The APE2 Exonuclease Is a Client of the Hsp70–Hsp90 Axis in Yeast and Mammalian Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Purification of HA-Tagged Apn2 from Yeast
2.3. Mammalian Cell Culture and Drug Treatment
2.4. Transfections and Co-Immunoprecipitation in Mammalian Cells
2.5. Western Blotting
3. Results
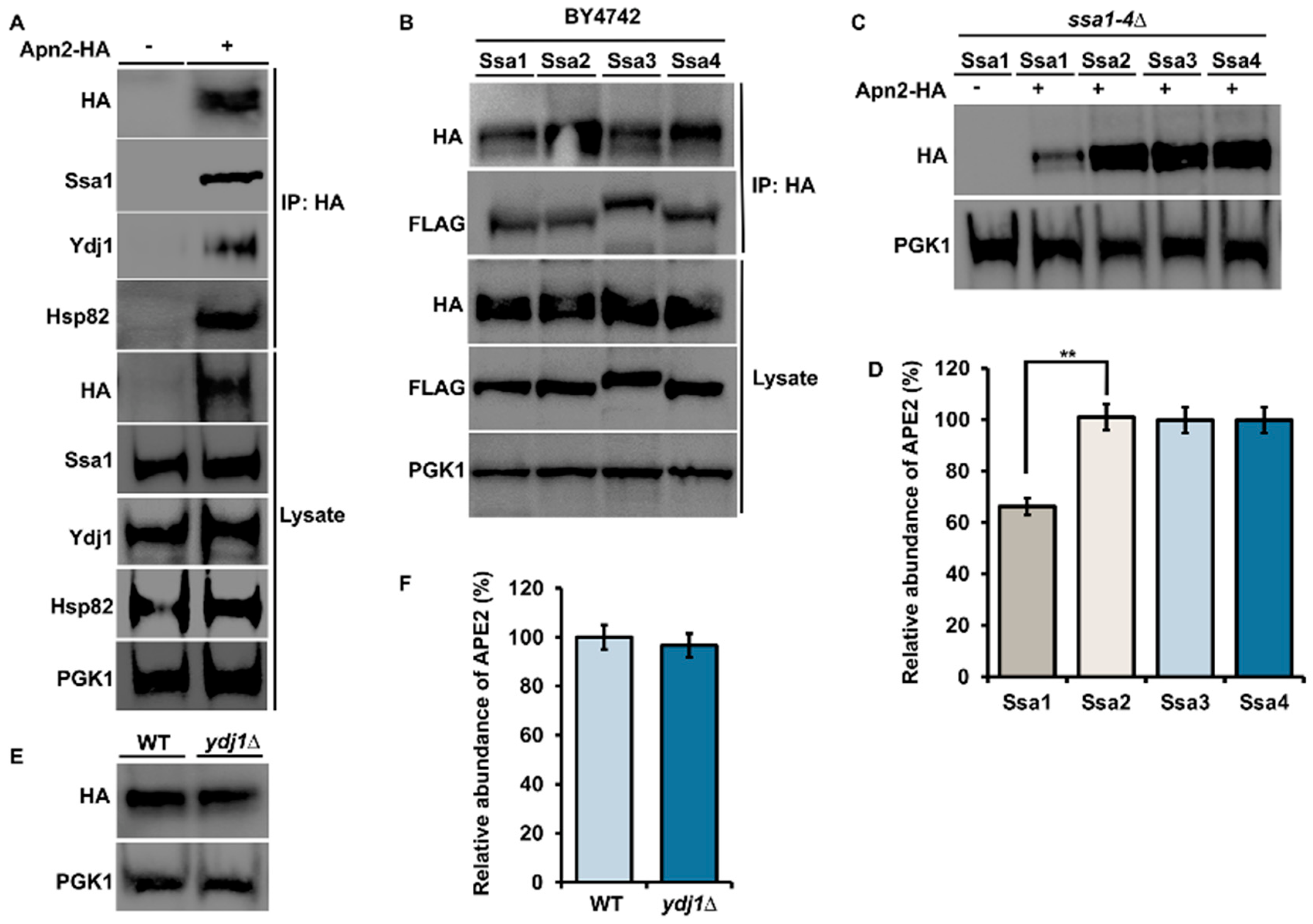
3.1. Apn2 Interacts with Ydj1, Hsp82 and Ssa1 in Yeast
3.2. Apn2 Interacts with Both Hsp82 and Is a Client of Hsp90 in Yeast
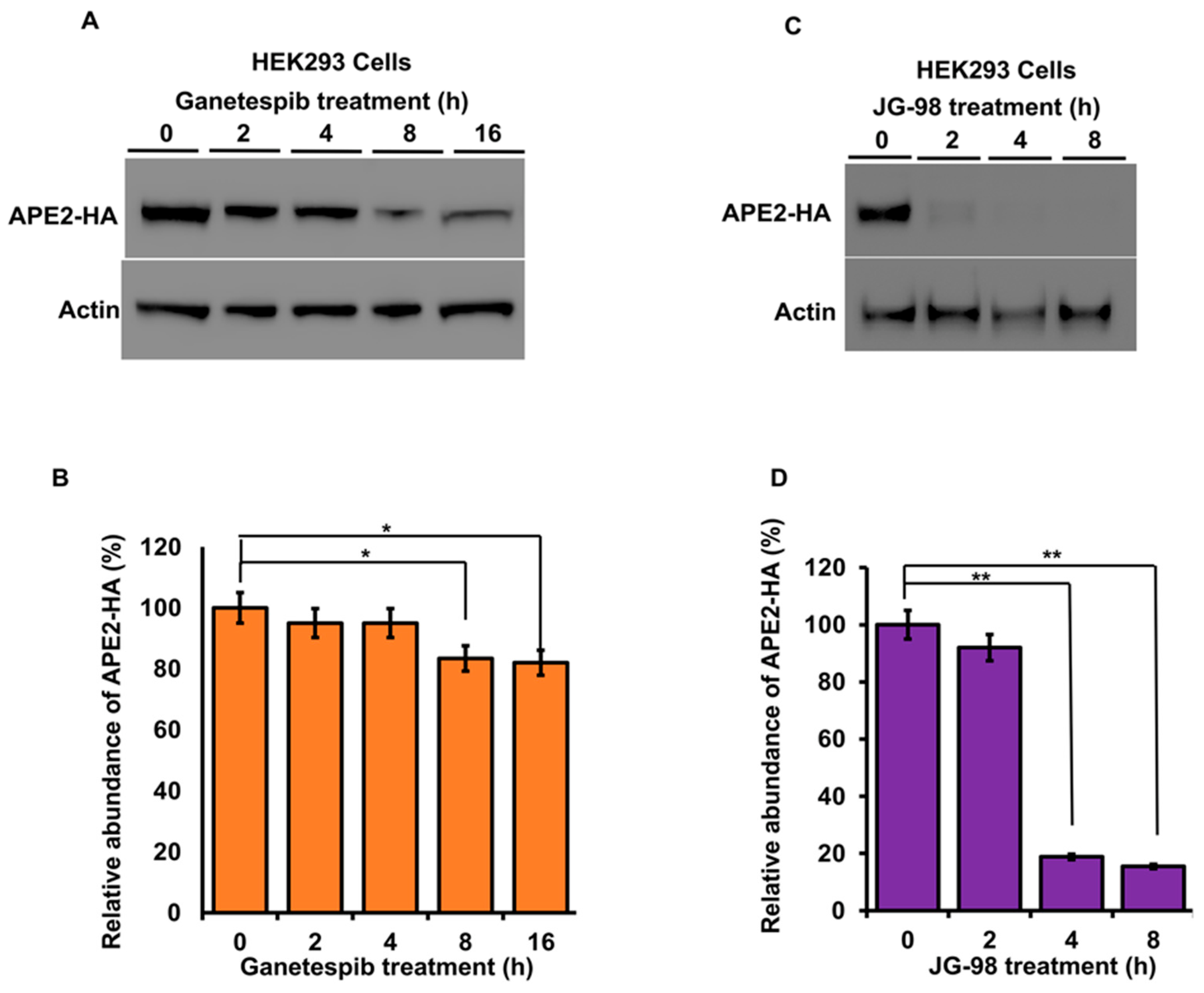
3.3. Mammalian APE2 Interacts with the Hsp90–Hsp70 Chaperone System
4. Discussion
4.1. APE2 and Apn2 Display Binding Preferences for Chaperone and Co-Chaperone Paralogs
4.2. Novel Anticancer Strategies Based on Fine-Tuning Chaperone Function
| Strain | Genotype | Reference/Source |
|---|---|---|
| yAT 685 | Hsc82 (PP30-HSC82-GLU (MAT a, trp1-289, leu2-3112, his3-200, ura3-52, ade2-101, lys2-801, hsc82::KANMX4, hsp82::KANMX4 LEU2-GPD-HSC82-GLU) | [51] |
| yAT 686 | Hsp82 PP30-HSP82-HIS (MAT a, trp1-289, leu2-3112, his3-200, ura3-52, ade2-101, lys2-801, hsc82::KANMX4, hsp82::KANMX4 LEU2-GPD-HSP82-HIS) | [51] |
| yAT01 | P82a W303–1a hsc82::LEU2 hsp82::LEU2 HIS3-GPD-HSP82a | [31] |
| yAT05 | G170D W303–1a hsc82::LEU2 hsp82::LEU2 HIS3-GPD-hsp82(G170D)a | [31] |
| yAT38 | MATα S288c (BY4742) his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| yAT414 | MATa (MH272) ssa1∆::trp1 ssa2::HisG ssa3::HisG ssa4::HisG (ssa1–4) [YCPlac33 SSA1] | [27] |
| yAT28 | MATα S288c (BY4742) ydj1∆::KanMX4 | Euroscarf |
| Plasmids | Description | Reference |
|---|---|---|
| pNK229 | GPD2-Apn2-HA | [18] |
| pAT778 | pRS315PSsa2-Flag-SSA1 (LEU2) | Vector Builder |
| pAT779 | pRS315PSsa2-Flag-SSA2 (LEU2) | Vector Builder |
| pAT780 | pRS315PSsa2-Flag-SSA3 (LEU2) | Vector Builder |
| pAT781 | pRS315PSsa2-Flag-SSA4 (LEU2) | Vector Builder |
| APE2-HA | [23] | |
| pAT758 | HSPA1A-V5 pcDNA5/FRT/TO | Harm Kampinga |
| pAT759 | HSPA1L-V5 pcDNA5/FRT/TO | Harm Kampinga |
| pAT763 | HSPA8-V5 pcDNA5/FRT/TO | Harm Kampinga |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Lotz, S.K.; Knighton, L.E.; Nitika; Jones, G.W.; Truman, A.W. Not quite the SSAme: Unique roles for the yeast cytosolic Hsp70s. Curr. Genet. 2019, 65, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Girstmair, H.; Tippel, F.; Lopez, A.; Tych, K.; Stein, F.; Haberkant, P.; Schmid, P.W.N.; Helm, D.; Rief, M.; Sattler, M.; et al. The Hsp90 isoforms from S. cerevisiae differ in structure, function and client range. Nat. Commun. 2019, 10, 3626. [Google Scholar] [CrossRef]
- Kabani, M.; Martineau, C.N. Multiple hsp70 isoforms in the eukaryotic cytosol: Mere redundancy or functional specificity? Curr. Genom. 2008, 9, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Werner-Washburne, M.; Stone, D.E.; Craig, E.A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell Biol. 1987, 7, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Werner-Washburne, M.; Becker, J.; Kosic-Smithers, J.; Craig, E.A. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 1989, 171, 2680–2688. [Google Scholar] [CrossRef] [Green Version]
- Werner-Washburne, M.; Craig, E.A. Expression of members of the Saccharomyces cerevisiae hsp70 multigene family. Genome 1989, 31, 684–689. [Google Scholar] [CrossRef]
- Hageman, J.; Kampinga, H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 2009, 14, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Daugaard, M.; Rohde, M.; Jaattela, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serlidaki, D.; van Waarde, M.; Rohland, L.; Wentink, A.S.; Dekker, S.L.; Kamphuis, M.J.; Boertien, J.M.; Brunsting, J.F.; Nillegoda, N.B.; Bukau, B.; et al. Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J. Biol. Chem. 2020, 295, 7301–7316. [Google Scholar] [CrossRef] [Green Version]
- Sreedhar, A.S.; Kalmar, E.; Csermely, P.; Shen, Y.F. Hsp90 isoforms: Functions, expression and clinical importance. FEBS Lett. 2004, 562, 11–15. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennisi, R.; Ascenzi, P.; di Masi, A. Hsp90: A New Player in DNA Repair? Biomolecules 2015, 5, 2589–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knighton, L.E.; Delgado, L.E.; Truman, A.W. Novel insights into molecular chaperone regulation of ribonucleotide reductase. Curr. Genet. 2019, 65, 477–482. [Google Scholar] [CrossRef]
- Knighton, L.E.; Truman, A.W. Role of the Molecular Chaperones Hsp70 and Hsp90 in the DNA Damage Response. In Heat Shock Proteins in Signaling Pathways; Springer: Cham, Switzerland, 2019; Volume 17, pp. 345–358. [Google Scholar]
- Whitaker, A.M.; Freudenthal, B.D. APE1: A skilled nucleic acid surgeon. DNA Repair 2018, 71, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.; Patel, Y.; Lentz, B.L.; Yan, S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 10592–10597. [Google Scholar] [CrossRef] [Green Version]
- McNeill, D.R.; Whitaker, A.M.; Stark, W.J.; Illuzzi, J.L.; McKinnon, P.J.; Freudenthal, B.D.; Wilson, D.M., 3rd. Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance. Mutagenesis 2020, 35, 27–38. [Google Scholar] [CrossRef]
- Lin, Y.; McMahon, A.; Driscoll, G.; Bullock, S.; Zhao, J.; Yan, S. Function and molecular mechanisms of APE2 in genome and epigenome integrity. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108347. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Lin, Y.; Yan, S. Single-Strand Break End Resection in Genome Integrity: Mechanism and Regulation by APE2. Int. J. Mol. Sci. 2018, 19, 2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boiteux, S.; Guillet, M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 2004, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Hao, J.J.; Zhang, Z.; Krane, L.S.; Hammerich, K.H.; Sanford, T.; Trepel, J.B.; Neckers, L.; Agarwal, P.K. Proteomic analysis of proteome and histone post-translational modifications in heat shock protein 90 inhibition-mediated bladder cancer therapeutics. Sci. Rep. 2017, 7, 201. [Google Scholar] [CrossRef] [Green Version]
- Mendez, F.; Sandigursky, M.; Kureekattil, R.P.; Kenny, M.K.; Franklin, W.A.; Bases, R. Specific stimulation of human apurinic/apyrimidinic endonuclease by heat shock protein 70. DNA Repair 2003, 2, 259–271. [Google Scholar] [CrossRef]
- Jaiswal, H.; Conz, C.; Otto, H.; Wolfle, T.; Fitzke, E.; Mayer, M.P.; Rospert, S. The chaperone network connected to human ribosome-associated complex. Mol. Cell Biol. 2011, 31, 1160–1173. [Google Scholar] [CrossRef] [Green Version]
- Sluder, I.T.; Nitika; Knighton, L.E.; Truman, A.W. The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity. PLoS Genet. 2018, 14, e1007462. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Quilon, A.; Wojtaszek, J.L.; Mathieu, M.C.; Patel, T.; Appel, C.D.; Hustedt, N.; Rossi, S.E.; Wallace, B.D.; Setiaputra, D.; Adam, S.; et al. Endogenous DNA 3’ Blocks Are Vulnerabilities for BRCA1 and BRCA2 Deficiency and Are Reversed by the APE2 Nuclease. Mol. Cell 2020, 78, 1152–1165 e1158. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Nathan, D.F.; Lindquist, S. Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol. Cell Biol. 1995, 15, 3917–3925. [Google Scholar] [CrossRef] [Green Version]
- Boorstein, W.R.; Ziegelhoffer, T.; Craig, E.A. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994, 38, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Knighton, L.E.; Nitika; Waller, S.J.; Strom, O.; Wolfgeher, D.; Reitzel, A.M.; Truman, A.W. Dynamic remodeling of the interactomes of Nematostella vectensis Hsp70 isoforms under heat shock. J. Proteom. 2019, 206, 103416. [Google Scholar] [CrossRef]
- Knighton, L.E.; Nitika; Omkar, S.; Truman, A.W. The C-terminal domain of Hsp70 is responsible for paralog-specific regulation of ribonucleotide reductase. PLoS Genet. 2022, 18, e1010079. [Google Scholar] [CrossRef]
- Sharma, D.; Masison, D.C. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. USA 2011, 108, 13665–13670. [Google Scholar] [CrossRef] [Green Version]
- Denney, A.S.; Weems, A.D.; McMurray, M.A. Selective functional inhibition of a tumor-derived p53 mutant by cytosolic chaperones identified using split-YFP in budding yeast. G3 2021, 11, jkab230. [Google Scholar] [CrossRef]
- Craig, E.A.; Marszalek, J. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef] [Green Version]
- Truman, A.W.; Kristjansdottir, K.; Wolfgeher, D.; Hasin, N.; Polier, S.; Zhang, H.; Perrett, S.; Prodromou, C.; Jones, G.W.; Kron, S.J. CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 2012, 151, 1308–1318. [Google Scholar] [CrossRef] [Green Version]
- Tutar, L.; Tutar, Y. Ydj1 but not Sis1 stabilizes Hsp70 protein under prolonged stress in vitro. Biopolymers 2008, 89, 171–174. [Google Scholar] [CrossRef]
- Walters, R.W.; Muhlrad, D.; Garcia, J.; Parker, R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 2015, 21, 1660–1671. [Google Scholar] [CrossRef] [Green Version]
- Wyszkowski, H.; Janta, A.; Sztangierska, W.; Obuchowski, I.; Chamera, T.; Klosowska, A.; Liberek, K. Class-specific interactions between Sis1 J-domain protein and Hsp70 chaperone potentiate disaggregation of misfolded proteins. Proc. Natl. Acad. Sci. USA 2021, 118, e2108163118. [Google Scholar] [CrossRef] [PubMed]
- Workman, P. Reflections and Outlook on Targeting HSP90, HSP70 and HSF1 in Cancer: A Personal Perspective. Adv. Exp. Med. Biol. 2020, 1243, 163–179. [Google Scholar] [CrossRef]
- Mosser, D.D.; Caron, A.W.; Bourget, L.; Meriin, A.B.; Sherman, M.Y.; Morimoto, R.I.; Massie, B. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol. Cell Biol. 2000, 20, 7146–7159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Colvin, T.; Rauch, J.N.; Acosta-Alvear, D.; Kampmann, M.; Dunyak, B.; Hann, B.; Aftab, B.T.; Murnane, M.; Cho, M.; et al. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol. Cancer Ther. 2015, 14, 642–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, M.E. The HSP70 family and cancer. Carcinogenesis 2013, 34, 1181–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Bai, L.; Cupello, S.; Hossain, M.A.; Deem, B.; McLeod, M.; Raj, J.; Yan, S. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Res. 2018, 46, 2479–2494. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Berman, Z.; Mueller, G.A.; Lin, Y.; Chang, T.; Andres, S.N.; Wojtaszek, J.L.; DeRose, E.F.; Appel, C.D.; London, R.E.; et al. APE2 Zf-GRF facilitates 3’-5’ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. USA 2017, 114, 304–309. [Google Scholar] [CrossRef] [Green Version]
- Shevtsov, M.; Huile, G.; Multhoff, G. Membrane heat shock protein 70: A theranostic target for cancer therapy. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2018, 373, 20160526. [Google Scholar] [CrossRef] [Green Version]
- Erlichman, C. Tanespimycin: The opportunities and challenges of targeting heat shock protein 90. Expert Opin. Investig. Drugs 2009, 18, 861–868. [Google Scholar] [CrossRef]
- Piper, P.W.; Millson, S.H.; Mollapour, M.; Panaretou, B.; Siligardi, G.; Pearl, L.H.; Prodromou, C. Sensitivity to Hsp90-targeting drugs can arise with mutation to the Hsp90 chaperone, cochaperones and plasma membrane ATP binding cassette transporters of yeast. Eur. J. Biochem. 2003, 270, 4689–4695. [Google Scholar] [CrossRef] [Green Version]
- Nitika; Blackman, J.S.; Knighton, L.E.; Takakuwa, J.E.; Calderwood, S.K.; Truman, A.W. Chemogenomic screening identifies the Hsp70 co-chaperone DNAJA1 as a hub for anticancer drug resistance. Sci. Rep. 2020, 10, 13831. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.A.; Kim, Y.S.; Rivera-Marquez, G.M.; Oshima, N.; Watson, M.J.; Beebe, K.E.; Wells, C.; Lee, S.; Zuehlke, A.D.; Shao, H.; et al. Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2018, 78, 4022–4035. [Google Scholar] [CrossRef] [Green Version]
- Wisen, S.; Bertelsen, E.B.; Thompson, A.D.; Patury, S.; Ung, P.; Chang, L.; Evans, C.G.; Walter, G.M.; Wipf, P.; Carlson, H.A.; et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem. Biol. 2010, 5, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Nitika; Porter, C.M.; Truman, A.W.; Truttmann, M.C. Post-translational modifications of Hsp70 family proteins: Expanding the chaperone code. J. Biol. Chem. 2020, 295, 10689–10708. [Google Scholar] [CrossRef] [PubMed]
- Backe, S.J.; Sager, R.A.; Woodford, M.R.; Makedon, A.M.; Mollapour, M. Post-translational modifications of Hsp90 and translating the chaperone code. J. Biol. Chem. 2020, 295, 11099–11117. [Google Scholar] [CrossRef] [PubMed]
- Truman, A.W.; Bourboulia, D.; Mollapour, M. Decrypting the chaperone code. J. Biol. Chem. 2021, 296, 100293. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omkar, S.; Wani, T.H.; Zheng, B.; Mitchem, M.M.; Truman, A.W. The APE2 Exonuclease Is a Client of the Hsp70–Hsp90 Axis in Yeast and Mammalian Cells. Biomolecules 2022, 12, 864. https://doi.org/10.3390/biom12070864
Omkar S, Wani TH, Zheng B, Mitchem MM, Truman AW. The APE2 Exonuclease Is a Client of the Hsp70–Hsp90 Axis in Yeast and Mammalian Cells. Biomolecules. 2022; 12(7):864. https://doi.org/10.3390/biom12070864
Chicago/Turabian StyleOmkar, Siddhi, Tasaduq H. Wani, Bo Zheng, Megan M. Mitchem, and Andrew W. Truman. 2022. "The APE2 Exonuclease Is a Client of the Hsp70–Hsp90 Axis in Yeast and Mammalian Cells" Biomolecules 12, no. 7: 864. https://doi.org/10.3390/biom12070864
APA StyleOmkar, S., Wani, T. H., Zheng, B., Mitchem, M. M., & Truman, A. W. (2022). The APE2 Exonuclease Is a Client of the Hsp70–Hsp90 Axis in Yeast and Mammalian Cells. Biomolecules, 12(7), 864. https://doi.org/10.3390/biom12070864






