A Thermodynamic Analysis of the Binding Specificity between Four Human PDZ Domains and Eight Host, Viral and Designed Ligands
Abstract
:1. Introduction
- Class I: X-(S/T)-X-ϕ-COOH;
- Class II: X-ϕ-X-ϕ-COOH;
- Class III: X-(D/E)-X-ϕ-COOH.
2. Materials and Methods
2.1. PDZ Domains Purification and Peptide Samples
2.2. Isothermal Titration Calorimetry (ITC)
2.3. Differential Scanning Calorimetry (DSC)
3. Results
3.1. Rationale of the Choice of the PDZ Domains and Binding Partners
3.2. ITC Experiments Between the PDZ Domains and Their Ligands
3.3. DSC Experiments for the ITC That Did Not Show Binding
3.4. Analysis of the Energetic Balance of the Interactions
3.5. Comparison With Other PDZ–Ligand Interactions in Bibliography
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nourry, C.; Grant, S.G.N.; Borg, J.-P. PDZ Domain Proteins: Plug and Play! Sci. STKE 2003, 2003, 1–12. [Google Scholar] [CrossRef]
- Kim, E.; Sheng, M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004, 5, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Schuetz, A. Structural Features of Tight-Junction Proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, C.D.; Roberts, S. Viral Interactions with PDZ Domain-Containing Proteins—An Oncogenic Trait? Pathogens 2016, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Torres-Flores, J.M.; Arias, C.F. Tight Junctions Go Viral! Viruses 2015, 7, 5145–5154. [Google Scholar] [CrossRef] [PubMed]
- Javier, R.T.; Rice, A.P. Emerging Theme: Cellular PDZ Proteins as Common Targets of Pathogenic Viruses. J. Virol. 2011, 85, 11544–11556. [Google Scholar] [CrossRef] [Green Version]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.-H.; Reva, B.; A Held, H.; Appleton, B.; Evangelista, M.; Wu, Y.; et al. A Specificity Map for the PDZ Domain Family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luck, K.; Charbonnier, S.; Travé, G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012, 586, 2648–2661. [Google Scholar] [CrossRef] [Green Version]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. JSBX 2020, 4, 100022–100040. [Google Scholar] [CrossRef]
- Chemes, L.B.; Gay, G.P.; Sánchez, I.E. Convergent evolution and mimicry of protein linear motifs in host–pathogen interactions. Curr. Opin. Struct. Biol. 2015, 32, 91–101. [Google Scholar] [CrossRef]
- Murciano-Calles, J.; Cobos, E.S.; Mateo, P.L.; Camara-Artigas, A.; Martinez, J.C. An Oligomeric Equilibrium Intermediate as the Precursory Nucleus of Globular and Fibrillar Supramacromolecular Assemblies in a PDZ Domain. Biophys. J. 2010, 99, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Murciano-Calles, J.; Guell-Bosch, J.; Villegas, S.; Martinez, J.C. Common features in the unfolding and misfolding of PDZ domains and beyond: The modulatory effect of domain swapping and extra-elements. Sci. Rep. 2016, 6, 19242–19248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Analytical biochemistry 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Murciano-Calles, J.; Coello, A.; Camara-Artigas, A.; Martinez, J.C. PDZ/PDZ interaction between PSD-95 and nNOS neuronal proteins. J. Mol. Recognit. 2019, 33, 1–11. [Google Scholar] [CrossRef]
- Sheng, M.; Sala, C. PDZ Domains and the Organization of Supramolecular Complexes. Annu. Rev. Neurosci. 2001, 24, 1–29. [Google Scholar] [CrossRef]
- A Chevalier, S.; Meertens, L.; Pise-Masison, C.; Calattini, S.; Park, H.; A Alhaj, A.; Zhou, M.; Gessain, A.; Kashanchi, F.; Brady, J.N.; et al. The tax protein from the primate T-cell lymphotropic virus type 3 is expressed in vivo and is functionally related to HTLV-1 Tax rather than HTLV-2 Tax. Oncogene 2006, 25, 4470–4482. [Google Scholar] [CrossRef] [Green Version]
- Rousset, R.; Fabre, S.; Desbois, C.; Bantignies, F.; Jalinot, P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene 1998, 16, 643–654. [Google Scholar] [CrossRef] [Green Version]
- Laura, R.P.; Witt, A.S.; Held, H.A.; Gerstner, R.; Deshayes, K.; Koehler, M.F.; Kosik, K.S.; Sidhu, S.S.; Lasky, L.A. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 2002, 277, 12906–12914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.Z.; Wang, Q.; Xiong, W.C.; Mei, L. Erbin Is a Protein Concentrated at Postsynaptic Membranes That Interacts with PSD-95. J. Biol. Chem. 2001, 276, 19318–19326. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Wang, W.; Li, M.; Liu, Y.; Zheng, D. Tax1 enhances cancer cell proliferation via Ras-Raf-MEK-ERK signaling pathway. IUBMB Life 2009, 61, 685–692. [Google Scholar] [CrossRef]
- Gassmann, M.; Lemke, G. Neuregulins and neuregulin receptors in neural development. Curr. Opin. Neurobiol. 1997, 7, 87–92. [Google Scholar] [CrossRef]
- Garcia, R.A.G.; Vasudevan, K.; Buonanno, A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc. Natl. Acad. Sci. USA 2000, 97, 3596–3601. [Google Scholar] [CrossRef] [PubMed]
- Skelton, N.J.; Koehler, M.F.; Zobel, K.; Wong, W.L.; Yeh, S.; Pisabarro, M.T.; Yin, J.P.; Lasky, L.A.; Sidhu, S.S. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 2003, 278, 7645–7654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillier, B.J.; Christopherson, K.S.; Prehoda, K.E.; Bredt, D.S.; Lim, W.A. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 1999, 284, 812–815. [Google Scholar] [CrossRef] [Green Version]
- E Brenman, J.; Chao, D.; Gee, S.H.; McGee, A.W.; E Craven, S.; Santillano, D.R.; Wu, Z.; Huang, F.; Xia, H.; Peters, M.F.; et al. Interaction of Nitric Oxide Synthase with the Postsynaptic Density Protein PSD-95 and α1-Syntrophin Mediated by PDZ Domains. Cell 1996, 84, 757–767. [Google Scholar] [CrossRef] [Green Version]
- Gracia, J.M.; Costas-Insua, C.; Canales, A.; Rodríguez-Crespo, I. Insights into the C-terminal Peptide Binding Specificity of the PDZ Domain of Neuronal Nitric-oxide Synthase. J. Biol. Chem. 2016, 291, 11581–11595. [Google Scholar] [CrossRef] [Green Version]
- Tochio, H.; Mok, Y.K.; Zhang, Q.; Kan, H.M.; Bredt, D.S.; Zhang, M. Formation of nNOS/PSD-95 PDZ dimer requires a preformed beta-finger structure from the nNOS PDZ domain. J. Mol. Biol. 2000, 303, 359–370. [Google Scholar] [CrossRef]
- Jaffrey, S.; Snowman, A.M.; Eliasson, M.J.; Cohen, N.; Snyder, S.H. CAPON: A Protein Associated with Neuronal Nitric Oxide Synthase that Regulates Its Interactions with PSD95. Neuron 1998, 20, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Tochio, H.; Zhang, Q.; Mandal, P.; Li, M.; Zhang, M. Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat. Genet. 1999, 6, 417–421. [Google Scholar] [CrossRef]
- Palencia, A.; Cobos, E.S.; Mateo, P.L.; Martı́nez, J.C.; Luque, I. Thermodynamic Dissection of the Binding Energetics of Proline-rich Peptides to the Abl-SH3 Domain: Implications for Rational Ligand Design. J. Mol. Biol. 2004, 336, 527–537. [Google Scholar] [CrossRef]
- Borg, J.-P.; Marchetto, S.; Le Bivic, A.; Ollendorff, V.; Jaulin-Bastard, F.; Saito, H.; Fournier, E.; Adélaïde, J.; Margolis, B.; Birnbaum, D. ERBIN: A basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat. Cell Biol. 2000, 2, 407–414. [Google Scholar] [CrossRef]
- Birrane, G.; Chung, J.; Ladias, J.A.A. Novel Mode of Ligand Recognition by the Erbin PDZ Domain. J. Biol. Chem. 2003, 278, 1399–1402. [Google Scholar] [CrossRef] [Green Version]
- Jelesarov, I.; Bosshard, H.R. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 1999, 12, 3–18. [Google Scholar] [CrossRef]
- Murciano-Calles, J.; Marin-Argany, M.; Cobos, E.S.; Villegas, S.; Martinez, J.C. The Impact of Extra-Domain Structures and Post-Translational Modifications in the Folding/Misfolding Behaviour of the Third PDZ Domain of MAGUK Neuronal Protein PSD-95. PLoS ONE 2014, 9, e98124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murciano-Calles, J.; Corbi-Verge, C.; Candel, A.M.; Luque, I.; Martinez, J.C. Post-Translational Modifications Modulate Ligand Recognition by the Third PDZ Domain of the MAGUK Protein PSD-95. PLoS ONE 2014, 9, e90030. [Google Scholar] [CrossRef]
- Jen-Jacobson, L.; Engler, L.E.; Jacobson, L.A. Structural and Thermodynamic Strategies for Site-Specific DNA Binding Proteins. Structure 2000, 8, 1015–1023. [Google Scholar] [CrossRef]
- Krug, R.R.; Hunter, W.G.; Grieger, R.A. Enthalpy-entropy compensation. 1. Some fundamental statistical problems associated with the analysis of van’t Hoff and Arrhenius data. J. Phys. Chem. 1976, 80, 2335–2341. [Google Scholar] [CrossRef]
- Krug, R.R.; Hunter, W.G.; Grieger, R.A. Enthalpy-entropy compensation. 2. Separation of the chemical from the statistical effect. J. Phys. Chem. 1976, 80, 2341–2351. [Google Scholar] [CrossRef]
- Takeda, Y.; Ross, P.D.; Mudd, C.P. Thermodynamics of Cro protein-DNA interactions. Proc. Natl. Acad. Sci. USA 1992, 89, 8180–8184. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Mori, T.; Inoue, Y. Supramolecular Enantiodifferentiating Photocyclodimerization of 2-Anthracenecarboxylate Mediated by Capped γ-Cyclodextrins: Critical Control of Enantioselectivity by Cap Rigidity. J. Org. Chem. 2008, 73, 5786–5794. [Google Scholar] [CrossRef] [PubMed]
- Leffler, J.E. The Enthalpy-Entropy Relationship and its Implications for Organic Chemestry. J. Org. Chem. 1955, 20, 1202–1231. [Google Scholar] [CrossRef]
- Saro, D.; Klosi, E.; Paredes, A.A.; Spaller, M.R. Thermodynamic Analysis of a Hydrophobic Binding Site: Probing the PDZ Domain with Nonproteinogenic Peptide Ligands. Org. Lett. 2004, 6, 3429–3432. [Google Scholar] [CrossRef] [PubMed]
- Saro, D.; Li, T.; Rupasinghe, C.; Paredes, A.; Caspers, N.; Spaller, M.R. A Thermodynamic Ligand Binding Study of the Third PDZ Domain (PDZ3) from the Mammalian Neuronal Protein PSD-95. Biochemistry 2007, 46, 6340–6352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivarsson, Y.; Arnold, R.; McLaughlin, M.; Nim, S.; Joshi, R.; Ray, D.; Liu, B.; Teyra, J.; Pawson, T.; Moffat, J.; et al. Large-scale interaction profiling of PDZ domains through proteomic peptide-phage display using human and viral phage peptidomes. Proc. Natl. Acad. Sci. USA 2014, 111, 2542–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babault, N.; Cordier, F.; Lafage, M.; Cockburn, J.; Haouz, A.; Prehaud, C.; Rey, F.; Delepierre, M.; Buc, H.; Lafon, M.; et al. Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death. Structure 2011, 19, 1518–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delhommel, F.; Chaffotte, A.; Terrien, E.; Raynal, B.; Buc, H.; Delepierre, M.; Cordier, F.; Wolff, N. Deciphering the unconventional peptide binding to the PDZ domain of MAST2. Biochem. J. 2015, 469, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Terrien, E.; Delhommel, F.; Lefebvre-Omar, C.; Bohl, D.; Vitry, S.; Bernard, C.; Ramirez, J.; Chaffotte, A.; Ricquier, K.; et al. Structure-based optimization of a PDZ-binding motif within a viral peptide stimulates neurite outgrowth. J. Biol. Chem. 2019, 294, 13755–13768. [Google Scholar] [CrossRef]
- Terrien, E.; Chaffotte, A.; Lafage, M.; Khan, Z.; Préhaud, C.; Cordier, F.; Simenel, C.; Delepierre, M.; Buc, H.; Lafon, M.; et al. Interference with the PTEN-MAST2 Interaction by a Viral Protein Leads to Cellular Relocalization of PTEN. Sci. Signal. 2012, 5, 1–12. [Google Scholar] [CrossRef]
- Sharma, S.C.; Rupasinghe, C.N.; Parisien, R.B.; Spaller, M.R. Design, Synthesis, and Evaluation of Linear and Cyclic Peptide Ligands for PDZ10 of the Multi-PDZ Domain Protein MUPP1. Biochemistry 2007, 46, 12709–12720. [Google Scholar] [CrossRef]
- Cascio, E.L.; Toto, A.; Babini, G.; De Maio, F.; Sanguinetti, M.; Mordente, A.; Della Longa, S.; Arcovito, A. Structural determinants driving the binding process between PDZ domain of wild type human PALS1 protein and SLiM sequences of SARS-CoV E proteins. Comput. Struct. Biotechnol. J. 2021, 19, 1838–1847. [Google Scholar] [CrossRef]
- Fournane, S.; Charbonnier, S.; Chapelle, A.; Kieffer, B.; Orfanoudakis, G.; Travé, G.; Masson, M.; Nominé, Y. Surface plasmon resonance analysis of the binding of high-risk mucosal HPV E6 oncoproteins to the PDZ1 domain of the tight junction protein MAGI-1. J. Mol. Recognit. 2010, 24, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.M.; Zhang, J.; Sapienza, P.J.; Fuentes, E.; Lee, A.L. Hidden dynamic allostery in a PDZ domain. Proc. Natl. Acad. Sci. USA 2009, 106, 18249–18254. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, K.; McLaughlin, R.; Ranganathan, R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell 2011, 147, 1564–1575. [Google Scholar] [CrossRef] [Green Version]
- Doyle, D.A.; Lee, A.; Lewis, J.; Kim, E.; Sheng, M.; MacKinnon, R. Crystal Structures of a Complexed and Peptide-Free Membrane Protein–Binding Domain: Molecular Basis of Peptide Recognition by PDZ. Cell 1996, 85, 1067–1076. [Google Scholar] [CrossRef] [Green Version]
- Calosci, N.; Chi, C.; Richter, B.; Camilloni, C.; Engstrom, A.; Eklund, L.; Travaglini-Allocatelli, C.; Gianni, S.; Vendruscolo, M.; Jemth, P. Comparison of successive transition states for folding reveals alternative early folding pathways of two homologous proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 19241–19246. [Google Scholar] [CrossRef] [Green Version]
- Gautier, C.; Visconti, L.; Jemth, P.; Gianni, S. Addressing the role of the α-helical extension in the folding of the third PDZ domain from PSD-95. Sci. Rep. 2017, 7, 12593–12600. [Google Scholar] [CrossRef]
- Murciano-Calles, J.; Martinez, J.C.; Marin-Argany, M.; Villegas, S.; Cobos, E.S. A thermodynamic study of the third PDZ domain of MAGUK neuronal protein PSD-95 reveals a complex three-state folding behavior. Biophys. Chem. 2014, 185, 1–7. [Google Scholar] [CrossRef]
- Kalia, L.V.; Salter, M.W. Interactions between Src family protein tyrosine kinases and PSD-95. Neuropharmacology 2003, 45, 720–728. [Google Scholar] [CrossRef]
- Long, J.-F.; Tochio, H.; Wang, P.; Fan, J.-S.; Sala, C.; Niethammer, M.; Sheng, M.; Zhang, M. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J. Mol. Biol. 2003, 327, 203–214. [Google Scholar] [CrossRef]
- Sainlos, M.; Tigaret, C.; Poujol, C.; Olivier, N.B.; Bard, L.; Breillat, C.; Thiolon, K.; Choquet, D.; Imperiali, B. Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Nat. Chem. Biol. 2010, 7, 81–91. [Google Scholar] [CrossRef]
- Ernst, A.; Gfeller, D.; Kan, Z.; Seshagiri, S.; Kim, P.M.; Bader, G.; Sidhu, S.S. Coevolution of PDZ domain–ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol. BioSyst. 2010, 6, 1782–1790. [Google Scholar] [CrossRef]
- Karlsson, O.A.; Sundell, G.N.; Andersson, E.; Ivarsson, Y.; Jemth, P. Improved affinity at the cost of decreased specificity: A recurring theme in PDZ-peptide interactions. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Bavagnoli, L.; Dundon, W.G.; Garbelli, A.; Zecchin, B.; Milani, A.; Parakkal, G.; Baldanti, F.; Paolucci, S.; Volmer, R.; Tu, Y.; et al. The PDZ-Ligand and Src-Homology Type 3 Domains of Epidemic Avian Influenza Virus NS1 Protein Modulate Human Src Kinase Activity during Viral Infection. PLoS ONE 2011, 6, e27789. [Google Scholar] [CrossRef] [Green Version]
- Belotti, E.; Polanowska, J.; Daulat, A.M.; Audebert, S.; Thomé, V.; Lissitzky, J.-C.; Lembo, F.; Blibek, K.; Omi, S.; Lenfant, N.; et al. The Human PDZome: A Gateway to PSD95-Disc Large-Zonula Occludens (PDZ)-mediated Functions. Mol. Cell. Proteom. 2013, 12, 2587–2603. [Google Scholar] [CrossRef] [Green Version]
- Genera, M.; Quioc-Salomon, B.; Nourisson, A.; Colcombet-Cazenave, B.; Haouz, A.; Mechaly, A.; Matondo, M.; Duchateau, M.; König, A.; Windisch, M.P.; et al. Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Melik, W.; Ellencrona, K.; Wigerius, M.; Hedström, C.; Elväng, A.; Johansson, M. Two PDZ binding motifs within NS5 have roles in Tick-borne encephalitis virus replication. Virus Res. 2012, 169, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.; Myers, M.P.; Massimi, P.; Guarnaccia, C.; Banks, L. Analysis of Multiple HPV E6 PDZ Interactions Defines Type-Specific PDZ Fingerprints That Predict Oncogenic Potential. PLoS Pathog. 2016, 12, e1005766. [Google Scholar] [CrossRef] [PubMed]
- Vincentelli, R.; Luck, K.; Poirson, J.; Polanowska, J.; Abdat, J.; Blémont, M.; Turchetto, J.; Iv, F.; Ricquier, K.; Straub, M.-L.; et al. Quantifying domain-ligand affinities and specificities by high-throughput holdup assay. Nat. Methods 2015, 12, 787–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murciano-Calles, J. The Conformational Plasticity Vista of PDZ Domains. Life 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Gordon, T.B.; Hayward, J.A.; Marsh, G.A.; Baker, M.L.; Tachedjian, G. Host and Viral Proteins Modulating Ebola and Marburg Virus Egress. Viruses 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]

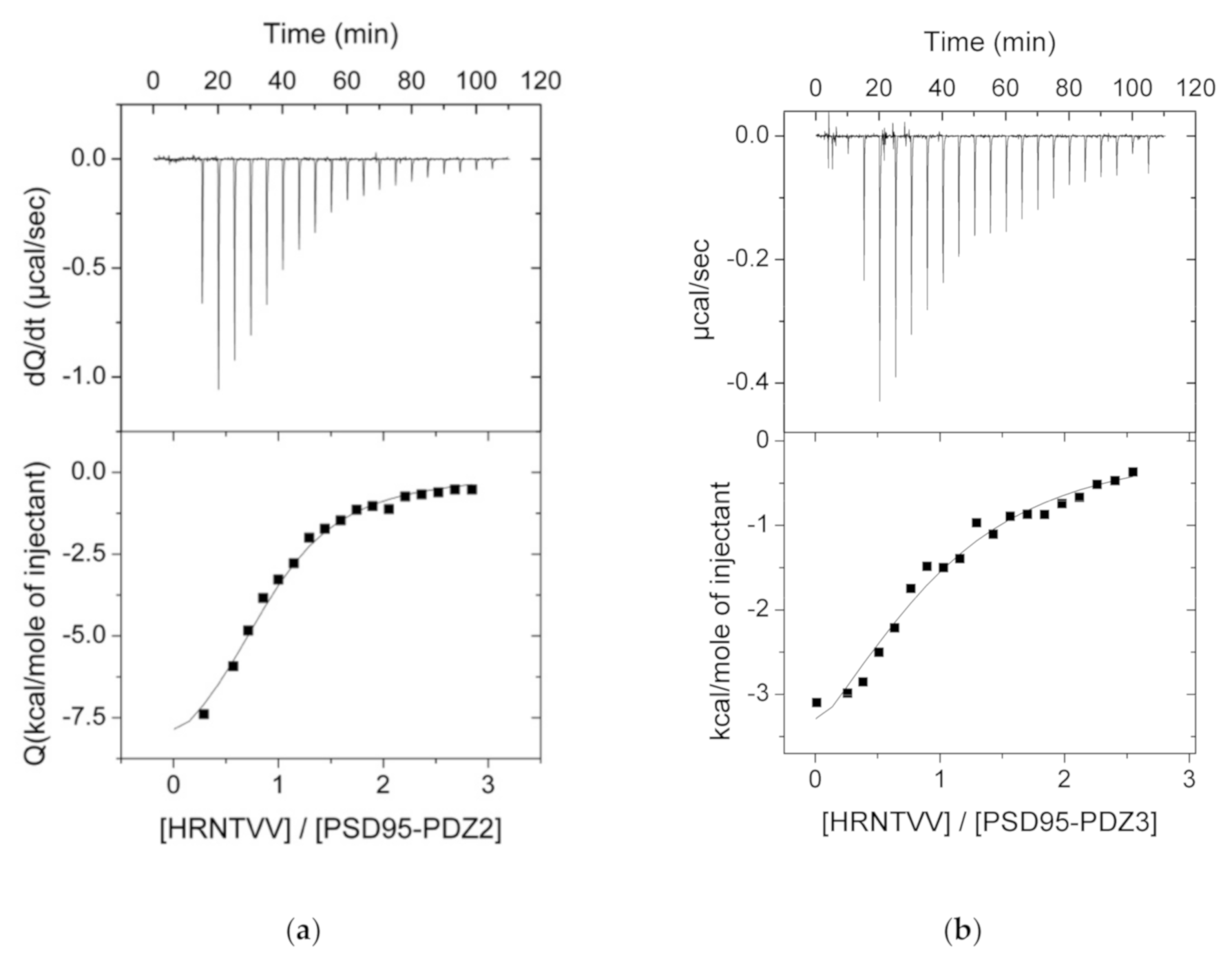
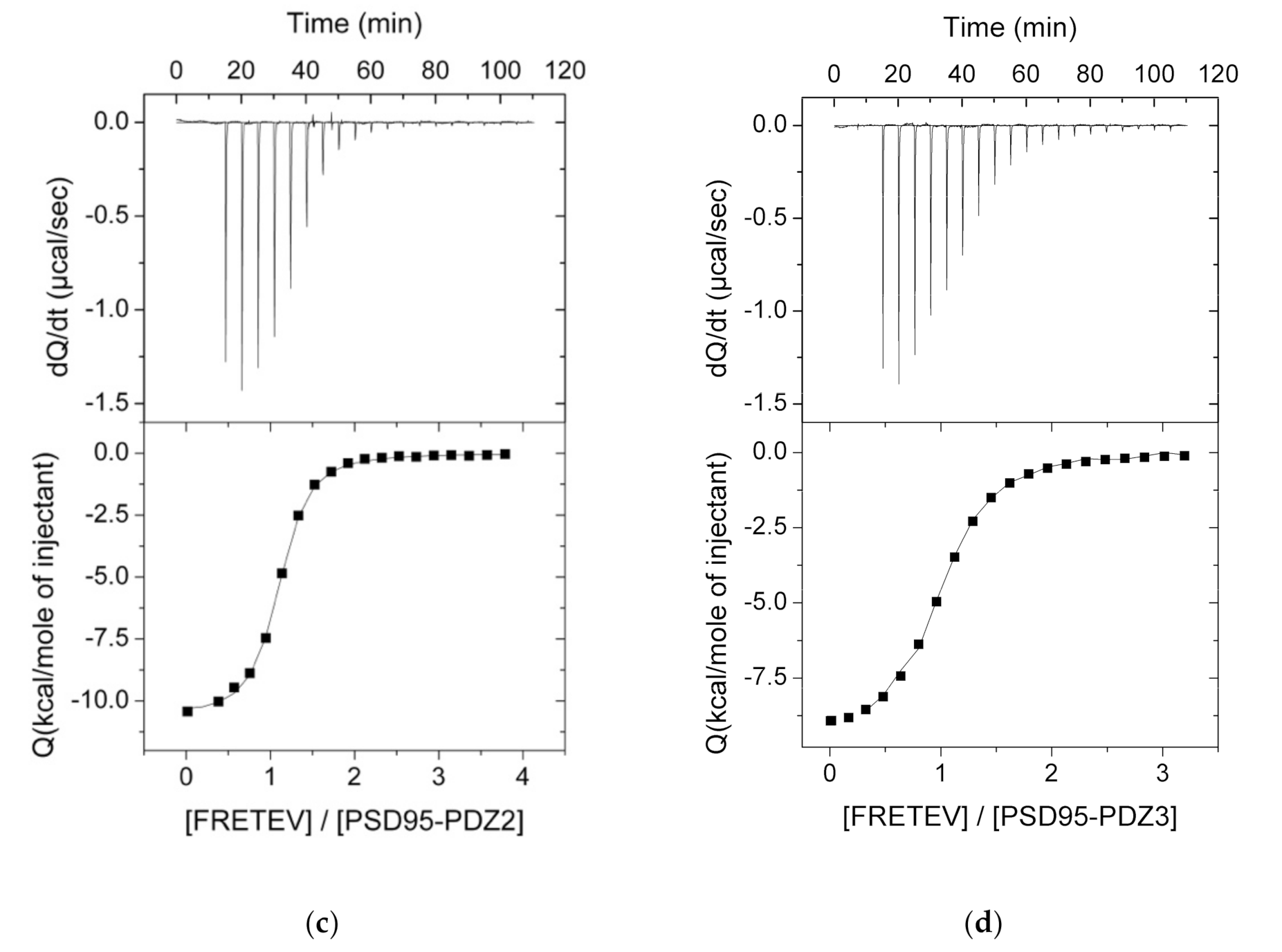
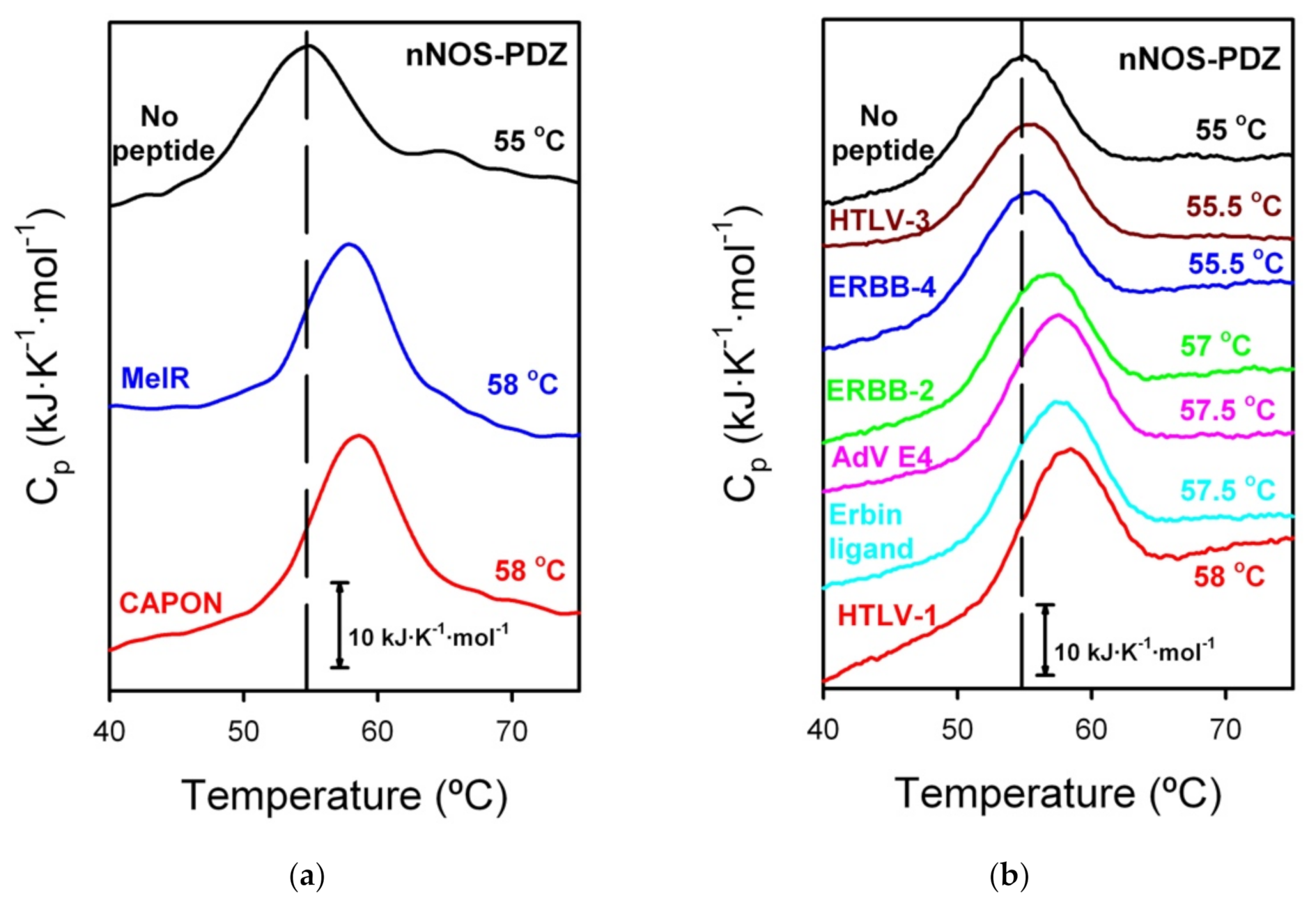

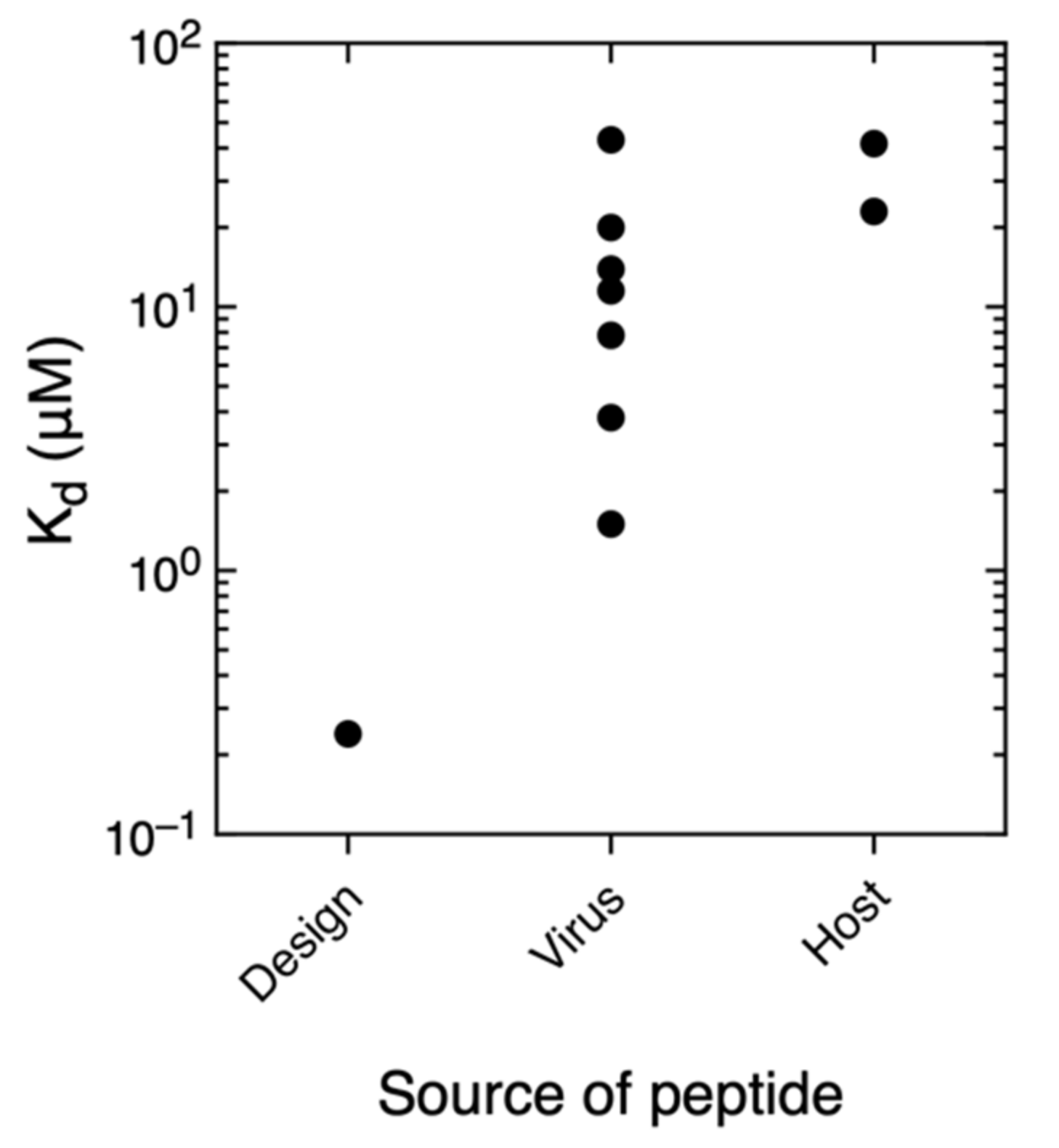
| Source | PDZ | Kd (μM) | ΔH (kcal·mol−1) | −Τ·ΔS (kcal·mol−1) | ΔG (kcal·mol−1) |
|---|---|---|---|---|---|
| Host | |||||
| ERBB4 | PSD95-PDZ2 | 13.9 ± 1.4 | −9.7 ± 0.3 | 3.1 | −6.6 ± 0.6 |
| PSD95-PDZ3 | 41.6 ± 10.2 | −5.5 ± 0.1 | −0.5 | −6.0 ± 1.2 | |
| Virus | |||||
| HTLV1 Tax | PSD95-PDZ2 | 1.5 ± 0.1 | −10.6 ± 0.1 | 2.6 | −8.0 ± 0.5 |
| PSD95-PDZ3 | 3.8 ± 0.2 | −9.5 ± 0.1 | 2.1 | −7.4 ± 0.5 | |
| Erbin-PDZ | 7.8 ± 0.9 | −9.3 ± 0.3 | 2.3 | −7.0 ± 0.8 | |
| HTLV3 Tax | PSD95-PDZ2 | 20.0 ± 3.8 | −5.5 ± 0.5 | −0.9 | −6.4 ± 1.3 |
| PSD95-PDZ3 | 43 ± 10 | −3.5 ± 0.7 | −2.4 | −5.9 ± 1.9 | |
| Erbin-PDZ | 23 ± 9 | −2.4 ± 0.3 | −3.8 | −6.2 ± 1.9 | |
| AdV E4 ORF1 | PSD95-PDZ2 | 11.5 ± 0.5 | −9.4 ± 0.2 | 2.6 | −6.8 ± 0.3 |
| Designed | |||||
| Erbin PDZ binder | PSD95-PDZ2 | 5.7 ± 0.2 | -9.6 ± 0.4 | 2.5 | −7.1 ± 0.3 |
| Erbin-PDZ | 0.24 ± 0.08 | -14.9 ± 0.1 | 5.8 | −9.1 ± 2.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cobos, E.S.; Sánchez, I.E.; Chemes, L.B.; Martinez, J.C.; Murciano-Calles, J. A Thermodynamic Analysis of the Binding Specificity between Four Human PDZ Domains and Eight Host, Viral and Designed Ligands. Biomolecules 2021, 11, 1071. https://doi.org/10.3390/biom11081071
Cobos ES, Sánchez IE, Chemes LB, Martinez JC, Murciano-Calles J. A Thermodynamic Analysis of the Binding Specificity between Four Human PDZ Domains and Eight Host, Viral and Designed Ligands. Biomolecules. 2021; 11(8):1071. https://doi.org/10.3390/biom11081071
Chicago/Turabian StyleCobos, Eva S., Ignacio E. Sánchez, Lucía B. Chemes, Jose C. Martinez, and Javier Murciano-Calles. 2021. "A Thermodynamic Analysis of the Binding Specificity between Four Human PDZ Domains and Eight Host, Viral and Designed Ligands" Biomolecules 11, no. 8: 1071. https://doi.org/10.3390/biom11081071






