In-Silico Analysis of pH-Dependent Liquid-Liquid Phase Separation in Intrinsically Disordered Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Generation
2.2. pH-Dependent Solubility and Disorder Analyses
3. Results
3.1. LLPS-DRs Present Maximum Solubility around Neutral pH
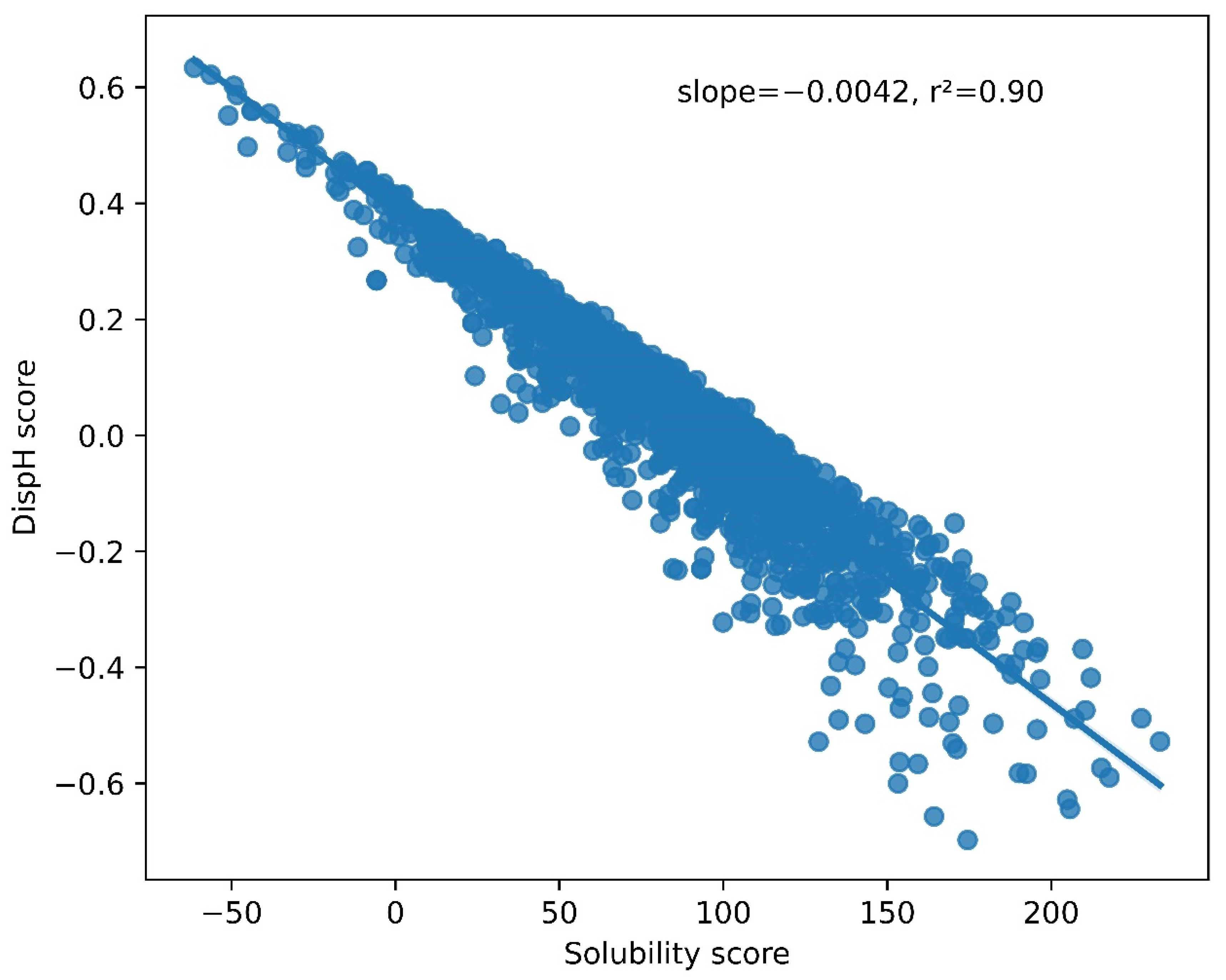
3.2. A Link between pH-Dependent Solubility and pH-Dependent Disorder in LLPS-DRs
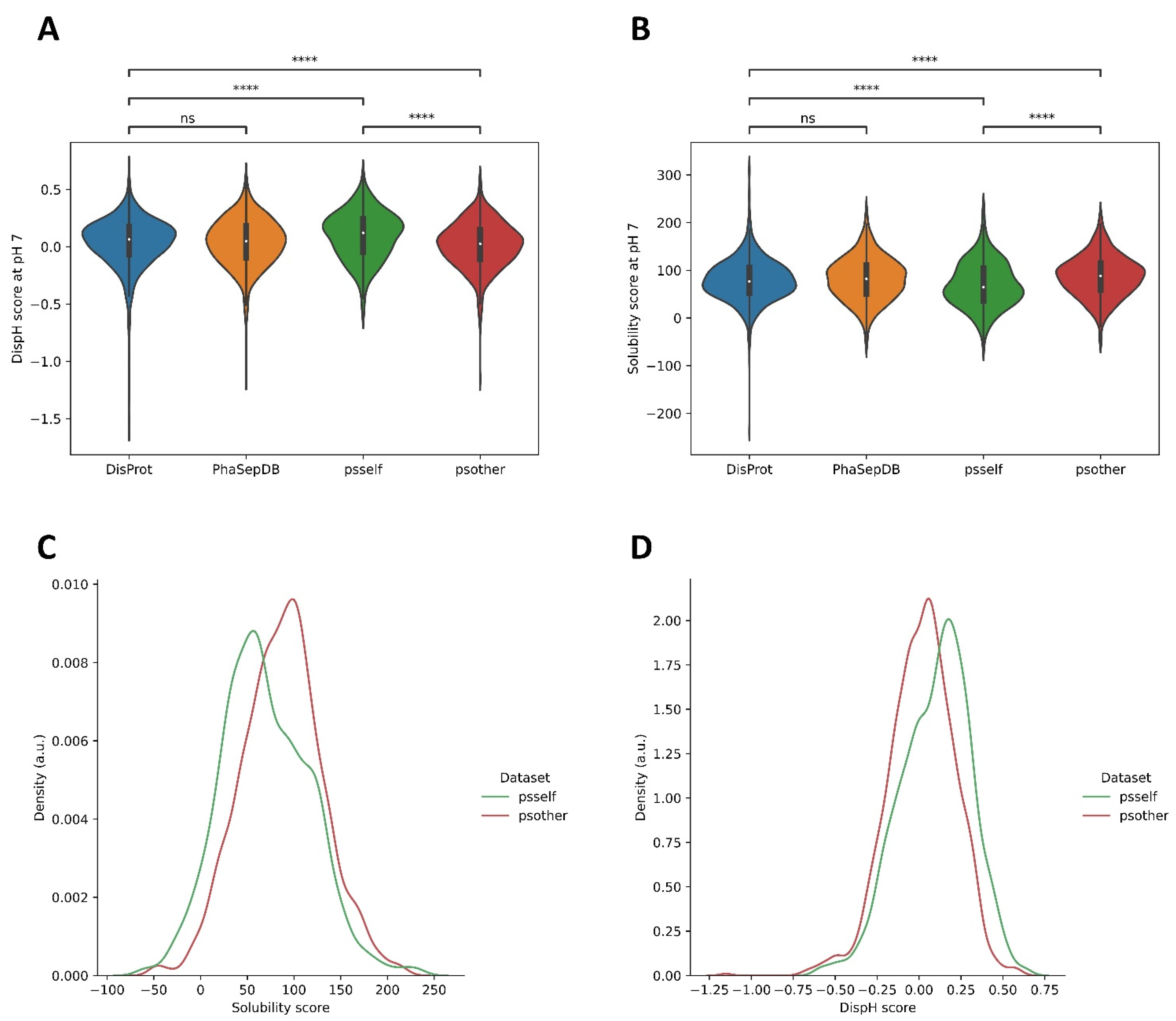
3.3. Different Datasets in LLPS-DRs Present Distinct Property Distributions
3.4. Psself Regions Present Lower Dispersion in Solubility and Disorder in the Physiological pH Range
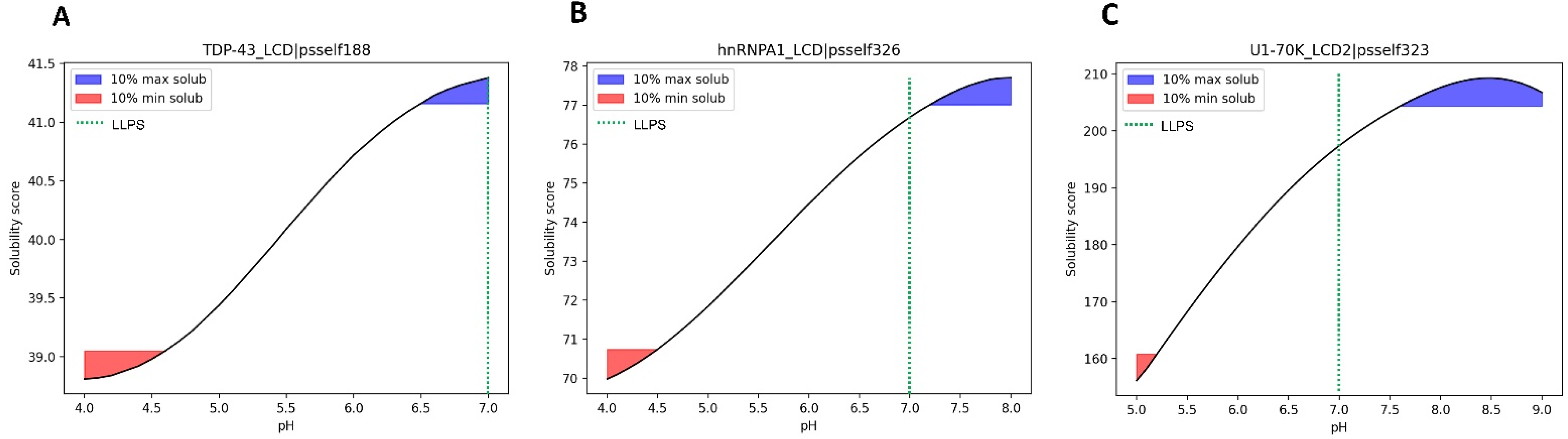
3.5. Case Study of Independent LLPS Happening at Physiological pH
3.6. Mutations in LLPS Formation and Disease
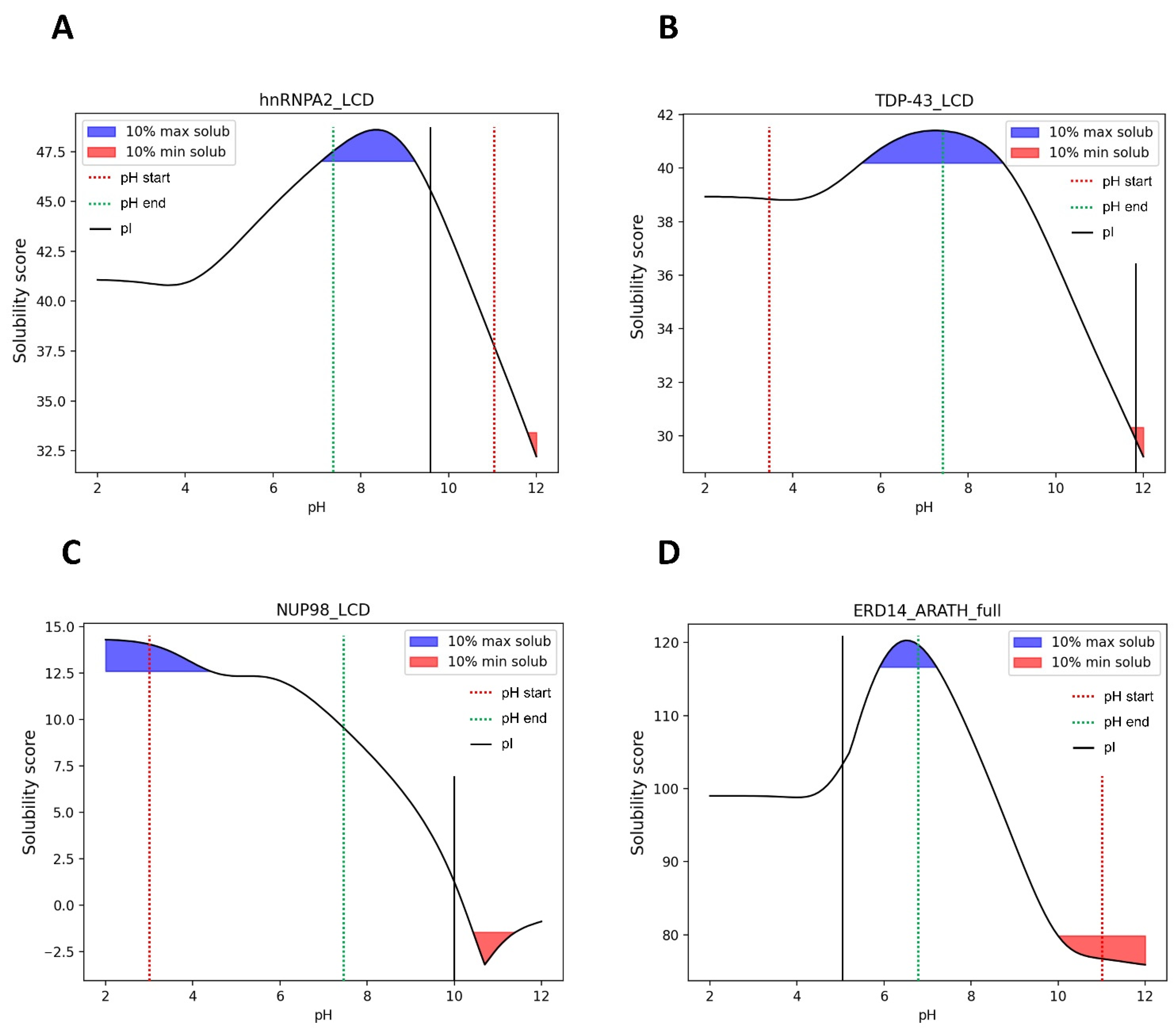
3.7. pH-Dependent LLPS: Optimal Condition Evaluation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| IDPs | Intrinsically disordered proteins |
| IDRs | Intrinsically disordered regions |
| LCD | Low complexity domain |
| LLPS | Liquid-liquid phase separation |
| LLPS-DRs | Liquid-liquid phase separation-disordered regions |
| MLOs | Membraneless organelles |
| psself | LLPS region that can phase-separate by itself |
| psother | LLPS region that requires a partner to phase-separate |
| ALS | Amyotrophic lateral sclerosis |
| FTD | Frontotemporal dementia |
| APRs | Aggregation-prone regions |
References
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. TIG 2011, 27, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Holehouse, A.S.; Pappu, R.V. Protein polymers: Encoding phase transitions. Nat. Mater. 2015, 14, 1083–1084. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef] [Green Version]
- Purice, M.D.; Taylor, J.P. Linking hnRNP Function to ALS and FTD Pathology. Front. Neurosci. 2018, 12, 326. [Google Scholar] [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, V.H.; Fawzi, N.L. Physiological, Pathological, and Targetable Membraneless Organelles in Neurons. Trends Neurosci. 2019, 42, 693–708. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Lin, Y.H.; Vernon, R.M.; Forman-Kay, J.D.; Chan, H.S. Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 28795–28805. [Google Scholar] [CrossRef] [PubMed]
- Borroni, B.; Bonvicini, C.; Alberici, A.; Buratti, E.; Agosti, C.; Archetti, S.; Papetti, A.; Stuani, C.; Di Luca, M.; Gennarelli, M.; et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 2009, 30, E974–E983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vance, C.; Rogelj, B.; Hortobagyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef] [Green Version]
- Batlle, C.; Ventura, S. Prion-like domain disease-causing mutations and misregulation of alternative splicing relevance in limb-girdle muscular dystrophy (LGMD) 1G. Neural Regen. Res. 2020, 15, 2239–2240. [Google Scholar] [CrossRef]
- Batlle, C.; Yang, P.; Coughlin, M.; Messing, J.; Pesarrodona, M.; Szulc, E.; Salvatella, X.; Kim, H.J.; Taylor, J.P.; Ventura, S. hnRNPDL Phase Separation Is Regulated by Alternative Splicing and Disease-Causing Mutations Accelerate Its Aggregation. Cell Rep. 2020, 30, 1117–1128. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef]
- Farahi, N.; Lazar, T.; Wodak, S.J.; Tompa, P.; Pancsa, R. Integration of Data from Liquid-Liquid Phase Separation Databases Highlights Concentration and Dosage Sensitivity of LLPS Drivers. Int. J. Mol. Sci. 2021, 22, 3017. [Google Scholar] [CrossRef]
- Santos, J.; Iglesias, V.; Santos-Suarez, J.; Mangiagalli, M.; Brocca, S.; Pallares, I.; Ventura, S. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells 2020, 9, 145. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.; Iglesias, V.; Pintado, C.; Santos-Suarez, J.; Ventura, S. DispHred: A Server to Predict pH-Dependent Order-Disorder Transitions in Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2020, 21, 5814. [Google Scholar] [CrossRef]
- Pintado, C.; Santos, J.; Iglesias, V.; Ventura, S. SolupHred: A server to predict the pH-dependent aggregation of intrinsically disordered proteins. Bioinformatics 2021, 37, 1602–1603. [Google Scholar] [CrossRef]
- Pintado-Grima, C.; Iglesias, V.; Santos, J.; Uversky, V.N.; Ventura, S. DispHScan: A Multi-Sequence Web Tool for Predicting Protein Disorder as a Function of pH. Biomolecules 2021, 11, 1596. [Google Scholar] [CrossRef]
- Jacoby, G.; Segal Asher, M.; Ehm, T.; Abutbul Ionita, I.; Shinar, H.; Azoulay-Ginsburg, S.; Zemach, I.; Koren, G.; Danino, D.; Kozlov, M.M.; et al. Order from Disorder with Intrinsically Disordered Peptide Amphiphiles. J. Am. Chem. Soc. 2021, 143, 11879–11888. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Millett, I.S.; Khodyakova, A.V.; Vasiliev, A.M.; Chernovskaya, T.V.; Vasilenko, R.N.; Kozlovskaya, G.D.; Dolgikh, D.A.; Fink, A.L.; et al. Natively unfolded human prothymosin alpha adopts partially folded collapsed conformation at acidic pH. Biochemistry 1999, 38, 15009–15016. [Google Scholar] [CrossRef]
- Wu, K.P.; Weinstock, D.S.; Narayanan, C.; Levy, R.M.; Baum, J. Structural reorganization of alpha-synuclein at low pH observed by NMR and REMD simulations. J. Mol. Biol. 2009, 391, 784–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, K.; Huang, Q.; Yu, C.; Shen, B.; Sevilla, C.; Shi, M.; Hermjakob, H.; Chen, Y.; Li, T. PhaSepDB: A database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 2020, 48, D354–D359. [Google Scholar] [CrossRef]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Quaglia, F.; Meszaros, B.; Salladini, E.; Hatos, A.; Pancsa, R.; Chemes, L.B.; Pajkos, M.; Lazar, T.; Pena-Diaz, S.; Santos, J.; et al. DisProt in 2022: Improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res. 2021, 50, D480–D487. [Google Scholar] [CrossRef]
- Iglesias, V.; Santos, J.; Santos-Suarez, J.; Pintado-Grima, C.; Ventura, S. SGnn: A Web Server for the Prediction of Prion-Like Domains Recruitment to Stress Granules Upon Heat Stress. Front. Mol. Biosci. 2021, 8, 718301. [Google Scholar] [CrossRef]
- Wallace, E.W.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babinchak, W.M.; Haider, R.; Dumm, B.K.; Sarkar, P.; Surewicz, K.; Choi, J.K.; Surewicz, W.K. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J. Biol. Chem. 2019, 294, 6306–6317. [Google Scholar] [CrossRef] [Green Version]
- Tsoi, P.S.; Quan, M.D.; Choi, K.J.; Dao, K.M.; Ferreon, J.C.; Ferreon, A.C.M. Electrostatic modulation of hnRNPA1 low-complexity domain liquid-liquid phase separation and aggregation. Protein Sci. Publ. Protein Soc. 2021, 30, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Gong, R.; He, F.; Li, Y.; Wang, Y.; Tan, T.; Luo, S.Z. Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Sci. Adv. 2019, 5, eaax5349. [Google Scholar] [CrossRef] [Green Version]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 2018, 7, e31486. [Google Scholar] [CrossRef]
- Van Lindt, J.; Lazar, T.; Pakravan, D.; Demulder, M.; Meszaros, A.; Van Den Bosch, L.; Maes, D.; Tompa, P. F/YGG-motif is an intrinsically disordered nucleic-acid binding motif. RNA Biol. 2022, 19, 622–635. [Google Scholar] [CrossRef]
- Iglesias, V.; Conchillo-Sole, O.; Batlle, C.; Ventura, S. AMYCO: Evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinform. 2019, 20, 24. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, N.C.; Wang, Y.D.; Scarborough, E.A.; Moore, J.; Diaz, Z.; MacLea, K.S.; Freibaum, B.; Li, S.; Molliex, A.; et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013, 495, 467–473. [Google Scholar] [CrossRef]
- Van Lindt, J.; Bratek-Skicki, A.; Nguyen, P.N.; Pakravan, D.; Duran-Armenta, L.F.; Tantos, A.; Pancsa, R.; Van Den Bosch, L.; Maes, D.; Tompa, P. A generic approach to study the kinetics of liquid-liquid phase separation under near-native conditions. Commun. Biol. 2021, 4, 77. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H., Jr.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pintado-Grima, C.; Bárcenas, O.; Ventura, S. In-Silico Analysis of pH-Dependent Liquid-Liquid Phase Separation in Intrinsically Disordered Proteins. Biomolecules 2022, 12, 974. https://doi.org/10.3390/biom12070974
Pintado-Grima C, Bárcenas O, Ventura S. In-Silico Analysis of pH-Dependent Liquid-Liquid Phase Separation in Intrinsically Disordered Proteins. Biomolecules. 2022; 12(7):974. https://doi.org/10.3390/biom12070974
Chicago/Turabian StylePintado-Grima, Carlos, Oriol Bárcenas, and Salvador Ventura. 2022. "In-Silico Analysis of pH-Dependent Liquid-Liquid Phase Separation in Intrinsically Disordered Proteins" Biomolecules 12, no. 7: 974. https://doi.org/10.3390/biom12070974
APA StylePintado-Grima, C., Bárcenas, O., & Ventura, S. (2022). In-Silico Analysis of pH-Dependent Liquid-Liquid Phase Separation in Intrinsically Disordered Proteins. Biomolecules, 12(7), 974. https://doi.org/10.3390/biom12070974







