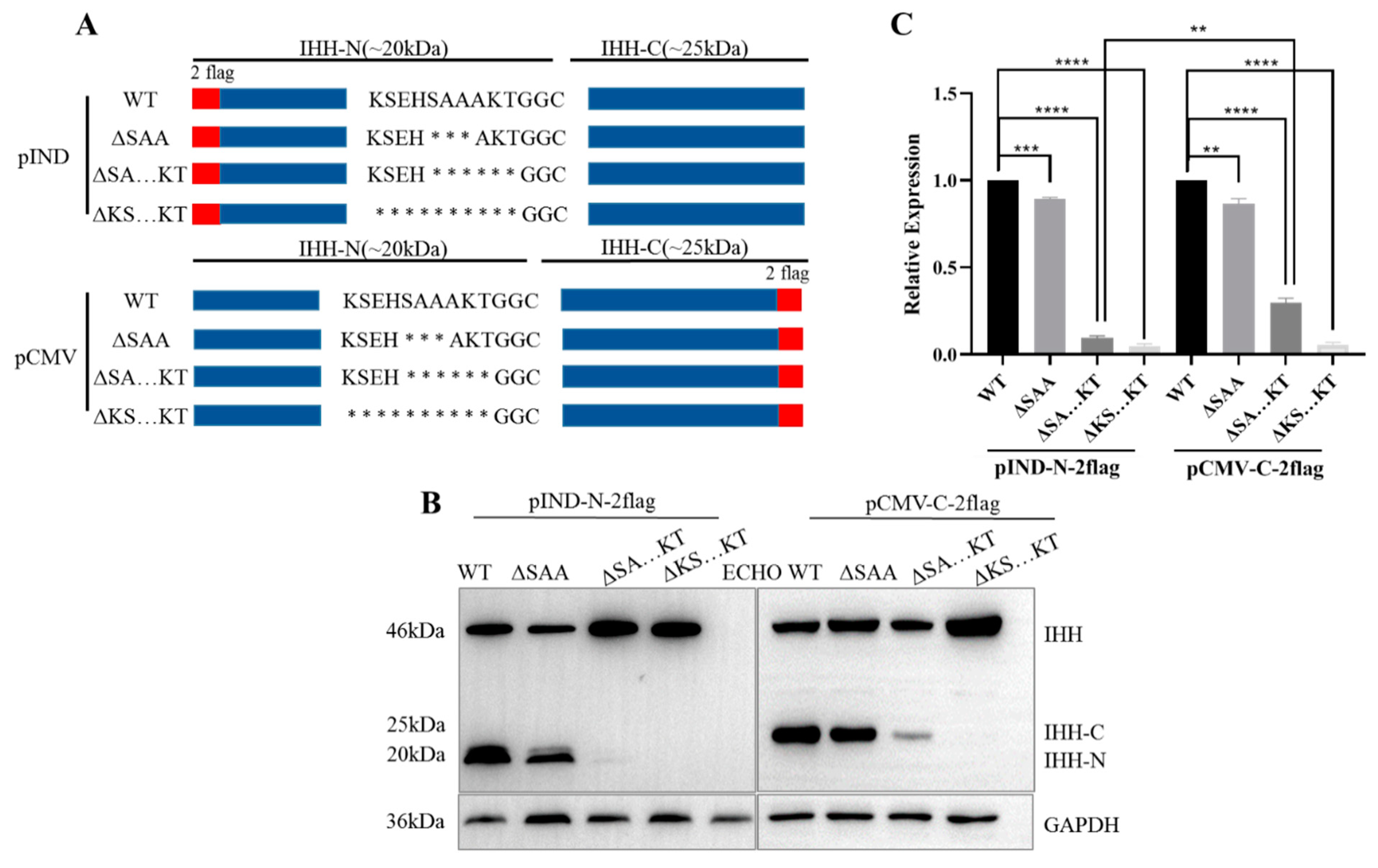
3.2. Conserved C-Terminus of IHH-N Fragment Affects Protein Self-Cleavage and Stability
3.3. K191, S192, E193, and H194 Synergistically Affect Self-Cleavage of IHH Precursor
3.4. K186, A187, E188, and N189 Also Severely Affect SHH Self-Cleavage by Obstructing the Presentation of the Cholesterol Modification Site
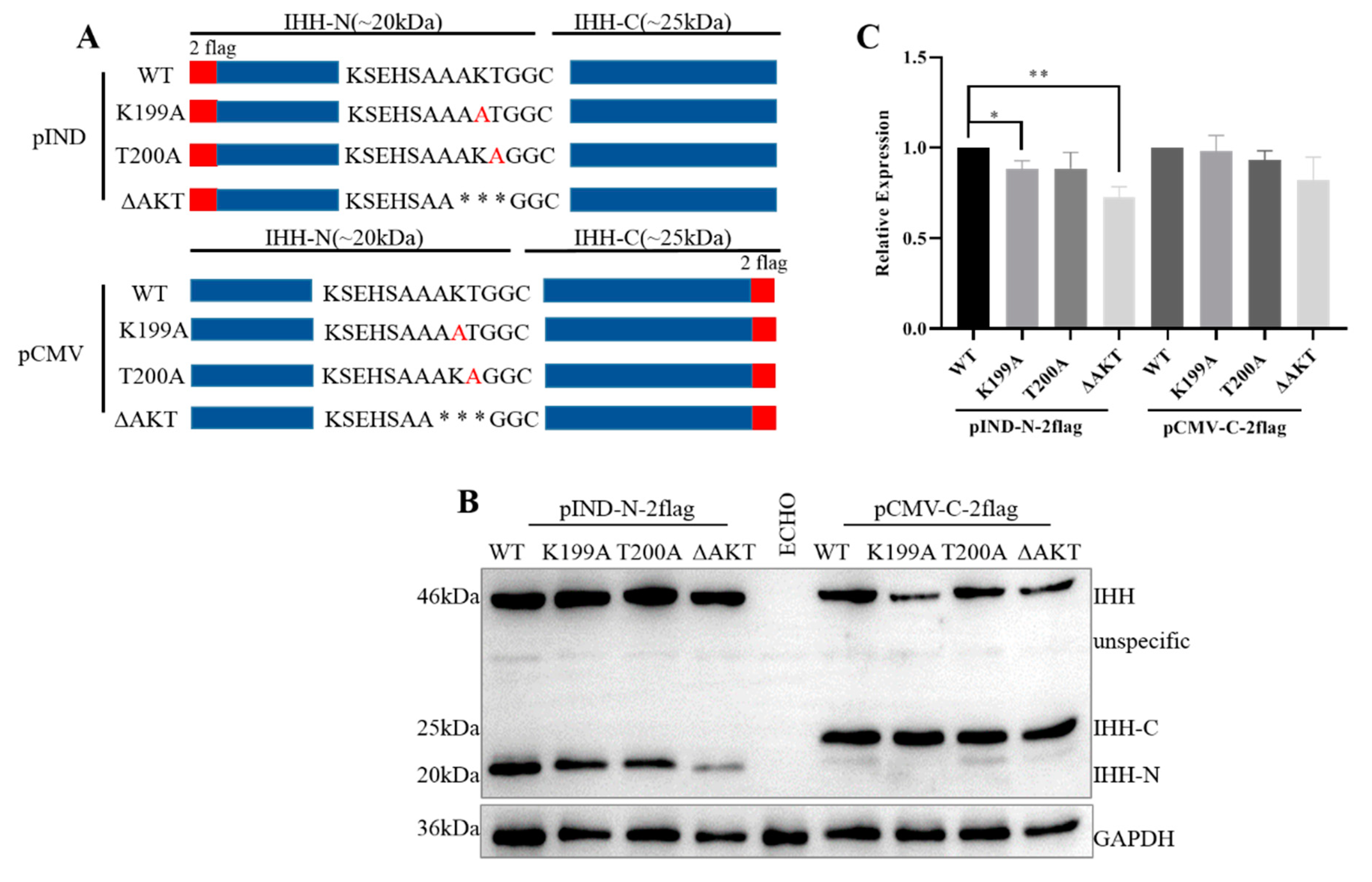
3.5. A198, K199, and T200 Evidently Affect the Stability of IHH-N
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Petrova, R.; Joyner, A.L. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhang, Y.; Sun, B.; McMahon, A.P.; Wang, Y. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem. Biol. 2017, 24, 252–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beachy, P.A.; Karhadkar, S.S.; Berman, D.M. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004, 432, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Andre, P.; Ye, L.; Yang, Y.Z. The Hedgehog signalling pathway in bone formation. Int. J. Oral Sci. 2015, 7, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.A.; Von Kessler, D.P.; Ekker, S.C.; Young, K.E.; Lee, J.J.; Moses, K.; Beachy, P.A. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 1995, 374, 363–366. [Google Scholar] [CrossRef]
- Lee, J.; Ekker, S.C.; Von Kessler, D.P.; Porter, J.A.; Sun, B.I.; Beachy, P.A. Autoproteolysis in hedgehog protein biogenesis. Science 1994, 266, 1528–1537. [Google Scholar] [CrossRef]
- Mann, R.K.; Beachy, P.A. Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 2004, 73, 891–923. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Li, Y.J.; Kawakami, T.; Xu, S.M.; Chuang, P.T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004, 18, 641–659. [Google Scholar] [CrossRef] [Green Version]
- Byrnes, A.M.; Racacho, L.; Grimsey, A.; Hudgins, L.; Kwan, A.C.; Sangalli, M.; Kidd, A.; Yaron, Y.; Lau, Y.L.; Nikkel, S.M.; et al. Brachydactyly A-1 mutations restricted to the central region of the N-terminal active fragment of Indian Hedgehog. Eur. J. Hum. Genet. 2009, 17, 1112–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, J.B.; Choi, Y.; Segal, R.A. Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 2002, 129, 2223–2232. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Petrov, K.; Watanabe, M.; Salic, A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. USA 2016, 113, E5866–E5875. [Google Scholar] [CrossRef] [Green Version]
- Goetz, J.A.; Singh, S.; Suber, L.M.; Kull, F.J.; Robbins, D.J. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J. Biol. Chem. 2006, 281, 4087–4093. [Google Scholar] [CrossRef] [Green Version]
- Traiffort, E.; Dubourg, C.; Faure, H.; Rognan, D.; Odent, S.; Durou, M.R.; David, V.; Ruat, M. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J. Biol. Chem. 2004, 279, 42889–42897. [Google Scholar] [CrossRef] [Green Version]
- Ma, G.; Yu, J.; Xiao, Y.; Chan, D.; Gao, B.; Hu, J.; He, Y.; Guo, S.; Zhou, J.; Zhang, L.; et al. Indian hedgehog mutations causing brachydactyly type A1 impair Hedgehog signal transduction at multiple levels. Cell Res. 2011, 21, 1343–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.M.T.; Porter, J.A.; Beachy, P.A.; Leahy, D.J. A potential catalytic site revealed by the 1.7-Å crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 1995, 378, 212–216. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Tang, J.J.; Peng, C.; Wang, Y.; Fu, L.; Qiu, Z.P.; Xiong, Y.; Yang, L.F.; Cui, H.W.; He, X.L.; et al. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol. Cell 2017, 66, 154–162.e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.M.T.; Porter, J.A.; Young, K.E.; Koonin, E.V.; Beachy, P.A.; Leahy, D.J. Crystal Structure of a Hedgehog Autoprocessing Domain: Homology between Hedgehog and Self-Splicing Proteins. Cell 1997, 91, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Vincent, S.M.; Thomas, A.; Brasher, B.B.; Benson, J.D. Targeting of proteins to membranes through hedgehog auto-processing. Nat. Biotechnol. 2003, 21, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Guo, J.; She, C.; Shu, A.; Yang, M.; Tan, Z.; Yang, X.; Guo, S.; Feng, G.; He, L. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat. Genet. 2001, 28, 386–388. [Google Scholar] [CrossRef]
- Hellemans, J.; Coucke, P.J.; Giedion, A.; De Paepe, A.; Kramer, P.; Beemer, F.; Mortier, G.R. Homozygous mutations in IHH cause acrocapitofemoral dysplasia, an autosomal recessive disorder with cone-shaped epiphyses in hands and hips. Am. J. Hum. Genet. 2003, 72, 1040–1046. [Google Scholar] [CrossRef] [Green Version]
- Fuse, N.; Maiti, T.; Wang, B.; Porter, J.A.; Hall, T.M.T.; Leahy, D.J.; Beachy, P.A. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for Patched. Proc. Natl. Acad. Sci. USA 1999, 96, 10992–10999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavran, J.M.; Ward, M.D.; Oladosu, O.O.; Mulepati, S.; Leahy, D.J. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J. Biol. Chem. 2010, 285, 24584–24590. [Google Scholar] [CrossRef] [Green Version]
- Bishop, B.; Aricescu, A.R.; Harlos, K.; O’Callaghan, C.A.; Jones, E.Y.; Siebold, C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat. Struct. Mol. Biol. 2009, 16, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Cao, P.; Hu, M.; Gao, S.; Yan, N.; Gong, X. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat. Commun. 2019, 10, 2320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, J.A.; Ekker, S.C.; Park, W.J.; Von Kessler, D.P.; Young, K.E.; Chen, C.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V. Hedgehog Patterning Activity: Role of a Lipophilic Modification Mediated by the Carboxy-Terminal Autoprocessing Domain. Cell 1996, 86, 21–34. [Google Scholar] [CrossRef] [Green Version]
- McLellan, J.S.; Zheng, X.; Hauk, G.; Ghirlando, R.; Beachy, P.A.; Leahy, D.J. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 2008, 455, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; White, B.; Tyurina, O.V.; Guner, B.; Larson, T.; Lee, H.Y.; Karlstrom, R.O.; Kohtz, J.D. Synergistic and antagonistic roles of the Sonic hedgehog N- and C-terminal lipids. Development 2004, 131, 4357–4370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietveld, A.; Neutz, S.; Simons, K.; Eaton, S. Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J. Biol. Chem. 1999, 274, 12049–12054. [Google Scholar] [CrossRef] [Green Version]
- Chamoun, Z.; Mann, R.K.; Nellen, D.; von Kessler, D.P.; Bellotto, M.; Beachy, P.A.; Basler, K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 2001, 293, 2080–2084. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, H.; Liu, Y.; Han, G.; Wang, Y.; Chen, H.; He, L.; Ma, G. Highly Conserved C-Terminal Region of Indian Hedgehog N-Fragment Contributes to Its Auto-Processing and Multimer Formation. Biomolecules 2021, 11, 792. https://doi.org/10.3390/biom11060792
Wang X, Liu H, Liu Y, Han G, Wang Y, Chen H, He L, Ma G. Highly Conserved C-Terminal Region of Indian Hedgehog N-Fragment Contributes to Its Auto-Processing and Multimer Formation. Biomolecules. 2021; 11(6):792. https://doi.org/10.3390/biom11060792
Chicago/Turabian StyleWang, Xiaoqing, Hao Liu, Yanfang Liu, Gefei Han, Yushu Wang, Haifeng Chen, Lin He, and Gang Ma. 2021. "Highly Conserved C-Terminal Region of Indian Hedgehog N-Fragment Contributes to Its Auto-Processing and Multimer Formation" Biomolecules 11, no. 6: 792. https://doi.org/10.3390/biom11060792






