Let-7 as a Promising Target in Aging and Aging-Related Diseases: A Promise or a Pledge
Abstract
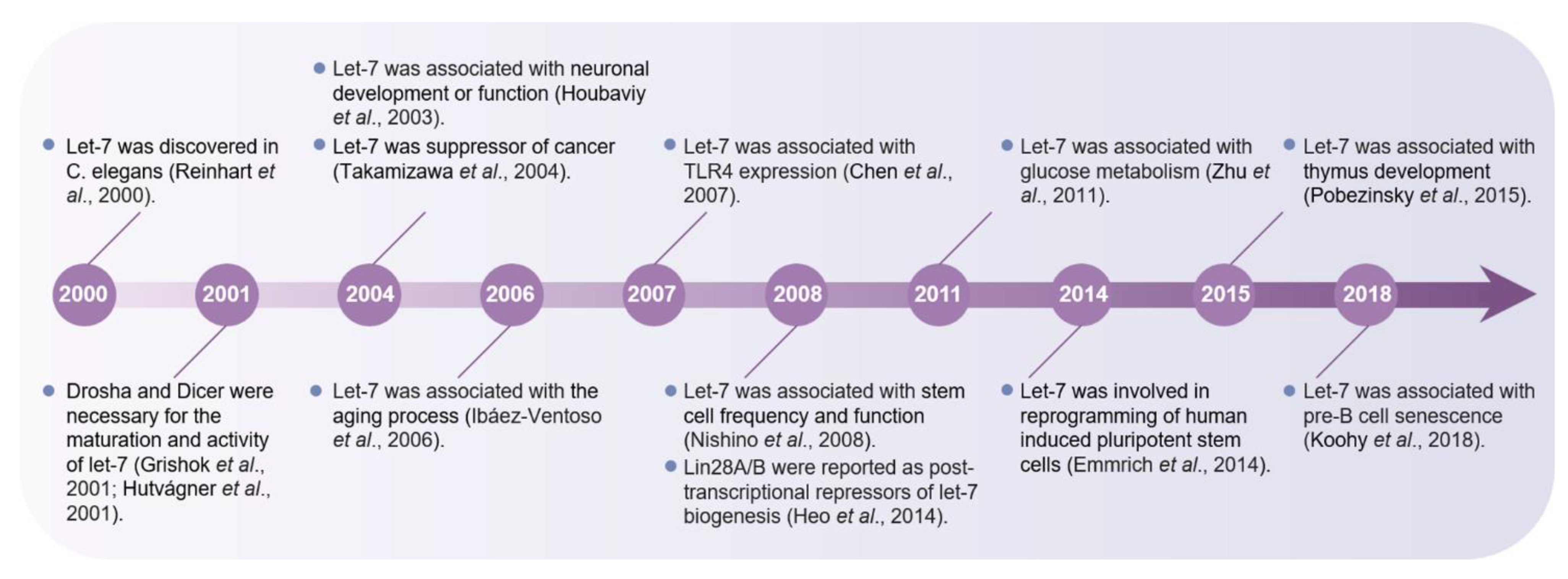
:1. Introduction
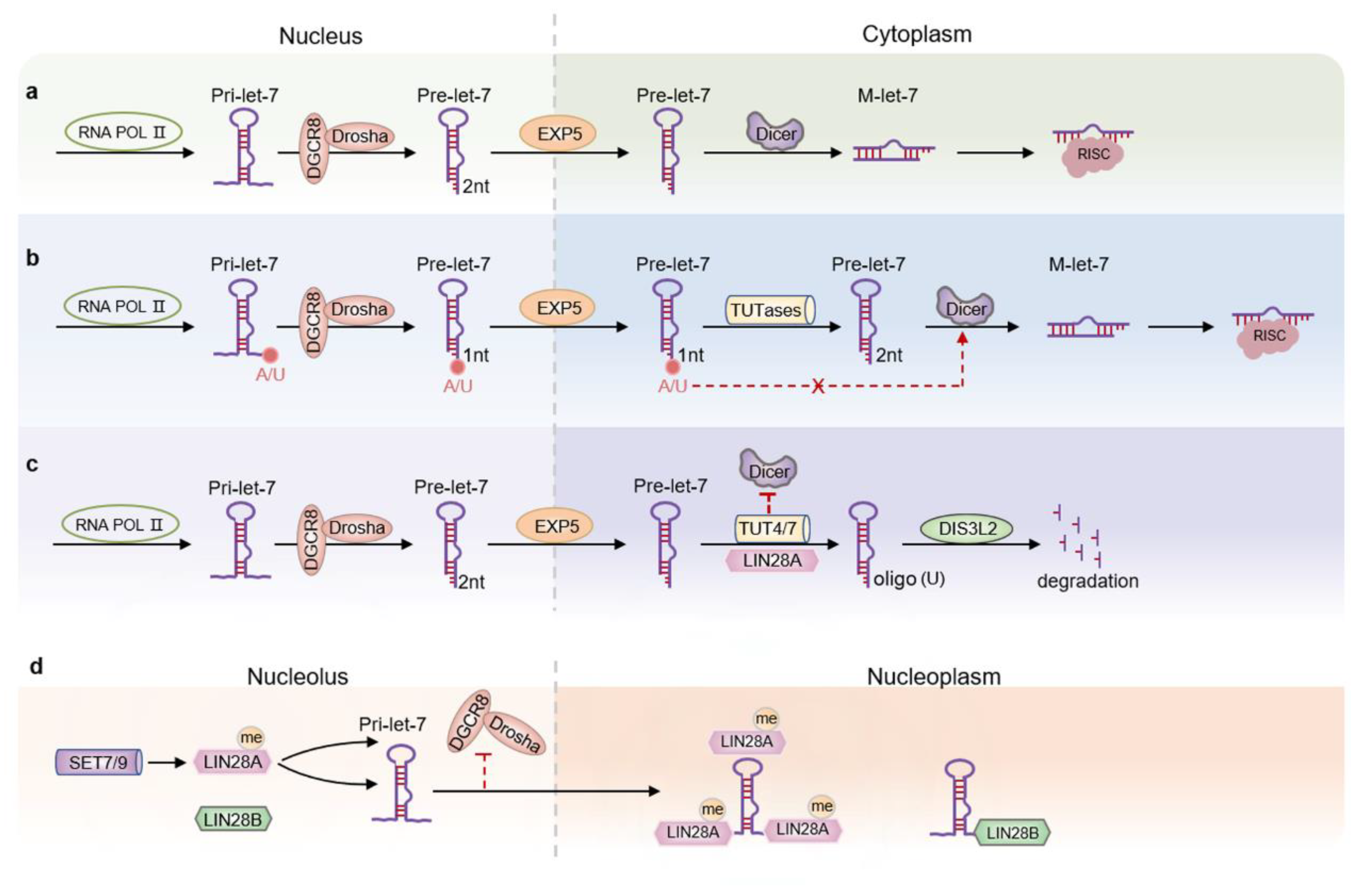
2. Let-7 Expression and Regulation Mechanism
3. Let-7 and Lifespan
3.1. Let-7—The Role in Tissue Aging and ARDs
3.1.1. Let-7 and the Nervous System
3.1.2. Let-7 and the Vision System
3.1.3. Let-7 and the Reproductive System
3.1.4. Let-7 and the Immune System
3.1.5. Let-7 and Other Organizations
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kinser, H.E.; Pincus, Z. MicroRNAs as modulators of longevity and the aging process. Hum. Genet. 2020, 139, 291–308. [Google Scholar] [CrossRef] [PubMed]
- da Costa, J.P.; Vitorino, R.; Silva, G.M.; Vogel, C.; Duarte, A.C.; Rocha-Santos, T. A synopsis on aging-Theories, mechanisms and future prospects. Ageing Res. Rev. 2016, 29, 90–112. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Wang, Y.; Grenell, A.; Zhong, F.; Yam, M.; Hauer, A.; Gregor, E.; Zhu, S.; Lohner, D.; Zhu, J.; Du, J. Metabolic signature of the aging eye in mice. Neurobiol. Aging 2018, 71, 223–233. [Google Scholar] [CrossRef]
- Chandra, A.; Rajawat, J. Skeletal Aging and Osteoporosis: Mechanisms and Therapeutics. Int. J. Mol. Sci. 2021, 22, 3553. [Google Scholar] [CrossRef]
- Campisi, J.; Kapahi, P.; Lithgow, G.J.; Melov, S.; Newman, J.C.; Verdin, E. From discoveries in ageing research to therapeutics for healthy ageing. Nature 2019, 571, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, F.; Godos, J.; Caruso, G.; Owczarek, M.; Jurek, J.; Castellano, S.; Ferri, R.; Caraci, F.; Grosso, G. Dietary Phytoestrogen Intake and Cognitive Status in Southern Italian Older Adults. Biomolecules 2022, 12, 760. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Hertel, J.; Bartschat, S.; Wintsche, A.; Otto, C. Students of the Bioinformatics Computer Lab, Stadler, PF. Evolution of the let-7 microRNA family. RNA Biol. 2012, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Chen, C.H.; Mercer, A.; Sokol, N.S. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev. Cell 2012, 23, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Bloom, O.; Unternaehrer, J.J.; Jiang, A.; Shin, J.S.; Delamarre, L.; Allen, P.; Mellman, I. Spinophilin participates in information transfer at immunological synapses. J. Cell Biol. 2008, 181, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef]
- Emmrich, S.; Rasche, M.; Schning, J.; Reimer, C.; Keihani, S.; Maroz, A.; Xie, Y.; Li, Z.; Schambach, A.; Reinhardt, D.; et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 2014, 28, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef]
- Worringer, K.A.; Rand, T.A.; Hayashi, Y.; Sami, S.; Takahashi, K.; Tanabe, K.; Narita, M.; Srivastava, D.; Yamanaka, S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 2014, 14, 40–52. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Ibáez-Ventoso, C.; Yang, M.; Guo, S.; Robins, H.; Padgett, R.W.; Driscoll, M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 2006, 5, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Akamine, P.S.; Lima, C.R.; Lustoza-Costa, G.J.; Fuziwara, C.S.; Del Debbio, C.B.; Kimura, E.T.; Santos, M.F.; Hamassaki, D.E. Age-related increase of let-7 family microRNA in rat retina and vitreous. Exp. Eye Res. 2021, 204, 108434. [Google Scholar] [CrossRef] [PubMed]
- Shamsuzzama, K.L.; Haque, R.; Nazir, A. Role of MicroRNA Let-7 in Modulating Multifactorial Aspect of Neurodegenerative Diseases: An Overview. Mol. Neurobiol. 2016, 53, 2787–2793. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xia, Q.; Zhao, P.; Cheng, T.; Hong, K.; Xiang, Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev. Biol. 2007, 7, 88. [Google Scholar] [CrossRef]
- Ali, A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. The Role of LIN28-let-7-ARID3B Pathway in Placental Development. Int. J. Mol. Sci. 2020, 21, 3637. [Google Scholar] [CrossRef]
- Joshi, S.; Wei, J.; Bishopric, N.H. A cardiac myocyte-restricted Lin28/let-7 regulatory axis promotes hypoxia-mediated apoptosis by inducing the AKT signaling suppressor PIK3IP1. Biochim. Biophys. Acta 2016, 1862, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Hennchen, M.; Stubbusch, J.; Abarchan-El Makhfi, I.; Kramer, M.; Deller, T.; Pierre-Eugene, C.; Janoueix-Lerosey, I.; Delattre, O.; Ernsberger, U.; Schulte, J.B.; et al. Lin28B and Let-7 in the Control of Sympathetic Neurogenesis and Neuroblastoma Development. J. Neurosci. 2015, 35, 16531–16544. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.; Feng, X.; Zhang, Y.W.; Lou, X.Y.; Cheng, Y.; Zhou, H.H. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int. J. Mol. Sci. 2013, 14, 23086–23102. [Google Scholar] [CrossRef]
- Corre, C.; Shinoda, G.; Zhu, H.; Cousminer, D.L.; Crossman, C.; Bellissimo, C.; Goldenberg, A.; Daley, G.Q.; Palmert, M.R. Sex-specific regulation of weight and puberty by the Lin28/let-7 axis. J. Endocrinol. 2016, 228, 179–191. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.J.; Park, J.E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, M.; Francis, P.; Bilke, S.; Li, X.L.; Hara, T.; Lu, X.; Jones, M.F.; Walker, R.L.; Zhu, Y.; Pineda, M.; et al. A mutant p53/let-7i-axis-regulated gene network drives cell migration, invasion and metastasis. Oncogene 2015, 34, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Cai, N.; Wu, X.L.; Cao, H.Z.; Xie, L.L.; Zheng, P.S. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013, 4, e760. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ni, S. Association between genetic polymorphisms in the promoters of let-7 and risk of cervical squamous cell carcinoma. Gene 2018, 642, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Mitra, D.; Das, P.M.; Huynh, F.C.; Jones, F.E. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J. Biol. Chem. 2011, 286, 40531–40535. [Google Scholar] [CrossRef] [PubMed]
- Ooki, A.; Del Carmen Rodriguez Pena, M.; Marchionni, L.; Dinalankara, W.; Begum, A.; Hahn, N.M.; VandenBussche, C.J.; Rasheed, Z.A.; Mao, S.; Netto, G.J.; et al. YAP1 and COX2 Coordinately Regulate Urothelial Cancer Stem-like. Cells Cancer Res. 2018, 78, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Cisneros-Soberanis, F.; Andonegui, M.A.; Herrera, L.A. miR-125b-1 is repressed by histone modifications in breast cancer cell lines. Springerplus 2016, 5, 959. [Google Scholar] [CrossRef]
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 2019, 74, 1278–1290.e9. [Google Scholar] [CrossRef] [PubMed]
- Hammell, C.M.; Karp, X.; Ambros, V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 18668–18673. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, H.; Han, K.; Kim, S.C.; Choi, Y.; Park, S.W.; Bak, G.; Lee, Y.; Choi, J.K.; Kim, T.K.; et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell 2014, 15, 735–749. [Google Scholar] [CrossRef]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; LaPierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Suzuki, H.I.; Katsura, A.; Miyazono, K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci. 2015, 106, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef]
- Thornton, J.E.; Chang, H.M.; Piskounova, E.; Gregory, R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef]
- Chang, H.M.; Triboulet, R.; Thornton, J.E.; Gregory, R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013, 497, 244–248. [Google Scholar] [CrossRef]
- Malecki, M.; Viegas, S.C.; Carneiro, T.; Golik, P.; Dressaire, C.; Ferreira, M.G.; Arraiano, C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013, 32, 1842–1854. [Google Scholar] [CrossRef]
- McCulloch, K.A.; Rougvie, A.E. Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proc. Natl. Acad. Sci. USA 2014, 111, 15450–15455. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Thomson, J.M.; Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, N.R.; Nowak, J.S.; Zuo, J.; Rappsilber, J.; Spoel, S.H.; Michlewski, G. Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation. Cell Rep. 2014, 9, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Ahmad, R.; Rajabi, H.; Kufe, D. MUC1-C Induces the LIN28B→LET-7→HMGA2 Axis to Regulate Self-Renewal in NSCLC. Mol. Cancer Res. 2015, 13, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.J.; Seo, J.B.; Kim, S.H.; Jeon, Y.J.; Suh, S.S. Fhit induces the reciprocal suppressions between Lin28/Let-7 and miR-17/92miR. Int. J. Med. Sci. 2021, 18, 706–714. [Google Scholar] [CrossRef]
- Sakamoto, S.; Aoki, K.; Higuchi, T.; Todaka, H.; Morisawa, K.; Tamaki, N.; Hatano, E.; Fukushima, A.; Taniguchi, T.; Agata, Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell Biol. 2009, 29, 3754–3769. [Google Scholar] [CrossRef]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010, 17, 1011–1018. [Google Scholar] [CrossRef]
- Haselmann, V.; Kurz, A.; Bertsch, U.; Hübner, S.; Olempska-Müller, M.; Fritsch, J.; Häsler, R.; Pickl, A.; Fritsche, H.; Annewanter, F.; et al. Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology 2014, 146, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Arase, M.; Matsuyama, H.; Choi, Y.L.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell 2011, 44, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Veksler-Lublinsky, I.; Morrissey, D.; Ambros, V. Staufen Negatively Modulates MicroRNA Activity in Caenorhabditis elegans. G3 2016, 6, 1227–1237. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, B.O.; Kim, K.; Park, B.C.; Park, S.G.; Kim, J.H.; Kim, S. Sjgren Syndrome antigen B regulates LIN28-let-7 axis in Caenorhabditis elegans and human. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194684. [Google Scholar] [CrossRef]
- Yu, B.; Bi, L.; Zheng, B.; Ji, L.; Chevalier, D.; Agarwal, M.; Ramachandran, V.; Li, W.; Lagrange, T.; Walker, J.C.; et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 10073–10078. [Google Scholar] [CrossRef]
- Kim, C.W.; Vo, M.T.; Kim, H.K.; Lee, H.H.; Yoon, N.A.; Lee, B.J.; Min, Y.J.; Joo, W.D.; Cha, H.J.; Park, J.W.; et al. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012, 40, 3856–3869. [Google Scholar] [CrossRef]
- Pilotte, J.; Dupont-Versteegden, E.E.; Vanderklish, P.W. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS ONE 2011, 6, e28446. [Google Scholar] [CrossRef]
- Kawai, S.; Amano, A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012, 197, 201–208. [Google Scholar] [CrossRef]
- Buratti, E.; De Conti, L.; Stuani, C.; Romano, M.; Baralle, M.; Baralle, F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010, 277, 2268–2281. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, S.; Kim, M.S.; Choi, K.; Cho, J.Y.; Gwak, H.S.; Kim, Y.J.; Yoo, H.; Lee, S.H.; Park, J.B.; et al. The ubiquitin ligase human TRIM71 regulates let-7 microRNA biogenesis via modulation of Lin28B protein. Biochim. Biophys. Acta 2014, 1839, 374–386. [Google Scholar] [CrossRef]
- Reinsborough, C.W.; Ipas, H.; Abell, N.S.; Gouws, E.B.; Williams, J.P.; Mercado, M.; Van Den Berg, C.; Xhemalçe, B. BCDIN3D RNA methyltransferase stimulates Aldolase C expression and glycolysis through let-7 microRNA in breast cancer cells. Oncogene 2021, 40, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chan, J.; Chen, W.; Li, J.; Sun, M.; Kannan, G.S.; Mok, Y.K.; Yuan, Y.A.; Jobichen, C. SYNCRIP: A new player in pri-let-7a processing. RNA 2020, 26, 290–305. [Google Scholar] [CrossRef]
- Bahn, J.H.; Ahn, J.; Lin, X.; Zhang, Q.; Lee, J.H.; Civelek, M.; Xiao, X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015, 6, 6355. [Google Scholar] [CrossRef]
- Zipeto, M.A.; Court, A.C.; Sadarangani, A.; Delos Santos, N.P.; Balaian, L.; Chun, H.J.; Pineda, G.; Morris, S.R.; Mason, C.N.; Geron, I.; et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell 2016, 19, 177–191. [Google Scholar] [CrossRef]
- Smith-Vikos, T.; Slack, F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012, 125 Pt 1, 7–17. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Zeng, L.; Shi, C.; Peng, Y.; Li, H.; Chen, H.; Yu, J.; Zhang, J.; Cheng, B.; et al. Tris(1,3-dichloro-2-propyl) phosphate reduces longevity through a specific microRNA-mediated DAF-16/FoxO in an unconventional insulin/insulin-like growth factor 1 signaling pathway. J. Hazard. Mater. 2022, 425, 128043. [Google Scholar] [CrossRef]
- Xu, F.; Pang, L.; Cai, X.; Liu, X.; Yuan, S.; Fan, X.; Jiang, B.; Zhang, X.; Dou, Y.; Gorospe, M.; et al. let-7-repressesed Shc translation delays replicative senescence. Aging Cell 2014, 13, 185–192. [Google Scholar] [CrossRef]
- Gendron, C.M.; Pletcher, S.D. MicroRNAs mir-184 and let-7 alter Drosophila metabolism and longevity. Aging Cell 2017, 16, 1434–1438. [Google Scholar] [CrossRef]
- Chawla, G.; Deosthale, P.; Childress, S.; Wu, Y.C.; Sokol, N.S. A let-7-to-miR-125 MicroRNA Switch Regulates Neuronal Integrity and Lifespan in Drosophila. PLoS Genet. 2016, 12, e1006247. [Google Scholar] [CrossRef]
- Kumar Dev, P.; Gray, A.J.; Scott-Hamilton, J.; Hagstrom, A.D.; Murphy, A.; Denham, J. Co-expression analysis identifies networks of miRNAs implicated in biological ageing and modulated by short-term interval training. Mech. Ageing Dev. 2021, 199, 111552. [Google Scholar] [CrossRef]
- Chiu, S.C.; Chung, H.Y.; Cho, D.Y.; Chan, T.M.; Liu, M.C.; Huang, H.M.; Li, T.Y.; Lin, J.Y.; Chou, P.C.; Fu, R.H.; et al. Therapeutic potential of microRNA let-7, tumor suppression or impeding normal stemness. Cell Transpl. 2014, 23, 459–469. [Google Scholar] [CrossRef]
- Wu, L.; Nguyen, L.H.; Zhou, K.; de Soysa, T.Y.; Li, L.; Miller, J.B.; Tian, J.; Locker, J.; Zhang, S.; Shinoda, G.; et al. Precise let-7 expression levels balance organ regeneration against tumor suppression. eLife 2015, 4, e09431. [Google Scholar] [CrossRef]
- Smith, A.M.; Dykeman, C.A.; King, B.L.; Yin, V.P. Modulation of TNFα Activity by the microRNA Let-7 Coordinates Zebrafish Heart Regeneration. iScience 2019, 15, 1–15. [Google Scholar] [CrossRef]
- Nishino, J.; Kim, I.; Chada, K.; Morrison, S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 2008, 135, 227–239. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- Zou, Y.; Chiu, H.; Zinovyeva, A.; Ambros, V.; Chuang, C.F.; Chang, C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 2013, 340, 372–376. [Google Scholar] [CrossRef]
- Armakola, M.; Ruvkun, G. Regulation of Caenorhabditis elegans neuronal polarity by heterochronic genes. Proc. Natl. Acad. Sci. USA 2019, 116, 12327–12336. [Google Scholar] [CrossRef]
- Zhao, J.; Yue, D.; Zhou, Y.; Jia, L.; Wang, H.; Guo, M.; Xu, H.; Chen, C.; Zhang, J.; Xu, L. The Role of MicroRNAs in Aβ Deposition and Tau Phosphorylation in Alzheimer’s Disease. Front. Neurol. 2017, 8, 342. [Google Scholar] [CrossRef]
- Hornsten, A.; Lieberthal, J.; Fadia, S.; Malins, R.; Ha, L.; Xu, X.; Daigle, I.; Markowitz, M.; O’Connor, G.; Plasterk, R.; et al. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc. Natl. Acad. Sci. USA 2007, 104, 1971–1976. [Google Scholar] [CrossRef]
- Niwa, R.; Zhou, F.; Li, C.; Slack, F.J. The expression of the Alzheimer’s amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev. Biol. 2008, 315, 418–425. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, T.; Yao, Q.; Zheng, L.; Zhang, Z.; Wang, J.; Hu, Z.; Cui, H.; Han, Y.; Han, X.; et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-κB-dependent manner. FEBS J. 2017, 284, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, T.; Xiu, W.; Cao, W.; He, M.; Sun, W.; Zhao, W. MiR-107 overexpression attenuates neurotoxicity induced by 6-hydroxydopamine both in vitro and in vivo. Chem. Biol. Interact. 2020, 315, 108908. [Google Scholar] [CrossRef]
- Chen, P.S.; Su, J.L.; Cha, S.T.; Tarn, W.Y.; Wang, M.Y.; Hsu, H.C.; Lin, M.T.; Chu, C.Y.; Hua, K.T.; Chen, C.N.; et al. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J. Clin. Investig. 2011, 121, 3442–3455. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, M.M.; Zheng, B.; Lu, T.; Yan, Z.; Py, B.F.; Ng, A.; Xavier, R.J.; Li, C.; Yankner, B.A.; Scherzer, C.R.; et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2010, 107, 14164–14169. [Google Scholar] [CrossRef]
- Nixon, R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007, 120, 4081–4091. [Google Scholar] [CrossRef]
- Liu, Q.Y.; Chang, M.N.; Lei, J.X.; Koukiekolo, R.; Smith, B.; Zhang, D.; Ghribi, O. Identification of microRNAs involved in Alzheimer’s progression using a rabbit model of the disease. Am. J. Neurodegener. Dis. 2014, 3, 33–44. [Google Scholar]
- Derkow, K.; Rössling, R.; Schipke, C.; Krüger, C.; Bauer, J.; Fähling, M.; Stroux, A.; Schott, E.; Ruprecht, K.; Peters, O.; et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer’s disease. PLoS ONE 2018, 13, e0200602. [Google Scholar] [CrossRef]
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef]
- Bordi, M.; Berg, M.J.; Mohan, P.S.; Peterhoff, C.M.; Alldred, M.J.; Che, S.; Ginsberg, S.D.; Nixon, R.A. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy 2016, 12, 2467–2483. [Google Scholar] [CrossRef]
- Gu, H.; Li, L.; Cui, C.; Zhao, Z.; Song, G. Overexpression of let-7a increases neurotoxicity in a PC12 cell model of Alzheimer’s disease via regulating autophagy. Exp. Ther. Med. 2017, 14, 3688–3698. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Romano, R.; Coelho-Júnior, H.J.; Bucci, C.; Marzetti, E. Mitochondrial Dysfunction, Protein Misfolding and Neuroinflammation in Parkinson’s Disease: Roads to Biomarker Discovery. Biomolecules 2021, 11, 1508. [Google Scholar] [CrossRef]
- Beitz, J.M. Parkinson’s disease: A review. Front. Biosci. 2014, 6, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, S.; Imai, Y.; Sokol, N.; Lu, B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 2010, 466, 637–641. [Google Scholar] [CrossRef]
- Shamsuzzama, K.L.; Nazir, A. Modulation of Alpha-synuclein Expression and Associated Effects by MicroRNA Let-7 in Transgenic, C. elegans. Front. Mol. Neurosci. 2017, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Xia, Y.; Zhang, J.; Chen, Y.; Yuan, J.; Niu, X.; Zhao, B.; Li, Q.; Wang, Y.; Deng, Z. ESC-sEVs Rejuvenate Senescent Hippocampal NSCs by Activating Lysosomes to Improve Cognitive Dysfunction in Vascular Dementia. Adv. Sci. 2020, 7, 1903330. [Google Scholar] [CrossRef] [PubMed]
- Wohl, S.G.; Reh, T.A. The microRNA expression profile of mouse Müller glia in vivo and in vitro. Sci. Rep. 2016, 6, 35423. [Google Scholar] [CrossRef] [PubMed]
- ElShelmani, H.; Wride, M.A.; Saad, T.; Rani, S.; Kelly, D.J.; Keegan, D. Identification of Novel Serum MicroRNAs in Age-Related Macular Degeneration. Transl. Vis. Sci. Technol. 2020, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.D.; Reibaldi, M.; Avitabile, T.; Bucolo, C.; Salomone, S.; Rejdak, R.; Nowomiejska, K.; Tripodi, S.; Posarelli, C.; Ragusa, M.; et al. MicroRNAs in the Vitreous Humor of Patients with Retinal Detachment and a Different Grading of Proliferative Vitreoretinopathy: A Pilot Study. Transl. Vis. Sci. Technol. 2020, 9, 23. [Google Scholar] [CrossRef]
- Liu, Y.C.; Wilkins, M.; Kim, T.; Malyugin, B.; Mehta, J.S. Cataracts. Lancet 2017, 390, 600–612. [Google Scholar] [CrossRef]
- Peng, C.H.; Liu, J.H.; Woung, L.C.; Lin, T.J.; Chiou, S.H.; Tseng, P.C.; Du, W.Y.; Cheng, C.K.; Hu, C.C.; Chien, K.H.; et al. MicroRNAs and cataracts: Correlation among let-7 expression, age and the severity of lens opacity. Br. J. Ophthalmol. 2012, 96, 747–751. [Google Scholar] [CrossRef]
- Dong, Y.; Zheng, Y.; Xiao, J.; Zhu, C.; Zhao, M. MicroRNA let-7b induces lens epithelial cell apoptosis by targeting leucine-rich repeat containing G protein-coupled receptor 4 (Lgr4) in age-related cataract. Exp. Eye Res. 2016, 147, 98–104. [Google Scholar] [CrossRef]
- Babizhayev, M.A.; Yegorov, Y.E. Reactive Oxygen Species and the Aging Eye: Specific Role of Metabolically Active Mitochondria in Maintaining Lens Function and in the Initiation of the Oxidation-Induced Maturity Onset Cataract—A Novel Platform of Mitochondria-Targeted Antioxidants With Broad Therapeutic Potential for Redox Regulation and Detoxification of Oxidants in Eye Diseases. Am. J. Ther. 2016, 23, e98–e117. [Google Scholar]
- Li, T.; Huang, Y.; Zhou, W.; Yan, Q. Let-7c-3p Regulates Autophagy under Oxidative Stress by Targeting ATG3 in Lens Epithelial Cells. BioMed Res Int. 2020, 2020, 6069390. [Google Scholar] [CrossRef]
- Kirkwood, T.B. Understanding the odd science of aging. Cell 2005, 120, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Arantes-Oliveira, N.; Apfeld, J.; Dillin, A.; Kenyon, C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 2002, 295, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Van Wynsberghe, P.M.; Kai, Z.S.; Massirer, K.B.; Burton, V.H.; Yeo, G.W.; Pasquinelli, A.E. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2011, 18, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hou, L.; Nakamura, S.; Su, M.; Li, F.; Chen, W.; Yan, Y.; Green, C.D.; Chen, D.; Zhang, H.; et al. LIN-28 balances longevity and germline stem cell number in Caenorhabditis elegans through let-7/AKT/DAF-16 axis. Aging Cell 2017, 16, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Sreejith, P.; Jang, W.; To, V.; Hun Jo, Y.; Biteau, B.; Kim, C. Lin28 is a critical factor in the function and aging of Drosophila testis stem cell niche. Aging 2019, 11, 855–873. [Google Scholar] [CrossRef]
- Toledano, H.; D’Alterio, C.; Czech, B.; Levine, E.; Jones, D.L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 2012, 485, 605–610. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, Y.; Peng, S.; Wu, L.; Lin, H.Y.; Wang, S.; Wang, H. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction 2012, 144, 235–244. [Google Scholar] [CrossRef]
- Cao, R.; Wu, W.J.; Zhou, X.L.; Xiao, P.; Wang, Y.; Liu, H.L. Expression and preliminary functional profiling of the let-7 family during porcine ovary follicle atresia. Mol. Cells 2015, 38, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wu, W.; Zhou, X.; Liu, K.; Li, B.; Huang, X.; Zhang, Y.; Liu, H. Let-7g induces granulosa cell apoptosis by targeting MAP3K1 in the porcine ovary. Int. J. Biochem. Cell Biol. 2015, 68, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, J.; Pan, Z.; Du, X.; Li, X.; Ma, B.; Yao, W.; Li, Q.; Liu, H. The let-7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor-β type 1 receptor. Mol. Cell Endocrinol. 2015, 409, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Jing, F.; Huang, P.; Geng, Z.; Xu, J.; Li, J.; Chen, D.; Zhu, Y.; Wang, Z.; Huang, W.; et al. Thymopentin alleviates premature ovarian failure in mice by activating YY2/Lin28A and inhibiting the expression of let-7 family microRNAs. Cell Prolif. 2021, 54, e13089. [Google Scholar] [CrossRef]
- Nikolich-Žugich, J. The twilight of immunity: Emerging concepts in aging of the immune system. Nat. Immunol. 2018, 19, 10–19. [Google Scholar] [CrossRef]
- Hu, L.; Zhou, Y.; Yang, J.; Zhao, X.; Mao, L.; Zheng, W.; Zhao, J.; Guo, M.; Chen, C.; He, Z.; et al. MicroRNA-7 overexpression positively regulates the CD8+ SP cell development via targeting PIK3R1. Exp. Cell Res. 2021, 407, 112824. [Google Scholar] [CrossRef]
- Zhao, J.; Chu, F.; Xu, H.; Guo, M.; Shan, S.; Zheng, W.; Tao, Y.; Zhou, Y.; Hu, Y.; Chen, C.; et al. C/EBPα/miR-7 Controls CD4+ T-Cell Activation and Function and Orchestrates Experimental Autoimmune Hepatitis in Mice. Hepatology 2021, 74, 379–396. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W.; Wang, S.E. MicroRNA Let-7 in B lymphocyte activation. Aging 2019, 11, 2547–2548. [Google Scholar] [CrossRef]
- Wells, A.C.; Daniels, K.A.; Angelou, C.C.; Fagerberg, E.; Burnside, A.S.; Markstein, M.; Alfandari, D.; Welsh, R.M.; Pobezinskaya, E.L.; Pobezinsky, L.A. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. eLife 2017, 6, e26398. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W.; Wang, S.E.; Baltimore, D. Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 12416–12421. [Google Scholar] [CrossRef]
- Pobezinsky, L.A.; Etzensperger, R.; Jeurling, S.; Alag, A.; Kadakia, T.; McCaughtry, T.M.; Kimura, M.Y.; Sharrow, S.O.; Guinter, T.I.; Feigenbaum, L.; et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat. Immunol. 2015, 16, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.P.; Sanders, V.M.; Witte, P.L. Stage-specific alterations in murine B lymphopoiesis with age. Int. Immunol. 1996, 8, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Cancro, M.P.; Hao, Y.; Scholz, J.L.; Riley, R.L.; Frasca, D.; Dunn-Walters, D.K.; Blomberg, B.B. B cells and aging: Molecules and mechanisms. Trends Immunol. 2009, 30, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Koohy, H.; Bolland, D.J.; Matheson, L.S.; Schoenfelder, S.; Stellato, C.; Dimond, A.; Várnai, C.; Chovanec, P.; Chessa, T.; Denizot, J.; et al. Genome organization and chromatin analysis identify transcriptional downregulation of insulin-like growth factor signaling as a hallmark of aging in developing B cells. Genome Biol. 2018, 19, 126. [Google Scholar] [CrossRef]
- Chung, H.Y.; Kim, D.H.; Lee, E.K.; Chung, K.W.; Chung, S.; Lee, B.; Seo, A.Y.; Chung, J.H.; Jung, Y.S.; Im, E.; et al. Redefining Chronic Inflammation in Aging and Age-Related Diseases: Proposal of the Senoinflammation Concept. Aging Dis. 2019, 10, 367–382. [Google Scholar] [CrossRef]
- Chen, H.; Guo, M.; Yue, D.; Zhao, J.; Zhou, Y.; Chen, C.; Liang, G.; Xu, L. MicroRNA-7 negatively regulates Toll-like receptor 4 signaling pathway through FAM177A. Immunology 2021, 162, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Zhao, J.; Chen, H.; Guo, M.; Chen, C.; Zhou, Y.; Xu, L. MicroRNA-7, synergizes with RORα, negatively controls the pathology of brain tissue inflammation. J. Neuroinflamm. 2020, 17, 28. [Google Scholar] [CrossRef]
- Chen, X.M.; Splinter, P.L.; O’Hara, S.P.; LaRusso, N.F. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 2007, 282, 28929–28938. [Google Scholar] [CrossRef]
- Diniz, G.P.; Wang, D.Z. Regulation of Skeletal Muscle by microRNAs. Compr. Physiol. 2016, 6, 1279–1294. [Google Scholar]
- Drummond, M.J.; McCarthy, J.J.; Sinha, M.; Spratt, H.M.; Volpi, E.; Esser, K.A.; Rasmussen, B.B. Aging and microRNA expression in human skeletal muscle: A microarray and bioinformatics analysis. Physiol. Genom. 2011, 43, 595–603. [Google Scholar] [CrossRef]
- Polesskaya, A.; Degerny, C.; Pinna, G.; Maury, Y.; Kratassiouk, G.; Mouly, V.; Morozova, N.; Kropp, J.; Frandsen, N.; Harel-Bellan, A. Genome-wide exploration of miRNA function in mammalian muscle cell differentiation. PLoS ONE 2013, 8, e71927. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.J.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080. [Google Scholar] [CrossRef] [PubMed]
- Luca, E.; Turcekova, K.; Hartung, A.; Mathes, S.; Rehrauer, H.; Krützfeldt, J. Genetic deletion of microRNA biogenesis in muscle cells reveals a hierarchical non-clustered network that controls focal adhesion signaling during muscle regeneration. Mol. Metab. 2020, 36, 100967. [Google Scholar] [CrossRef]
- Cappelletti, C.; Galbardi, B.; Bruttini, M.; Salerno, F.; Canioni, E.; Pasanisi, M.B.; Rodolico, C.; Brizzi, T.; Mora, M.; Renieri, A.; et al. Aging-associated genes and let-7 microRNAs: A contribution to myogenic program dysregulation in oculopharyngeal muscular dystrophy. FASEB J. 2019, 33, 7155–7167. [Google Scholar] [CrossRef]
- Tokuzawa, Y.; Yagi, K.; Yamashita, Y.; Nakachi, Y.; Nikaido, I.; Bono, H.; Ninomiya, Y.; Kanesaki-Yatsuka, Y.; Akita, M.; Motegi, H.; et al. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS Genet. 2010, 6, e1001019. [Google Scholar] [CrossRef]
- Wei, J.; Li, H.; Wang, S.; Li, T.; Fan, J.; Liang, X.; Li, J.; Han, Q.; Zhu, L.; Fan, L.; et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014, 23, 1452–1463. [Google Scholar] [CrossRef]
- Chen, R.; Liao, X.; Chen, F.; Wang, B.; Huang, J.; Jian, G.; Huang, Z.; Yin, G.; Liu, H.; Jin, D. Circulating microRNAs, miR-10b-5p, miR-328-3p, miR-100 and let-7, are associated with osteoblast differentiation in osteoporosis. Int. J. Clin. Exp. Pathol. 2018, 11, 1383–1390. [Google Scholar]
- Jin, B.; Wang, W.; Meng, X.X.; Du, G.; Li, J.; Zhang, S.Z.; Zhou, B.H.; Fu, Z.H. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer 2016, 16, 863. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef]
- Borgoni, S.; Kudryashova, K.S.; Burka, K.; de Magalhães, J.P. Targeting immune dysfunction in aging. Ageing Res. Rev. 2021, 70, 101410. [Google Scholar] [CrossRef]
- Arsenis, N.C.; You, T.; Ogawa, E.F.; Tinsley, G.M.; Zuo, L. Physical activity and telomere length: Impact of aging and potential mechanisms of action. Oncotarget 2017, 8, 45008–45019. [Google Scholar] [CrossRef] [PubMed]
- Kubben, N.; Misteli, T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 2017, 18, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.Q.; Franck, N.; Egan, B.; Sjgren, R.J.; Katayama, M.; Duque-Guimaraes, D.; Arner, P.; Zierath, J.R.; Krook, A. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1359–E1366. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Q.; Yao, J.; Jiang, H.; Xiao, C.; Wu, F. MicroRNA let-7g and let-7i inhibit hepatoma cell growth concurrently via downregulation of the anti-apoptotic protein B-cell lymphoma-extra large. Oncol. Lett. 2015, 9, 213–218. [Google Scholar] [CrossRef]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Chen, C.; Zhao, J.; Wang, H.; Guo, M.; Zhou, Y.; Luo, J.; Zhang, J.; Xu, L. Targeted Expression of miR-7 Operated by TTF-1 Promoter Inhibited the Growth of Human Lung Cancer through the NDUFA4 Pathway. Mol. Ther. Nucleic Acids 2017, 6, 183–197. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Schofield, A.L.; Brown, J.P.; Brown, J.; Wilczynska, A.; Bell, C.; Glaab, W.E.; Hackl, M.; Howell, L.; Lee, S.; Dear, J.W.; et al. Systems analysis of miRNA biomarkers to inform drug safety. Arch. Toxicol. 2021, 95, 3475–3495. [Google Scholar] [CrossRef]


| Inhibitory Regulatory Protein | Family Member | Mechanism | Ref. |
| Lin42 | let-7a, 7b | Suppresses let-7 transcriptionally by binding to the pri-let-7 3′UTR | [8,52] |
| Lin28A-TUTases4/7 | let-7a, 7b, 7d, 7g, 7i | Represses let-7 through TUTase-dependent uridylation of pre-let-7 | [48,49] |
| Lin28B | let-7a, 7d, 7f, 7g, 7i | Represses let-7 by sequestering pri-let-7 into the nucleolus | [45,53] |
| TRIM25 | let-7a | Activates TuT4, allowing for more efficient Lin28A-mediated uridylation | [54] |
| MUC1-C | let-7c | Activates Lin28B and synergistically represses let-7 | [55] |
| MSI1 | let-7b, 7g, miR-98 | Recruits Lin28 to the nucleus and represses let-7 | [56] |
| FHIT | let-7a, 7b, 7d, 7f, 7g | Induces Lin28B protein, consequently inhibiting let-7 | [57] |
| NF90/NF45 | let-7a | Directly binds to pri-let-7 and interacts with Drosha complex to inhibit pri-let-7 processing | [58] |
| YAP | Let-7g | Translocates into the nucleus and sequesters DDX17 and interferes with Drosha processing | [59] |
| hnRNPA1 | let-7a | Reduces Drosha processing | [60] |
| TRAIL-R2 | let-7a, 7b, 7c, 7d, 7e, 7g | Interacts with Drosha and DGCR8 to inhibit pri-let-7 processing | [61] |
| MCPIP1 | let-7g | Cleaves terminal loops on the pre-let-7 leading to degradation | [62] |
| STAUFEN | let-7s | Likely binds to pri-let-7 3′UTR and negatively modulates let-7 | [63] |
| SSB | let-7a, 7b, 7c, 7d, 7e, 7f, 7g, 7i | Positively regulates Lin28 to suppress the maturation of let-7 | [64] |
| Activating Regulatory Protein | Family Member | Mechanism | Ref. |
| METTL1 | let-7e | METTL1-mediated methylation augments let-7 processing by disrupting an inhibitory secondary structure within the pri-let-7 transcript | [41] |
| TUTases2/4/7 | let-7a, 7b, 7d, 7f, 7g, 7i, miR-98 | Specific mono-uridylation of pre-let-7 for preferential binding and cleavage by Dicer | [34] |
| SNIP1 | let-7i | Likely binds pri-let-7 and enhances Drosha processing | [65] |
| TTP | Let-7a, 7b, 7f, 7g | Enhances let-7 expression by down-regulation of Lin28A expression | [66] |
| KSRP | let-7a | Promotes let-7 maturation as part of Drosha and Dicer complexes | [60] |
| RBM3 | let-7a, 7g, 7i | Binds pre-let-7s/enhance Dicer | [67] |
| BRCA1 | let-7a | Enhances pri-let-7s processing mediated by Drosha complex | [68] |
| TDP-43 | let-7b | Promotes microRNA biogenesis as a component of the Drosha and Dicer complexes | [69] |
| TRIM71 | let-7a, 7b, 7c, 7d, 7e, 7f, 7g, 7i, miR-98 | Negatively regulates Lin28B through polyubiquitination | [70] |
| BCDIN3D | let-7b, 7d, 7e, 7f, 7g, 7i, miR-98 | Methylates pre-let-7s and enhances Dicer processing | [71] |
| SYNCRIP | let-7a | Binds to pri-let-7 terminal loop and enhances Drosha processing | [72] |
| Inhibitory/Activating Regulatory Protein | Family Member | Mechanism | Ref. |
| DAF-12 | let-7 family | Unliganded DAF-12 represses let-7 and liganded DAF-12 promotes let-7 transcriptionally through binding to pri-let-7 3′-UTR | [42] |
| MYC | let-7a, 7d, 7f | Inhibited let-7 promoter activity via binding to the noncanonical E-box 3 downstream of the transcription start sites Enhanced promoter activity by binding to the canonical E-box 2 upstream of the transcription start sites | [43] |
| ADAR1 | let-7 family | Directly binds and edits pri-let-7d transcripts thereby reducing the expression of mature let-7d Enhances Drosha and Dicer processing through direct interactions | [73,74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhao, J.; Chen, S.; Li, D.; Yang, J.; Zhao, X.; Qin, M.; Guo, M.; Chen, C.; He, Z.; et al. Let-7 as a Promising Target in Aging and Aging-Related Diseases: A Promise or a Pledge. Biomolecules 2022, 12, 1070. https://doi.org/10.3390/biom12081070
Wang Y, Zhao J, Chen S, Li D, Yang J, Zhao X, Qin M, Guo M, Chen C, He Z, et al. Let-7 as a Promising Target in Aging and Aging-Related Diseases: A Promise or a Pledge. Biomolecules. 2022; 12(8):1070. https://doi.org/10.3390/biom12081070
Chicago/Turabian StyleWang, Ya, Juanjuan Zhao, Shipeng Chen, Dongmei Li, Jing Yang, Xu Zhao, Ming Qin, Mengmeng Guo, Chao Chen, Zhixu He, and et al. 2022. "Let-7 as a Promising Target in Aging and Aging-Related Diseases: A Promise or a Pledge" Biomolecules 12, no. 8: 1070. https://doi.org/10.3390/biom12081070






