Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Subjects, and Sample Collection
2.2. Sample Processing and Microbiome Analysis
2.3. 16S rRNA Gene Sequence Analysis
2.4. Biochemical Analysis
2.5. Statistics
3. Results
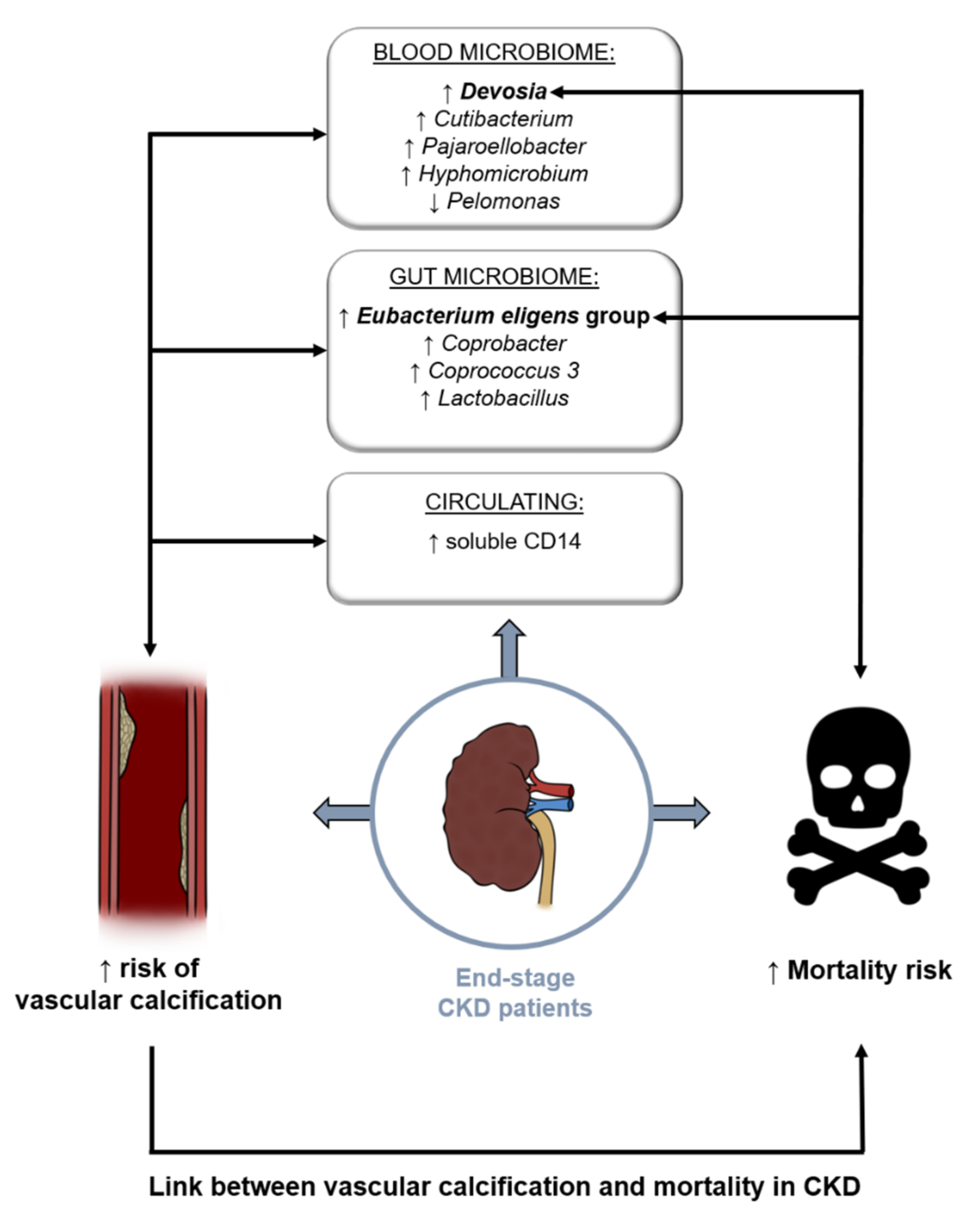
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johansen, K.L.; Chertow, G.M.; Foley, R.N.; Gilbertson, D.T.; Herzog, C.A.; Ishani, A.; Israni, A.K.; Ku, E.; Kurella Tamura, M.; Li, S.; et al. US renal data system 2020 annual data report: Epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 2021, 77, A7–A8. [Google Scholar] [CrossRef] [PubMed]
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.E.; Tang, K.; Yuan, C.W.; et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Sarnak, M.J. Epidemiology: The global burden of reduced GFR: ESRD, CVD and mortality. Nat. Rev. Nephrol. 2017, 13, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, J.; Chonchol, M.B. Nontraditional risk factors for cardiovascular disease in patients with chronic kidney disease. Nat. Clin. Pract. Nephrol. 2008, 4, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Mihai, S.; Codrici, E.; Popescu, I.D.; Enciu, A.M.; Albulescu, L.; Necula, L.G.; Mambet, C.; Anton, G.; Tanase, C. Inflammation-related mechanisms in chronic kidney disease prediction, progression, and outcome. J. Immunol. Res. 2018, 2018, 2180373. [Google Scholar] [CrossRef]
- Amdur, R.L.; Feldman, H.I.; Dominic, E.A.; Anderson, A.H.; Beddhu, S.; Rahman, M.; Wolf, M.; Reilly, M.; Ojo, A.; Townsend, R.R.; et al. Use of Measures of inflammation and kidney function for prediction of atherosclerotic vascular disease events and death in patients with CKD: Findings from the CRIC study. Am. J. Kidney Dis. 2019, 73, 344–353. [Google Scholar] [CrossRef] [Green Version]
- Canyelles, M.; Tondo, M.; Cedo, L.; Farras, M.; Escola-Gil, J.C.; Blanco-Vaca, F. Trimethylamine N-oxide: A link among diet, gut microbiota, gene regulation of liver and intestine cholesterol homeostasis and HDL function. Int. J. Mol. Sci. 2018, 19, 3228. [Google Scholar] [CrossRef] [Green Version]
- Evenepoel, P.; Dejongh, S.; Verbeke, K.; Meijers, B. The role of gut dysbiosis in the bone-vascular axis in chronic kidney disease. Toxins 2020, 12, 285. [Google Scholar] [CrossRef]
- Jovanovich, A.; Isakova, T.; Stubbs, J. Microbiome and cardiovascular disease in CKD. Clin. J. Am. Soc. Nephrol. 2018, 13, 1598–1604. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Sampaio-Maia, B.; Simoes-Silva, L.; Pestana, M.; Araujo, R.; Soares-Silva, I.J. The role of the gut microbiome on chronic kidney disease. Adv. Appl. Microbiol. 2016, 96, 65–94. [Google Scholar] [CrossRef]
- Shimizu, H.; Hirose, Y.; Nishijima, F.; Tsubakihara, Y.; Miyazaki, H. ROS and PDGF-beta [corrected] receptors are critically involved in indoxyl sulfate actions that promote vascular smooth muscle cell proliferation and migration. Am. J. Physiol. Cell Physiol. 2009, 297, C389–C396. [Google Scholar] [CrossRef]
- Meijers, B.; Evenepoel, P.; Anders, H.J. Intestinal microbiome and fitness in kidney disease. Nat. Rev. Nephrol. 2019, 15, 531–545. [Google Scholar] [CrossRef]
- Lau, W.L.; Kalantar-Zadeh, K.; Vaziri, N.D. The gut as a source of inflammation in chronic kidney disease. Nephron 2015, 130, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The healthy human blood microbiome: Fact or fiction? Front. Cell Infect. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef] [Green Version]
- D’Aquila, P.; Giacconi, R.; Malavolta, M.; Piacenza, F.; Burkle, A.; Villanueva, M.M.; Dolle, M.E.T.; Jansen, E.; Grune, T.; Gonos, E.S.; et al. Microbiome in blood samples from the general population recruited in the MARK-AGE project: A pilot study. Front. Microbiol. 2021, 12, 707515. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.; Lange, C.; Payros, G.; Garret, C.; Chabo, C.; Lantieri, O.; Courtney, M.; Marre, M.; Charles, M.A.; Balkau, B.; et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: The D.E.S.I.R. study. PLoS ONE 2013, 8, e54461. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef] [Green Version]
- Schierwagen, R.; Alvarez-Silva, C.; Madsen, M.S.A.; Kolbe, C.C.; Meyer, C.; Thomas, D.; Uschner, F.E.; Magdaleno, F.; Jansen, C.; Pohlmann, A.; et al. Circulating microbiome in blood of different circulatory compartments. Gut 2019, 68, 578–580. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.B.; Allegretti, A.S.; Nigwekar, S.U.; Kalim, S.; Zhao, S.; Lelouvier, B.; Servant, F.; Serena, G.; Thadhani, R.I.; Raj, D.S.; et al. Blood microbiome profile in CKD: A pilot study. Clin. J. Am. Soc. Nephrol. 2019, 14, 692–701. [Google Scholar] [CrossRef]
- Evenepoel, P.; Opdebeeck, B.; David, K.; D’Haese, P.C. Bone-vascular axis in chronic kidney disease. Adv. Chronic Kidney Dis. 2019, 26, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Longenecker, C.T.; Jiang, Y.; Orringer, C.E.; Gilkeson, R.C.; Debanne, S.; Funderburg, N.T.; Lederman, M.M.; Storer, N.; Labbato, D.E.; McComsey, G.A. Soluble CD14 is independently associated with coronary calcification and extent of subclinical vascular disease in treated HIV infection. AIDS 2014, 28, 969–977. [Google Scholar] [CrossRef]
- Rodrigues, F.G.; Ormanji, M.S.; Heilberg, I.P.; Bakker, S.J.L.; de Borst, M.H. Interplay between gut microbiota, bone health and vascular calcification in chronic kidney disease. Eur. J. Clin. Invest. 2021, 51, e13588. [Google Scholar] [CrossRef] [PubMed]
- Adragao, T.; Pires, A.; Lucas, C.; Birne, R.; Magalhaes, L.; Goncalves, M.; Negrao, A.P. A simple vascular calcification score predicts cardiovascular risk in haemodialysis patients. Nephrol. Dial. Transpl. 2004, 19, 1480–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlson, M. Charlson Comorbidity Index (CCI). Available online: https://www.mdcalc.com/charlson-comorbidity-index-cci (accessed on 7 August 2020).
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic. Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Simoes-Silva, L.; Araujo, R.; Pestana, M.; Soares-Silva, I.; Sampaio-Maia, B. Peritoneal microbiome in end-stage renal disease patients and the impact of peritoneal dialysis therapy. Microorganisms 2020, 8, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escudie, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, rapidly, OTUs with galaxy solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef]
- Calaf, R.; Cerini, C.; Genovesio, C.; Verhaeghe, P.; Jourde-Chiche, N.; Berge-Lefranc, D.; Gondouin, B.; Dou, L.; Morange, S.; Argiles, A.; et al. Determination of uremic solutes in biological fluids of chronic kidney disease patients by HPLC assay. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011, 879, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.; Dunlap, C.; Franco, C.M.M. Analogous wheat root rhizosphere microbial successions in field and greenhouse trials in the presence of biocontrol agents Paenibacillus peoriae SP9 and Streptomyces fulvissimus FU14. Mol. Plant Pathol. 2020, 21, 622–635. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Zhang, C.; Feng, H.; Yuan, J.; Ding, L.; Fang, W.; Gu, A.; Huang, J.; Li, N.; Gu, L.; et al. Clinical characteristics associated with the properties of gut microbiota in peritoneal dialysis patients. Perit. Dial. Int. 2021, 41, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 2019, 8, e00678. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Zhao, W.; Lin, Z.; Wu, J.; Lin, H.; Li, Y.; Song, J.; Zhang, J.; Peng, H. The effects of hemodialysis and peritoneal dialysis on the gut microbiota of end-stage renal disease patients, and the relationship between gut microbiota and patient prognoses. Front. Cell Infect. Microbiol. 2021, 11, 579386. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Roach, J.; Marsh, A.; Chey, W.D.; Sandborn, W.J.; Ritter, A.J.; Savaiano, D.A.; Klaenhammer, T.R. A double-blind, 377-subject randomized study identifies Ruminococcus, Coprococcus, Christensenella, and Collinsella as long-term potential key players in the modulation of the gut microbiome of lactose intolerant individuals by galacto-oligosaccharides. Gut Microbes 2021, 13, 1957536. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.L.; Kable, M.E.; Marco, M.; Keim, N.L. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models. Part II: Results. Adv. Nutr. 2019, 10, 979–998. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.B.; Alderete, T.L.; Kim, J.S.; Millstein, J.; Gilliland, F.D.; Goran, M.I. High intake of dietary fructose in overweight/obese teenagers associated with depletion of Eubacterium and Streptococcus in gut microbiome. Gut Microbes 2019, 10, 712–719. [Google Scholar] [CrossRef]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, D.D.; Satija, A.; Ivey, K.L.; Li, J.; Wilkinson, J.E.; Li, R.; Baden, M.; Chan, A.T.; Huttenhower, C.; et al. Plant-based diet index and metabolic risk in men: Exploring the role of the gut microbiome. J. Nutr. 2021, 151, 2780–2789. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, W.; Liu, X.; Cheng, L. Metagenomic analysis of the gut microbiome in atherosclerosis patients identify cross-cohort microbial signatures and potential therapeutic target. FASEB J. 2020, 34, 14166–14181. [Google Scholar] [CrossRef]
- Nie, X.; Chen, J.; Ma, X.; Ni, Y.; Shen, Y.; Yu, H.; Panagiotou, G.; Bao, Y. A metagenome-wide association study of gut microbiome and visceral fat accumulation. Comput. Struct. Biotechnol. J. 2020, 18, 2596–2609. [Google Scholar] [CrossRef]
- Kossenas, K.; Constantinou, C. Epidemiology, molecular mechanisms, and clinical trials: An update on research on the association between red meat consumption and colorectal cancer. Curr. Nutr. Rep. 2021, 10, 435–467. [Google Scholar] [CrossRef]
- Paisse, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossola, M.; Sanguinetti, M.; Scribano, D.; Zuppi, C.; Giungi, S.; Luciani, G.; Torelli, R.; Posteraro, B.; Fadda, G.; Tazza, L. Circulating bacterial-derived DNA fragments and markers of inflammation in chronic hemodialysis patients. Clin. J. Am. Soc. Nephrol. 2009, 4, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Mair, R.D.; Sirich, T.L. Blood microbiome in CKD: Should we care? Clin. J. Am. Soc. Nephrol. 2019, 14, 648–649. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, C.; Magnussen, K.; Enevold, C.; Nilsson, M.; Tolker-Nielsen, T.; Holmstrup, P.; Nielsen, C.H. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS ONE 2015, 10, e0120826. [Google Scholar] [CrossRef] [Green Version]
- Bay, L.; Barnes, C.J.; Fritz, B.G.; Thorsen, J.; Restrup, M.E.M.; Rasmussen, L.; Sorensen, J.K.; Hesselvig, A.B.; Odgaard, A.; Hansen, A.J.; et al. Universal dermal microbiome in human skin. mBio 2020, 11, e02945-19. [Google Scholar] [CrossRef] [Green Version]
- De, R.; Mukhopadhyay, A.K.; Dutta, S. Metagenomic analysis of gut microbiome and resistome of diarrheal fecal samples from Kolkata, India, reveals the core and variable microbiota including signatures of microbial dark matter. Gut Pathog. 2020, 12, 32. [Google Scholar] [CrossRef]
- Rudney, J.D.; Xie, H.; Rhodus, N.L.; Ondrey, F.G.; Griffin, T.J. A metaproteomic analysis of the human salivary microbiota by three-dimensional peptide fractionation and tandem mass spectrometry. Mol. Oral Microbiol. 2010, 25, 38–49. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Chang, Y.; Zheng, Q.; Zhang, R.; Hu, C.; Jia, W. Altered intestinal microbiota associated with colorectal cancer. Front. Med. 2019, 13, 461–470. [Google Scholar] [CrossRef]
- Bai, X.; Shi, Y.; Tang, L.; Chen, L.; Fan, H.; Wang, H.; Wang, J.; Jia, X.; Chen, S.; Lai, S. Heat stress affects faecal microbial and metabolic alterations of rabbits. Front. Microbiol. 2021, 12, 817615. [Google Scholar] [CrossRef] [PubMed]
- Balla, J.; Balla, G.; Zarjou, A. Ferritin in kidney and vascular related diseases: Novel roles for an old player. Pharmaceuticals 2019, 12, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.R.; Lee, Y.K.; Cho, A.J.; Park, H.C.; Han, C.H.; Choi, M.J.; Koo, J.R.; Yoon, J.W.; Noh, J.W. Malnutrition, inflammation, progression of vascular calcification and survival: Inter-relationships in hemodialysis patients. PLoS ONE 2019, 14, e0216415. [Google Scholar] [CrossRef] [PubMed]
- Henze, L.A.; Luong, T.T.D.; Boehme, B.; Masyout, J.; Schneider, M.P.; Brachs, S.; Lang, F.; Pieske, B.; Pasch, A.; Eckardt, K.U.; et al. Impact of C-reactive protein on osteo-/chondrogenic transdifferentiation and calcification of vascular smooth muscle cells. Aging 2019, 11, 5445–5462. [Google Scholar] [CrossRef]
- Bas, S.; Gauthier, B.R.; Spenato, U.; Stingelin, S.; Gabay, C. CD14 is an acute-phase protein. J. Immunol. 2004, 172, 4470–4479. [Google Scholar] [CrossRef] [Green Version]
- Reiner, A.P.; Lange, E.M.; Jenny, N.S.; Chaves, P.H.; Ellis, J.; Li, J.; Walston, J.; Lange, L.A.; Cushman, M.; Tracy, R.P. Soluble CD14: Genomewide association analysis and relationship to cardiovascular risk and mortality in older adults. Arter. Thromb. Vasc. Biol. 2013, 33, 158–164. [Google Scholar] [CrossRef] [Green Version]
- Poesen, R.; Ramezani, A.; Claes, K.; Augustijns, P.; Kuypers, D.; Barrows, I.R.; Muralidharan, J.; Evenepoel, P.; Meijers, B.; Raj, D.S. Associations of soluble CD14 and endotoxin with mortality, cardiovascular disease, and progression of kidney disease among patients with CKD. Clin. J. Am. Soc. Nephrol. 2015, 10, 1525–1533. [Google Scholar] [CrossRef] [Green Version]
- Raj, D.S.; Shah, V.O.; Rambod, M.; Kovesdy, C.P.; Kalantar-Zadeh, K. Association of soluble endotoxin receptor CD14 and mortality among patients undergoing hemodialysis. Am. J. Kidney Dis. 2009, 54, 1062–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seabra, V.F.; Thomas, G.; Jaber, B.L. Soluble CD14 and endotoxin levels in hemodialysis patients: A tale of 2 molecules. Am. J. Kidney Dis. 2009, 54, 990–992. [Google Scholar] [CrossRef]
- Niu, Q.; Zhao, H.; Wu, B.; Tsai, S.; Wu, J.; Zhang, M.; Lu, L.; Qiao, J.; Men, C.; Zuo, L.; et al. Study on the prevalence of vascular calcification in different types of arteries and influencing factors in maintenance peritoneal dialysis patients. Blood Purif. 2019, 47, 8–16. [Google Scholar] [CrossRef]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Visseren, F.L.J.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Back, M.; Benetos, A.; Biffi, A.; Boavida, J.M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef] [PubMed]
- Dimkovic, N.; Schlieper, G.; Jankovic, A.; Djuric, Z.; Ketteler, M.; Damjanovic, T.; Djuric, P.; Marinkovic, J.; Radojcic, Z.; Markovic, N.; et al. Prognostic value of cardiovascular calcifications in hemodialysis patients: A longitudinal study. Int. Urol. Nephrol. 2018, 50, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Floege, J.; Ketteler, M. Vascular calcification in patients with end-stage renal disease. Nephrol. Dial. Transpl. 2004, 19, V59–V66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group. KDIGO clinical practice guideline for the care of kidney transplant recipients. Am. J. Transpl. 2009, 9, S1–S155. [Google Scholar] [CrossRef]
- Cozzolino, M.; Ciceri, P.; Galassi, A.; Mangano, M.; Carugo, S.; Capelli, I.; Cianciolo, G. The Key Role of Phosphate on Vascular Calcification. Toxins 2019, 11, 213. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Tandon, S.; Tandon, C. An update on vascular calcification and potential therapeutics. Mol. Biol. Rep. 2021, 48, 887–896. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.J.; Robertson, G.R.; Lee, V.W. Vitamin D in vascular calcification: A double-edged sword? Nutrients 2018, 10, 652. [Google Scholar] [CrossRef] [Green Version]
- Mathew, S.; Lund, R.J.; Chaudhary, L.R.; Geurs, T.; Hruska, K.A. Vitamin D receptor activators can protect against vascular calcification. J. Am. Soc. Nephrol. 2008, 19, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Sandoval, J.C.; Casanova, I.; Villar, A.; Gomez, F.E.; Cruz, C.; Correa-Rotter, R. Biomarkers associated with vascular calcification in peritoneal dialysis. Perit. Dial. Int. 2016, 36, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Ghorbanihaghjo, A.; Argani, H.; Golmohamadi, Z.; Rashtchizadeh, N.; Abbasi, M.M.; Bargahi, N.; Vatankhah, A.M.; Sanajou, D. Linkage of fibroblast growth factor 23 and phosphate in serum: Phosphate and fibroblast growth factor 23 reduction by increasing dose of sevelamer. J. Bone Metab. 2018, 25, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Qiu, Y.; Yu, J.; Lin, T.; Lu, M.; Yi, C.; Lin, J.; Ye, H.; Chen, W.; Mao, H.; et al. Ten-year survival of patients treated with peritoneal dialysis: A prospective observational cohort study. Perit. Dial. Int. 2020, 40, 573–580. [Google Scholar] [CrossRef] [PubMed]
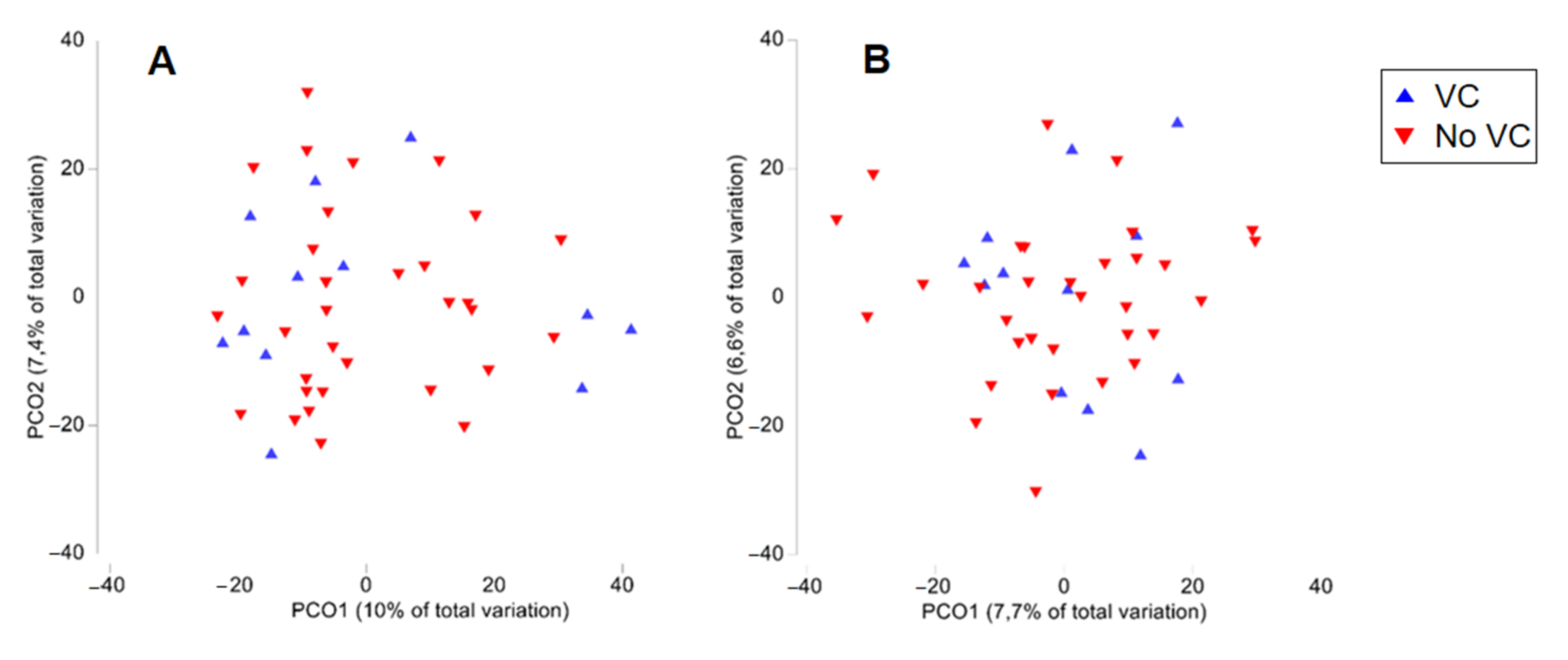



| CKD-PD (n = 44) | CKD-PD With no VC (n = 12) | CKD-PD with VC (n = 32) | p-Value | |
|---|---|---|---|---|
| Demographic data | ||||
| Age, years | 56.1 ± 10.9 | 47.7 ± 11.5 | 59.4 ± 8.8 | <0.001 a |
| Sex, % male | 65.9% | 33.3% | 78.1% | 0.011 d |
| PD parameters | ||||
| PD duration, months | 33.4 ± 30.0 | 36.3 ± 43.4 | 30.9 ± 23.8 | 0.668 b |
| PD type, % | >0.999 d | |||
| APD | 52.3% | 50.0% | 53.1% | |
| CAPD | 47.7% | 50.0% | 46.9% | |
| Ccreat, L/week | 114.8 ± 56.8 | 105.7 ± 45.1 | 118.2 ± 60.8 | 0.668 b |
| Residual renal function, mL/min | 5.6 ± 4.0 | 5.8 ± 3.8 | 5.6 ± 4.1 | 0.706 b |
| Kt/V (urea) | 2.2 ± 0.5 | 2.6 ± 0.6 | 2.1 ± 0.4 | 0.004 b |
| Charlson Index, % | 0.003 c | |||
| Low (≤2) | 18.2% | 50.0% | 6.3% | |
| Moderate (3–4) | 31.8% | 25.0% | 34.4% | |
| Severe (≥5) | 50.0% | 25.0% | 59.4% | |
| Biochemical parameters | ||||
| Urea, mg/dL | 125.0 ± 37.0 | 127.6 ± 20.1 | 124.0 ± 41.8 | 0.780 a |
| Proteinuria mg/24 h | 1.0 ± 1.2 | 0.9 ± 1.0 | 1.0 ± 1.2 | 0.342 b |
| Albumin, g/L | 37.1 ± 3.3 | 37.0 ± 2.6 | 37.1 ± 3.6 | 0.944 a |
| Hemoglobin, g/dL | 11.5 ± 1.4 | 11.0 ± 0.9 | 11.7 ± 1.6 | 0.133 a |
| Cholesterol, mg/dL | 171.0 ± 56.8 | 169.9 ± 42.8 | 171.4 ± 61.8 | 0.825 b |
| LDL, mg/dL | 95.7 ± 42.6 | 99.9 ± 33.7 | 94.0 ± 46.1 | 0.547 b |
| HDL, mg/dL | 45.6 ± 10.7 | 47.4 ± 9.3 | 45.0 ± 11.3 | 0.267 b |
| Triglycerides, mg/dL | 158.6 ± 68.4 | 129.8 ± 42.9 | 169.4 ± 73.5 | 0.169 b |
| P, mg/dL | 5.0 ± 1.1 | 5.72 ± 1.05 | 4.73 ± 1.02 | 0.011 a |
| Ca, mg/dL | 9.02 ± 0.89 | 9.39 ± 0.85 | 8.84 ± 0.89 | 0.073 a |
| Ca • P product | 43.83 ± 10.63 | 52.08 ± 9.32 | 40.67 ± 9.70 | 0.002 b |
| Ferritin, ng/mL | 361.3 ± 222.9 | 316.1 ± 221.3 | 378.3 ± 224.6 | 0.419 a |
| BNP, pg/mL | 143.1 ± 119.2 | 87.0 ± 36.6 | 163.1 ± 131.9 | 0.124 b |
| PTH, pg/mL | 462.5 ± 280.0 | 485.5 ± 366.4 | 453.9 ± 246.7 | 0.866 b |
| SV, mm | 64.2 ± 25.6 | 67.2 ± 18.7 | 63.1 ± 27.9 | 0.644 a |
| CRP, mg/L | 5.3 ± 8.5 | 4.8 ± 7.7 | 5.5 ± 8.9 | 0.907 b |
| TNF-α, pg/mL | 11.4 ± 4.3 | 10.4 ± 2.8 | 11.7 ± 4.7 | 0.524 b |
| IL-1β, pg/mL | 1.3 ± 0.93 | 1.3 ± 1.0 | 1.3 ± 0.9 | 0.969 b |
| IL-10, pg/mL | 17.7 ± 14.7 | 17.5 ± 16.7 | 17.8 ± 14.2 | 0.825 b |
| IL-6, pg/mL | 2.9 ± 6.3 | 5.4 ± 10.3 | 2.0 ± 3.8 | 0.687 b |
| Endotoxins, EU/mL | 3.8 ± 0.8 | 3.8 ± 0.4 | 3.7 ± 0.8 | 0.978 a |
| LPS-BP, µg/mL | 39.9 ± 17.1 | 32.2 ± 13.4 | 41.2 ± 18.3 | 0.442 b |
| TLR-4, pg/mL | 624.4 ± 439.2 | 699.1 ± 464.5 | 596.4 ± 433.7 | 0.630 b |
| sCD14, µg/mL | 5.0 ± 2.1 | 4.4 ± 2.0 | 5.3 ± 2.1 | 0.224 b |
| T-MAO | 0.52 ± 0.62 | 0.47 ± 0.40 | 0.57 ± 0.70 | 0.854 b |
| PCS, mg/L | 33.5 ± 19.1 | 36.4 ± 18.0 | 32.3 ± 19.7 | 0.341 b |
| 3-INDS, mg/L | 23.7 ± 14.6 | 24.1 ± 9.6 | 23.5 ± 16.22 | 0.442 b |
| 3-IAA, mg/L | 1.1 ± 1.2 | 1.0 ± 0.5 | 1.1 ± 1.4 | 0.169 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merino-Ribas, A.; Araujo, R.; Pereira, L.; Campos, J.; Barreiros, L.; Segundo, M.A.; Silva, N.; Costa, C.F.F.A.; Quelhas-Santos, J.; Trindade, F.; et al. Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study. Biomolecules 2022, 12, 867. https://doi.org/10.3390/biom12070867
Merino-Ribas A, Araujo R, Pereira L, Campos J, Barreiros L, Segundo MA, Silva N, Costa CFFA, Quelhas-Santos J, Trindade F, et al. Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study. Biomolecules. 2022; 12(7):867. https://doi.org/10.3390/biom12070867
Chicago/Turabian StyleMerino-Ribas, Ana, Ricardo Araujo, Luciano Pereira, Joana Campos, Luísa Barreiros, Marcela A. Segundo, Nádia Silva, Carolina F. F. A. Costa, Janete Quelhas-Santos, Fábio Trindade, and et al. 2022. "Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study" Biomolecules 12, no. 7: 867. https://doi.org/10.3390/biom12070867
APA StyleMerino-Ribas, A., Araujo, R., Pereira, L., Campos, J., Barreiros, L., Segundo, M. A., Silva, N., Costa, C. F. F. A., Quelhas-Santos, J., Trindade, F., Falcão-Pires, I., Alencastre, I., Dumitrescu, I. B., & Sampaio-Maia, B. (2022). Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study. Biomolecules, 12(7), 867. https://doi.org/10.3390/biom12070867











