Protein Function Analysis through Machine Learning
Abstract
1. Introduction
2. Foundational Concepts of Protein Function
2.1. Elucidating Protein Function Using Computational Biology: A Short History
2.2. Convergence of Machine Learning and Computational Biology
3. Selected Applications of Machine Learning in Computational Biology
3.1. Protein Structure Prediction
3.2. Conformational Ensembles
3.2.1. Enhanced Sampling Methods
3.2.2. Identifying Collective Variables
3.2.3. Automated Potential Biasing
3.2.4. Clustering Conformations and Markov Model State Space Partitioning
3.3. Protein Stability
3.3.1. Role of Environment
3.3.2. Protein Engineering Through Mutagenesis
3.3.3. Protein–Protein Interactions
| Paradigm | Method | Year | Model | Stand Alone | Webserver |
|---|---|---|---|---|---|
| Sequence | DPPI [254] | 2018 | CNN | Yes | No |
| EnsDNN [255] | 2019 | DNN | Yes | No | |
| MDPN [256] | 2019 | DPN | No | No | |
| CNN-FSRF [257] | 2019 | CNN + RF | No | No | |
| S-VGAE [258] | 2020 | GCNN + VAE | Yes | No | |
| EnAmDNN [259] | 2020 | DNN + Att | Yes | No | |
| PCPIP [260] | 2021 | SVM | No | Yes | |
| CAMP [261] | 2021 | CNN | Yes | No | |
| Balogh et al. [262] | 2022 | GAN | Yes | No | |
| TAGPPI [263] | 2022 | GCNN | Yes | No | |
| Structure | Daberdaku et al. [264] | 2018 | SVM | Yes | No |
| BIPSPI-structure [265] | 2018 | DT | Yes | Yes | |
| IntPred [266] | 2020 | RF | No | No | |
| MaSIF [267] | 2021 | ANN | Yes | No | |
| GraphPPIS [268] | 2021 | GCNN | Yes | Yes |
3.3.4. Role of Rigidity in Disordered Proteins
| Method | Year | Model | Stand Alone | Webserver |
|---|---|---|---|---|
| MoRFchibi [278] | 2016 | SVM | Yes | Yes |
| AUCpreD [279] | 2016 | CNF | Yes | Yes |
| Predict-MoRFs [280] | 2016 | SVM | Yes | No |
| SPOT-Disorder1 [281] | 2017 | RNN | Yes | Yes |
| MoRFPred-plus [282] | 2018 | SVM | Yes | No |
| OPAL+ [283] | 2018 | SVM | Yes | Yes |
| rawMSA [284] | 2019 | CNN + RNN | Yes | No |
| SPOT-Disorder2 [285] | 2019 | CNN + RNN | Yes | Yes |
| ODiNPred [286] | 2020 | SNN | No | Yes |
| IDP-Seq2Seq [287] | 2020 | RNN + Att | No | Yes |
| flDPnn [288] | 2021 | DNN + RF | Yes | Yes |
| flDPlr [288] | 2021 | RF | No | No |
| RFPR-IDP [289] | 2021 | CNN + RNN | No | Yes |
| Metapredict [290] | 2021 | RNN | Yes | No |
| DeepDISOBind [291] | 2022 | MTNN | No | Yes |
| MoRF-FUNCpred [292] | 2022 | BR + SVM + RF | Yes | No |
| DisoMine [293] | 2022 | RNN | No | Yes |
3.4. Protein Dynamics
3.4.1. Protein Flexibility and Conformational Dynamics
3.4.2. Dynamic Allostery
3.4.3. Potential Energy and Force Field Calculations
3.5. Drug Design
3.5.1. Molecular Docking
3.5.2. Binding Affinity
4. Conclusions
Future Opportunities
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jarvis, R.A.; Patrick, E.A. Clustering Using a Similarity Measure Based on Shared Near Neighbors. IEEE Trans. Comput. 1973, C-22, 1025–1034. [Google Scholar] [CrossRef]
- Sturm, B.L.; Ben-Tal, O.; Monaghan, Ú.; Collins, N.; Herremans, D.; Chew, E.; Hadjeres, G.; Deruty, E.; Pachet, F. Machine learning research that matters for music creation: A case study. J. New Music Res. 2019, 48, 36–55. [Google Scholar] [CrossRef]
- Rodolfa, K.T.; Lamba, H.; Ghani, R. Empirical observation of negligible fairness–accuracy trade-offs in machine learning for public policy. Nat. Mach. Intell. 2021, 3, 896–904. [Google Scholar] [CrossRef]
- Brook, T. Music, Art, Machine Learning, and Standardization. Leonardo 2021, 1–11. [Google Scholar] [CrossRef]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Alquraishi, M. ProteinNet: A standardized data set for machine learning of protein structure. BMC Bioinform. 2019, 20, 311. [Google Scholar] [CrossRef]
- Robertson, A.D.; Murphy, K.P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97, 1251–1268. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that Govern the Folding of Protein Chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef]
- Orengo, C.A.; Michie, A.D.; Jones, S.; Jones, D.T.; Swindells, M.B.; Thornton, J.M. CATH—A hierarchic classification of protein domain structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef]
- Chandonia, J.M.; Guan, L.; Lin, S.; Yu, C.; Fox, N.K.; Brenner, S.E. SCOPe: Improvements to the structural classification of proteins—Extended database to facilitate variant interpretation and machine learning. Nucleic Acids Res. 2022, 50, D553–D559. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Pawson, T.; Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 2003, 300, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Nooren, I.M.A. NEW EMBO MEMBER’S REVIEW: Diversity of protein-protein interactions. EMBO J. 2003, 22, 3486–3492. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Heald, R.; Johnson, A.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 7th ed.; Garland Science, Taylor and Francis Group: New York, NY, USA, 2022. [Google Scholar]
- Liberles, D.A.; Teichmann, S.A.; Bahar, I.; Bastolla, U.; Bloom, J.; Bornberg-Bauer, E.; Colwell, L.J.; de Koning, A.P.J.; Dokholyan, N.V.; Echave, J.; et al. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012, 21, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Livesay, D.R.; Jacobs, D.J. Conserved quantitative stability/flexibility relationships (QSFR) in an orthologous RNase H pair. Proteins Struct. Funct. Bioinform. 2006, 62, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Guerois, R.; Nielsen, J.E.; Serrano, L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More Than 1000 Mutations. J. Mol. Biol. 2002, 320, 369–387. [Google Scholar] [CrossRef]
- Jacobs, D.J.; Livesay, D.R.; Hules, J.; Tasayco, M.L. Elucidating Quantitative Stability/Flexibility Relationships Within Thioredoxin and its Fragments Using a Distance Constraint Model. J. Mol. Biol. 2006, 358, 882–904. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Dayhoff, M.O. Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Waltham, MA, USA, 1972. [Google Scholar]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Aloy, P.; Russell, R.B. Structural systems biology: Modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006, 7, 188–197. [Google Scholar] [CrossRef]
- Good, M.C.; Zalatan, J.G.; Lim, W.A. Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science 2011, 332, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Schwab, D.J. Energetic costs of cellular computation. Proc. Natl. Acad. Sci. USA 2012, 109, 17978–17982. [Google Scholar] [CrossRef] [PubMed]
- Fall, C.P.; Marland, E.S.; Wagner, J.M.; Tyson, J.J. Computational Cell Biology; Springer: New York, NY, USA, 2004. [Google Scholar]
- Wilke, C.O. Bringing Molecules Back into Molecular Evolution. PLoS Comput. Biol. 2012, 8, e1002572. [Google Scholar] [CrossRef] [PubMed]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Levitt, M. The birth of computational structural biology. Nat. Struct. Biol. 2001, 8, 392–393. [Google Scholar] [CrossRef]
- Dill, K.A.; Bromberg, S.; Yue, K.; Chan, H.S.; Ftebig, K.M.; Yee, D.P.; Thomas, P.D. Principles of protein folding—A perspective from simple exact models. Protein Sci. 2008, 4, 561–602. [Google Scholar] [CrossRef]
- Takada, S. Gō model revisited. Biophys. Physicobiol. 2019, 16, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Ettayapuram Ramaprasad, A.S.; Uddin, S.; Casas-Finet, J.; Jacobs, D.J. Decomposing Dynamical Couplings in Mutated scFv Antibody Fragments into Stabilizing and Destabilizing Effects. J. Am. Chem. Soc. 2017, 139, 17508–17517. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Additivity Principles in Biochemistry. J. Biol. Chem. 1997, 272, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Mark, A.E.; van Gunsteren, W.F. Decomposition of the free energy of a system in terms of specific interactions. Implications for theoretical and experimental studies. J. Mol. Biol. 1994, 240, 167–176. [Google Scholar] [CrossRef]
- Jacobs, D.J.; Dallakyan, S.; Wood, G.G.; Heckathorne, A. Network rigidity at finite temperature: Relationships between thermodynamic stability, the nonadditivity of entropy, and cooperativity in molecular systems. Phys. Rev. E 2003, 68. [Google Scholar] [CrossRef]
- Jacobs, D.J.; Dallakyan, S. Elucidating Protein Thermodynamics from the Three-Dimensional Structure of the Native State Using Network Rigidity. Biophys. J. 2005, 88, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Livesay, D.R.; Dallakyan, S.; Wood, G.G.; Jacobs, D.J. A flexible approach for understanding protein stability. FEBS Lett. 2004, 576, 468–476. [Google Scholar] [CrossRef]
- Li, T.; Tracka, M.B.; Uddin, S.; Casas-Finet, J.; Jacobs, D.J.; Livesay, D.R. Rigidity Emerges during Antibody Evolution in Three Distinct Antibody Systems: Evidence from QSFR Analysis of Fab Fragments. PLoS Comput. Biol. 2015, 11, e1004327. [Google Scholar] [CrossRef]
- Jacobs, D.J.; Wood, G.G. Understanding the α-helix to coil transition in polypeptides using network rigidity: Predicting heat and cold denaturation in mixed solvent conditions. Biopolymers 2004, 75, 1–31. [Google Scholar] [CrossRef]
- Jackel, C.; Kast, P.; Hilvert, D. Protein design by directed evolution. Annu. Rev. Biophys. 2008, 37, 153–173. [Google Scholar] [CrossRef]
- James, L.C.; Tawfik, D.S. Conformational diversity and protein evolution—A 60-year-old hypothesis revisited. Trends Biochem. Sci. 2003, 28, 361–368. [Google Scholar] [CrossRef]
- Glasner, M.E.; Gerlt, J.A.; Babbitt, P.C. Mechanisms of protein evolution and their application to protein engineering. Adv. Enzym. Relat. Areas Mol. Biol. 2007, 75, 193–239. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Samuel, A. Computing Bit by Bit or Digital Computers Made Easy. Proc. IRE 1953, 41, 1223–1230. [Google Scholar] [CrossRef]
- Samuel, A.L. Artificial Intelligence: A Frontier of Automation. ANNALS Am. Acad. Political Soc. Sci. 1962, 340, 10–20. [Google Scholar] [CrossRef]
- Rosenblatt, F. Perceptron Simulation Experiments. Proc. IRE 1960, 48, 301–309. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N., Weinberger, K., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. In Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 1877–1901. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); Association for Computational Linguistics: Minneapolis, MN, USA, 2019; pp. 4171–4186. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.U.; Polosukhin, I. Attention is All you Need. In Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Sun, X.; Jia, L.; Ma, J.; Xing, H.; Wu, J.; Gao, H.; Sun, Y.; Boulnois, F.; Fan, J. Chemi-Net: A Molecular Graph Convolutional Network for Accurate Drug Property Prediction. Int. J. Mol. Sci. 2019, 20, 3389. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. On Bias, Variance, 0/1—Loss, and the Curse-of-Dimensionality. Data Min. Knowl. Discov. 1997, 1, 55–77. [Google Scholar] [CrossRef]
- Wu, F.; Xu, J. Deep template-based protein structure prediction. PLoS Comput. Biol. 2021, 17, e1008954. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Muhammed, M.T.; Aki-Yalcin, E. Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019, 93, 12–20. [Google Scholar] [CrossRef]
- Seffernick, J.T.; Lindert, S. Hybrid methods for combined experimental and computational determination of protein structure. J. Chem. Phys. 2020, 153, 240901. [Google Scholar] [CrossRef]
- Burley, S.K.; Joachimiak, A.; Montelione, G.T.; Wilson, I.A. Contributions to the NIH-NIGMS Protein Structure Initiative from the PSI Production Centers. Structure 2008, 16, 5–11. [Google Scholar] [CrossRef]
- Bolje, A.; Gobec, S. Analytical Techniques for Structural Characterization of Proteins in Solid Pharmaceutical Forms: An Overview. Pharmaceutics 2021, 13, 534. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Cheng, T.; Liu, Z.; Wang, R. Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J. Comput. Chem. 2010, 31, 2109–2125. [Google Scholar] [CrossRef]
- Dhingra, S.; Sowdhamini, R.; Cadet, F.; Offmann, B. A glance into the evolution of template-free protein structure prediction methodologies. Biochimie 2020, 175, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Bystroff, C.; Baker, D. Prediction of local structure in proteins using a library of sequence-structure motifs. J. Mol. Biol. 1998, 281, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Rohl, C.A.; Strauss, C.E.; Misura, K.M.; Baker, D. Protein structure prediction using Rosetta. Methods Enzym. 2004, 383, 66–93. [Google Scholar] [CrossRef]
- Noé, F.; De Fabritiis, G.; Clementi, C. Machine learning for protein folding and dynamics. Curr. Opin. Struct. Biol. 2020, 60, 77–84. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins 2019, 87, 1011–1020. [Google Scholar] [CrossRef]
- Heo, L.; Feig, M. High-accuracy protein structures by combining machine-learning with physics-based refinement. Proteins 2020, 88, 637–642. [Google Scholar] [CrossRef]
- Ovchinnikov, S.; Park, H.; Kim, D.E.; Dimaio, F.; Baker, D. Protein structure prediction using Rosetta in CASP12. Proteins Struct. Funct. Bioinform. 2018, 86, 113–121. [Google Scholar] [CrossRef]
- Hong, S.H.; Joung, I.; Flores-Canales, J.C.; Manavalan, B.; Cheng, Q.; Heo, S.; Kim, J.Y.; Lee, S.Y.; Nam, M.; Joo, K.; et al. Protein structure modeling and refinement by global optimization in CASP12. Proteins Struct. Funct. Bioinform. 2018, 86, 122–135. [Google Scholar] [CrossRef]
- Zhang, C.; Mortuza, S.M.; He, B.; Wang, Y.; Zhang, Y. Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins Struct. Funct. Bioinform. 2018, 86, 136–151. [Google Scholar] [CrossRef]
- Olechnovič, K.; Venclovas, Č. VoroMQA: Assessment of protein structure quality using interatomic contact areas. Proteins Struct. Funct. Bioinform. 2017, 85, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Alquraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zidek, A.; Nelson, A.; Bridgland, A.; Penedones, H.; et al. De novo structure prediction with deeplearning based scoring. Annu. Rev. Biochem. 2018, 77, 6. [Google Scholar]
- Li, Y.; Zhang, C.; Bell, E.W.; Yu, D.; Zhang, Y. Ensembling multiple raw coevolutionary features with deep residual neural networks for contact-map prediction in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1082–1091. [Google Scholar] [CrossRef]
- Hou, J.; Wu, T.; Cao, R.; Cheng, J. Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1165–1178. [Google Scholar] [CrossRef]
- Zheng, W.; Li, Y.; Zhang, C.; Pearce, R.; Mortuza, S.M.; Zhang, Y. Deep-learning contact-map guided protein structure prediction in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1149–1164. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Anishchenko, I.; Baek, M.; Park, H.; Hiranuma, N.; Kim, D.E.; Dauparas, J.; Mansoor, S.; Humphreys, I.R.; Baker, D. Protein tertiary structure prediction and refinement using deep learning and Rosetta in CASP14. Proteins Struct. Funct. Bioinform. 2021, 89, 1722–1733. [Google Scholar] [CrossRef]
- Baek, M.; Anishchenko, I.; Park, H.; Humphreys, I.R.; Baker, D. Protein oligomer modeling guided by predicted interchain contacts in CASP14. Proteins Struct. Funct. Bioinform. 2021, 89, 1824–1833. [Google Scholar] [CrossRef]
- Heo, L.; Janson, G.; Feig, M. Physics-based protein structure refinement in the era of artificial intelligence. Proteins Struct. Funct. Bioinform. 2021, 89, 1870–1887. [Google Scholar] [CrossRef]
- Zheng, W.; Li, Y.; Zhang, C.; Zhou, X.; Pearce, R.; Bell, E.W.; Huang, X.; Zhang, Y. Protein structure prediction using deep learning distance and hydrogen-bonding restraints in CASP14. Proteins 2021, 89, 1734–1751. [Google Scholar] [CrossRef]
- Fersht, A.R. AlphaFold—A Personal Perspective on the Impact of Machine Learning. J. Mol. Biol. 2021, 433, 167088. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. Machine learning in protein structure prediction. Curr. Opin. Chem. Biol. 2021, 65, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Torrisi, M.; Pollastri, G.; Le, Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020, 18, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, S.H.P.; Shi, J.; Deane, C.M. Comparing co-evolution methods and their application to template-free protein structure prediction. Bioinformatics 2016, 33, 373–381. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Ahdritz, G.; Bouatta, N.; Kadyan, S.; Xia, Q.; Gerecke, W.; AlQuraishi, M. OpenFold. Zenodo 2021. [Google Scholar] [CrossRef]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Berger, B.; et al. High-resolution de novo structure prediction from primary sequence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef]
- Kokh, D.B.; Kaufmann, T.; Kister, B.; Wade, R.C. Machine Learning Analysis of tauRAMD Trajectories to Decipher Molecular Determinants of Drug-Target Residence Times. Front. Mol. Biosci. 2019, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.N.; Philot, E.A.; Trossini, G.H.G.; Scott, L.P.B.; Maltarollo, V.G.; Honorio, K.M. Use of machine learning approaches for novel drug discovery. Expert Opin. Drug Discov. 2016, 11, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Shala, A.; Bezginov, A.; Sljoka, A.; Audette, G.; Wilson, D.J. Hyperphosphorylation of Intrinsically Disordered Tau Protein Induces an Amyloidogenic Shift in Its Conformational Ensemble. PLoS ONE 2015, 10, e0120416. [Google Scholar] [CrossRef]
- Joshi, S.Y.; Deshmukh, S.A. A review of advancements in coarse-grained molecular dynamics simulations. Mol. Simul. 2021, 47, 786–803. [Google Scholar] [CrossRef]
- Liwo, A.; Czaplewski, C.; Sieradzan, A.K.; Lipska, A.G.; Samsonov, S.A.; Murarka, R.K. Theory and Practice of Coarse-Grained Molecular Dynamics of Biologically Important Systems. Biomolecules 2021, 11, 1347. [Google Scholar] [CrossRef]
- Singh, N.; Li, W. Recent Advances in Coarse-Grained Models for Biomolecules and Their Applications. Int. J. Mol. Sci. 2019, 20, 3774. [Google Scholar] [CrossRef]
- Togashi, Y.; Flechsig, H. Coarse-Grained Protein Dynamics Studies Using Elastic Network Models. Int. J. Mol. Sci. 2018, 19, 3899. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Marrink, S.J.; Monticelli, L.; Melo, M.N.; Alessandri, R.; Tieleman, D.P.; Souza, P.C.T. Two decades of Martini: Better beads, broader scope. WIREs Comput. Mol. Sci. 2022, e1620. [Google Scholar] [CrossRef]
- Rojas, A.; Czaplewski, C.; Liwo, A.; Makowski, M.; O_dziej, S.; Kaz, R.; Scheraga, H.; Murarka, R.; Voth, G. Simulation of Protein Structure and Dynamics with the Coarse-Grained UNRES Force Field. Coarse-Graining Condens. Phase Biomol. Syst. 2008, 1, 1391–1411. [Google Scholar]
- Liwo, A.; Baranowski, M.; Czaplewski, C.; Gołaś, E.; He, Y.; Jagieła, D.; Krupa, P.; Maciejczyk, M.; Makowski, M.; Mozolewska, M.A.; et al. A unified coarse-grained model of biological macromolecule based on mean-field multipole–multipole interactions. J. Mol. Model. 2014, 20, 2306. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yuan, C.; Ma, R.; Zhang, Z. Backmapping from Multiresolution Coarse-Grained Models to Atomic Structures of Large Biomolecules by Restrained Molecular Dynamics Simulations Using Bayesian Inference. J. Chem. Theory Comput. 2019, 15, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, J.; Wang, H.; Car, R.; E, W. DeePCG: Constructing coarse-grained models via deep neural networks. J. Chem. Phys. 2018, 149, 034101. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Olsson, S.; Wehmeyer, C.; Perez, A.; Charron, N.E.; de Fabritiis, G.; Noe, F.; Clementi, C. Machine Learning of Coarse-Grained Molecular Dynamics Force Fields. ACS Cent. Sci. 2019, 5, 755–767. [Google Scholar] [CrossRef]
- Husic, B.E.; Charron, N.E.; Lemm, D.; Wang, J.; Perez, A.; Majewski, M.; Kramer, A.; Chen, Y.; Olsson, S.; de Fabritiis, G.; et al. Coarse graining molecular dynamics with graph neural networks. J. Chem. Phys. 2020, 153, 194101. [Google Scholar] [CrossRef]
- Wang, J.; Chmiela, S.; Muller, K.R.; Noe, F.; Clementi, C. Ensemble learning of coarse-grained molecular dynamics force fields with a kernel approach. J. Chem. Phys. 2020, 152, 194106. [Google Scholar] [CrossRef]
- Zhou, R. Replica exchange molecular dynamics method for protein folding simulation. Methods Mol. Biol. 2007, 350, 205–223. [Google Scholar] [CrossRef]
- Mori, T.; Miyashita, N.; Im, W.; Feig, M.; Sugita, Y. Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochim. Biophys. Acta 2016, 1858, 1635–1651. [Google Scholar] [CrossRef]
- Affentranger, R.; Tavernelli, I.; Di Iorio, E.E. A Novel Hamiltonian Replica Exchange MD Protocol to Enhance Protein Conformational Space Sampling. J. Chem. Theory Comput. 2006, 2, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.C.; Melo, M.C.R.; Schulten, K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta 2015, 1850, 872–877. [Google Scholar] [CrossRef]
- Melo, M.C.; Bernardi, R.C.; Fernandes, T.V.; Pascutti, P.G. GSAFold: A new application of GSA to protein structure prediction. Proteins 2012, 80, 2305–2310. [Google Scholar] [CrossRef] [PubMed]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef] [PubMed]
- Barducci, A.; Bussi, G.; Parrinello, M. Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008, 100, 020603. [Google Scholar] [CrossRef] [PubMed]
- Comer, J.; Gumbart, J.C.; Henin, J.; Lelievre, T.; Pohorille, A.; Chipot, C. The adaptive biasing force method: Everything you always wanted to know but were afraid to ask. J. Phys. Chem. B 2015, 119, 1129–1151. [Google Scholar] [CrossRef] [PubMed]
- Hénin, J.; Chipot, C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J. Chem. Phys. 2004, 121, 2904–2914. [Google Scholar] [CrossRef]
- Liphardt, J.; Dumont, S.; Smith, S.B.; Tinoco, I.; Bustamante, C. Equilibrium Information from Nonequilibrium Measurements in an Experimental Test of Jarzynski’s Equality. Science 2002, 296, 1832–1835. [Google Scholar] [CrossRef]
- Shamsi, Z.; Moffett, A.S.; Shukla, D. Enhanced unbiased sampling of protein dynamics using evolutionary coupling information. Sci. Rep. 2017, 7, 12700. [Google Scholar] [CrossRef]
- Palazzesi, F.; Valsson, O.; Parrinello, M. Conformational Entropy as Collective Variable for Proteins. J. Phys. Chem. Lett. 2017, 8, 4752–4756. [Google Scholar] [CrossRef]
- Fiorin, G.; Klein, M.L.; Hénin, J. Using collective variables to drive molecular dynamics simulations. Mol. Phys. 2013, 111, 3345–3362. [Google Scholar] [CrossRef]
- Chen, M. Collective variable-based enhanced sampling and machine learning. Eur. Phys. J. B 2021, 94, 1–17. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J. Essential dynamics of proteins. Proteins 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- David, C.C.; Avery, C.S.; Jacobs, D.J. JEDi: Java essential dynamics inspector—A molecular trajectory analysis toolkit. BMC Bioinform. 2021, 22, 226. [Google Scholar] [CrossRef] [PubMed]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.; Nizami, B.; Glenister, M.; Sheik Amamuddy, O.; Atilgan, A.R.; Atilgan, C.; Tastan Bishop, O. MODE-TASK: Large-scale protein motion tools. Bioinformatics 2018, 34, 3759–3763. [Google Scholar] [CrossRef]
- Peng, J.; Zhang, Z. Simulating Large-Scale Conformational Changes of Proteins by Accelerating Collective Motions Obtained from Principal Component Analysis. J. Chem. Theory Comput. 2014, 10, 3449–3458. [Google Scholar] [CrossRef]
- Shkurti, A.; Styliari, I.D.; Balasubramanian, V.; Bethune, I.; Pedebos, C.; Jha, S.; Laughton, C.A. CoCo-MD: A Simple and Effective Method for the Enhanced Sampling of Conformational Space. J. Chem. Theory Comput. 2019, 15, 2587–2596. [Google Scholar] [CrossRef]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef]
- Spiwok, V.; Kralova, B. Metadynamics in the conformational space nonlinearly dimensionally reduced by Isomap. J. Chem. Phys. 2011, 135, 224504. [Google Scholar] [CrossRef]
- Tribello Gareth, A.; Ceriotti, M.; Parrinello, M. Using sketch-map coordinates to analyze and bias molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, 5196–5201. [Google Scholar] [CrossRef]
- Rohrdanz, M.A.; Zheng, W.; Maggioni, M.; Clementi, C. Determination of reaction coordinates via locally scaled diffusion map. J. Chem. Phys. 2011, 134, 124116. [Google Scholar] [CrossRef]
- Sultan, M.M.; Pande, V.S. Automated design of collective variables using supervised machine learning. J. Chem. Phys. 2018, 149, 094106. [Google Scholar] [CrossRef] [PubMed]
- Naritomi, Y.; Fuchigami, S. Slow dynamics in protein fluctuations revealed by time-structure based independent component analysis: The case of domain motions. J. Chem. Phys. 2011, 134, 065101. [Google Scholar] [CrossRef] [PubMed]
- Hyvarinen, A.; Oja, E. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Perez-Hernandez, G.; Noe, F. Hierarchical Time-Lagged Independent Component Analysis: Computing Slow Modes and Reaction Coordinates for Large Molecular Systems. J. Chem. Theory Comput. 2016, 12, 6118–6129. [Google Scholar] [CrossRef]
- M, M.S.; Pande, V.S. tICA-Metadynamics: Accelerating Metadynamics by Using Kinetically Selected Collective Variables. J. Chem. Theory Comput. 2017, 13, 2440–2447. [Google Scholar] [CrossRef]
- Perez-Hernandez, G.; Paul, F.; Giorgino, T.; De Fabritiis, G.; Noe, F. Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 2013, 139, 015102. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.K.; Trendelkamp-Schroer, B.; Paul, F.; Perez-Hernandez, G.; Hoffmann, M.; Plattner, N.; Wehmeyer, C.; Prinz, J.H.; Noe, F. PyEMMA 2: A Software Package for Estimation, Validation, and Analysis of Markov Models. J. Chem. Theory Comput. 2015, 11, 5525–5542. [Google Scholar] [CrossRef]
- Harrigan, M.P.; Sultan, M.M.; Hernandez, C.X.; Husic, B.E.; Eastman, P.; Schwantes, C.R.; Beauchamp, K.A.; McGibbon, R.T.; Pande, V.S. MSMBuilder: Statistical Models for Biomolecular Dynamics. Biophys. J. 2017, 112, 10–15. [Google Scholar] [CrossRef]
- Ma, A.; Dinner, A.R. Automatic method for identifying reaction coordinates in complex systems. J. Phys. Chem. B 2005, 109, 6769–6779. [Google Scholar] [CrossRef]
- Chen, W.; Ferguson, A.L. Molecular enhanced sampling with autoencoders: On-the-fly collective variable discovery and accelerated free energy landscape exploration. J. Comput. Chem. 2018, 39, 2079–2102. [Google Scholar] [CrossRef]
- Chen, W.; Tan, A.R.; Ferguson, A.L. Collective variable discovery and enhanced sampling using autoencoders: Innovations in network architecture and error function design. J. Chem. Phys. 2018, 149, 072312. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, G.; Vishal, V.; Pande, V.S. Using massively parallel simulation and Markovian models to study protein folding: Examining the dynamics of the villin headpiece. J. Chem. Phys. 2006, 124, 164902. [Google Scholar] [CrossRef] [PubMed]
- Chodera, J.D.; Swope, W.C.; Pitera, J.W.; Dill, K.A. Long-Time Protein Folding Dynamics from Short-Time Molecular Dynamics Simulations. Multiscale Model. Simul. 2006, 5, 1214–1226. [Google Scholar] [CrossRef]
- Wehmeyer, C.; Noe, F. Time-lagged autoencoders: Deep learning of slow collective variables for molecular kinetics. J. Chem. Phys. 2018, 148, 241703. [Google Scholar] [CrossRef] [PubMed]
- Lamim Ribeiro, J.M.; Provasi, D.; Filizola, M. A combination of machine learning and infrequent metadynamics to efficiently predict kinetic rates, transition states, and molecular determinants of drug dissociation from G protein-coupled receptors. J. Chem. Phys. 2020, 153, 124105. [Google Scholar] [CrossRef]
- Ravindra, P.; Smith, Z.; Tiwary, P. Automatic mutual information noise omission (AMINO): Generating order parameters for molecular systems. Mol. Syst. Des. Eng. 2020, 5, 339–348. [Google Scholar] [CrossRef]
- Ribeiro, J.M.L.; Bravo, P.; Wang, Y.; Tiwary, P. Reweighted autoencoded variational Bayes for enhanced sampling (RAVE). J. Chem. Phys. 2018, 149, 072301. [Google Scholar] [CrossRef]
- Wu, H.; Noé, F. Variational Approach for Learning Markov Processes from Time Series Data. J. Nonlinear Sci. 2020, 30, 23–66. [Google Scholar] [CrossRef]
- Koopman, B.O. Hamiltonian Systems and Transformation in Hilbert Space. Proc. Natl. Acad. Sci. USA 1931, 17, 315–318. [Google Scholar] [CrossRef]
- Koopman, B.O.; Neumann, J.V. Dynamical Systems of Continuous Spectra. Proc. Natl. Acad. Sci. USA 1932, 18, 255–263. [Google Scholar] [CrossRef]
- Williams, M.O.; Kevrekidis, I.G.; Rowley, C.W. A Data–Driven Approximation of the Koopman Operator: Extending Dynamic Mode Decomposition. J. Nonlinear Sci. 2015, 25, 1307–1346. [Google Scholar] [CrossRef]
- Mardt, A.; Pasquali, L.; Wu, H.; Noe, F. VAMPnets for deep learning of molecular kinetics. Nat. Commun. 2018, 9, 5. [Google Scholar] [CrossRef]
- Sidky, H.; Chen, W.; Ferguson, A.L. High-Resolution Markov State Models for the Dynamics of Trp-Cage Miniprotein Constructed Over Slow Folding Modes Identified by State-Free Reversible VAMPnets. J. Phys. Chem. B 2019, 123, 7999–8009. [Google Scholar] [CrossRef] [PubMed]
- Konovalov, K.A.; Unarta, I.C.; Cao, S.; Goonetilleke, E.C.; Huang, X. Markov State Models to Study the Functional Dynamics of Proteins in the Wake of Machine Learning. JACS Au 2021, 1, 1330–1341. [Google Scholar] [CrossRef]
- Laio, A.; Gervasio, F.L. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008, 71, 126601. [Google Scholar] [CrossRef]
- Galvelis, R.; Sugita, Y. Neural Network and Nearest Neighbor Algorithms for Enhancing Sampling of Molecular Dynamics. J. Chem. Theory Comput. 2017, 13, 2489–2500. [Google Scholar] [CrossRef]
- Guo, A.Z.; Sevgen, E.; Sidky, H.; Whitmer, J.K.; Hubbell, J.A.; Pablo, J.J.d. Adaptive enhanced sampling by force-biasing using neural networks. J. Chem. Phys. 2018, 148, 134108. [Google Scholar] [CrossRef]
- Sidky, H.; Whitmer, J.K. Learning free energy landscapes using artificial neural networks. J. Chem. Phys. 2018, 148, 104111. [Google Scholar] [CrossRef]
- Salawu, E.O. DESP: Deep Enhanced Sampling of Proteins’ Conformation Spaces Using AI-Inspired Biasing Forces. Front. Mol. Biosci. 2021, 8, 587151. [Google Scholar] [CrossRef]
- Ezugwu, A.E.; Ikotun, A.M.; Oyelade, O.O.; Abualigah, L.; Agushaka, J.O.; Eke, C.I.; Akinyelu, A.A. A comprehensive survey of clustering algorithms: State-of-the-art machine learning applications, taxonomy, challenges, and future research prospects. Eng. Appl. Artif. Intell. 2022, 110, 104743. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Holm, L.; Sander, C. Protein Structure Comparison by Alignment of Distance Matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Shindyalov, I.N.; Bourne, P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998, 11, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef] [PubMed]
- Shirkhorshidi, A.S.; Aghabozorgi, S.; Wah, T.Y. A Comparison Study on Similarity and Dissimilarity Measures in Clustering Continuous Data. PLoS ONE 2015, 10, e0144059. [Google Scholar] [CrossRef]
- Mehta, V.; Bawa, S.; Singh, J. Analytical review of clustering techniques and proximity measures. Artif. Intell. Rev. 2020, 53, 5995–6023. [Google Scholar] [CrossRef]
- Bowman, G.R. Improved coarse-graining of Markov state models via explicit consideration of statistical uncertainty. J. Chem. Phys. 2012, 137, 134111. [Google Scholar] [CrossRef]
- Baek, M.; Kim, C. A review on spectral clustering and stochastic block models. J. Korean Stat. Soc. 2021, 50, 818–831. [Google Scholar] [CrossRef]
- Röblitz, S.; Weber, M. Fuzzy spectral clustering by PCCA+: Application to Markov state models and data classification. Adv. Data Anal. Classif. 2013, 7, 147–179. [Google Scholar] [CrossRef]
- Deuflhard, P.; Weber, M. Robust Perron cluster analysis in conformation dynamics. Linear Algebra Its Appl. 2005, 398, 161–184. [Google Scholar] [CrossRef]
- Huang, R.; Lo, L.T.; Wen, Y.; Voter, A.F.; Perez, D. Cluster analysis of accelerated molecular dynamics simulations: A case study of the decahedron to icosahedron transition in Pt nanoparticles. J. Chem. Phys. 2017, 147, 152717. [Google Scholar] [CrossRef]
- Huang, X.; Yao, Y.; Bowman, G.R.; Sun, J.; Guibas, L.J.; Carlsson, G.; Pande, V.S. Constructing multi-resolution Markov State Models (MSMs) to elucidate RNA hairpin folding mechanisms. Pac. Symp. Biocomput. 2010, 2010, 228–239. [Google Scholar] [CrossRef]
- Yao, Y.; Cui, R.Z.; Bowman, G.R.; Silva, D.A.; Sun, J.; Huang, X. Hierarchical Nyström methods for constructing Markov state models for conformational dynamics. J. Chem. Phys. 2013, 138, 174106. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Stock, G. Identifying Metastable States of Folding Proteins. J. Chem. Theory Comput. 2012, 8, 3810–3819. [Google Scholar] [CrossRef]
- Wang, W.; Cao, S.; Zhu, L.; Huang, X. Constructing Markov State Models to elucidate the functional conformational changes of complex biomolecules. WIREs Comput. Mol. Sci. 2018, 8, e1343. [Google Scholar] [CrossRef]
- Orioli, S.; Faccioli, P. Dimensional reduction of Markov state models from renormalization group theory. J. Chem. Phys. 2016, 145, 124120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sheong, F.K.; Zeng, X.; Huang, X. Elucidation of the conformational dynamics of multi-body systems by construction of Markov state models. Phys. Chem. Chem. Phys. 2016, 18, 30228–30235. [Google Scholar] [CrossRef]
- Cocina, F.; Vitalis, A.; Caflisch, A. Sapphire-Based Clustering. J. Chem. Theory Comput. 2020, 16, 6383–6396. [Google Scholar] [CrossRef] [PubMed]
- Mallet, V.; Nilges, M.; Bouvier, G. quicksom: Self-Organizing Maps on GPUs for clustering of molecular dynamics trajectories. Bioinformatics 2021, 37, 2064–2065. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef]
- Rao, S.J.A.; Shetty, N.P. Evolutionary selectivity of amino acid is inspired from the enhanced structural stability and flexibility of the folded protein. Life Sci. 2021, 281, 119774. [Google Scholar] [CrossRef] [PubMed]
- Walport, L.J.; Low, J.K.K.; Matthews, J.M.; Mackay, J.P. The characterization of protein interactions—What, how and how much? Chem. Soc. Rev. 2021, 50, 12292–12307. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Bioinform. 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, S.; Zhu, Q.; Zhou, Y. A Knowledge-Based Energy Function for Protein–Ligand, Protein–Protein, and Protein–DNA Complexes. J. Med. Chem. 2005, 48, 2325–2335. [Google Scholar] [CrossRef]
- Dodge, C.; Schneider, R.; Sander, C. The HSSP database of protein structure—Sequence alignments and family profiles. Nucleic Acids Res. 1998, 26, 313–315. [Google Scholar] [CrossRef]
- Lobry, J.; Gautier, C. Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res. 1994, 22, 3174–3180. [Google Scholar] [CrossRef]
- Huang, P.; Chu, S.K.S.; Frizzo, H.N.; Connolly, M.P.; Caster, R.W.; Siegel, J.B. Evaluating Protein Engineering Thermostability Prediction Tools Using an Independently Generated Dataset. ACS Omega 2020, 5, 6487–6493. [Google Scholar] [CrossRef]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of Molecular Recognition Features (MoRFs). J. Mol. Biol. 2006, 362, 1043–1059. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Rother, D.; Sapiro, G.; Pande, V. Statistical characterization of protein ensembles. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008, 5, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, G.; Desdouits, N.; Ferber, M.; Blondel, A.; Nilges, M. An automatic tool to analyze and cluster macromolecular conformations based on self-organizing maps. Bioinformatics 2015, 31, 1490–1492. [Google Scholar] [CrossRef]
- Bhattacharyya, M.; Bhat, C.R.; Vishveshwara, S. An automated approach to network features of protein structure ensembles. Protein Sci. 2013, 22, 1399–1416. [Google Scholar] [CrossRef]
- Jo, T.; Hou, J.; Eickholt, J.; Cheng, J. Improving Protein Fold Recognition by Deep Learning Networks. Sci. Rep. 2015, 5, 17573. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Misiura, M.; Shroff, R.; Thyer, R.; Kolomeisky, A.B. DLPacker: Deep learning for prediction of amino acid side chain conformations in proteins. Proteins Struct. Funct. Bioinform. 2022, 90, 1278–1290. [Google Scholar] [CrossRef]
- King, J.E.; Koes, D.R. SidechainNet: An all-atom protein structure dataset for machine learning. Proteins Struct. Funct. Bioinform. 2021, 89, 1489–1496. [Google Scholar] [CrossRef]
- Igashov, I.; Olechnovič, K.; Kadukova, M.; Venclovas, Č.; Grudinin, S. VoroCNN: Deep convolutional neural network built on 3D Voronoi tessellation of protein structures. Bioinformatics 2021, 37, 2332–2339. [Google Scholar] [CrossRef]
- Luttrell, J.; Liu, T.; Zhang, C.; Wang, Z. Predicting protein residue-residue contacts using random forests and deep networks. BMC Bioinform. 2019, 20, 100. [Google Scholar] [CrossRef] [PubMed]
- Audagnotto, M.; Czechtizky, W.; De Maria, L.; Käck, H.; Papoian, G.; Tornberg, L.; Tyrchan, C.; Ulander, J. Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble. Sci. Rep. 2022, 12, 10018. [Google Scholar] [CrossRef]
- Duong, V.T.; Diessner, E.M.; Grazioli, G.; Martin, R.W.; Butts, C.T. Neural Upscaling from Residue-Level Protein Structure Networks to Atomistic Structures. Biomolecules 2021, 11, 1788. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.H.; Kuhn, L.T.; Goez, M.; Day, I.J.; Lin, J.C.; Andersen, N.H.; Hore, P.J. A pre-existing hydrophobic collapse in the unfolded state of an ultrafast folding protein. Nature 2007, 447, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Nassar, R.; Brini, E.; Parui, S.; Liu, C.; Dignon, G.L.; Dill, K.A. Accelerating Protein Folding Molecular Dynamics Using Inter-Residue Distances from Machine Learning Servers. J. Chem. Theory Comput. 2022, 18, 1929–1935. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Pande, V.S. Note: Variational encoding of protein dynamics benefits from maximizing latent autocorrelation. J. Chem. Phys. 2018, 149, 216101. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J.; Green, S.B.; Jacobs, D.J. Distribution of volume, microvoid percolation, and packing density in globular proteins. arXiv 2018, arXiv:1810.08745. [Google Scholar]
- Fried, S.D.; Boxer, S.G. Electric Fields and Enzyme Catalysis. Annu. Rev. Biochem. 2017, 86, 387–415. [Google Scholar] [CrossRef]
- Jamasb, A.R.; Viñas, R.; Ma, E.J.; Harris, C.; Huang, K.; Hall, D.; Lió, P.; Blundell, T.L. Graphein—A Python Library for Geometric Deep Learning and Network Analysis on Protein Structures and Interaction Networks. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Guo, D.; Ott, M.; Zitnick, C.L.; Ma, J.; et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 2021, 118, e2016239118. [Google Scholar] [CrossRef]
- Kawano, K.; Koide, S.; Imamura, C. Seq2seq Fingerprint with Byte-Pair Encoding for Predicting Changes in Protein Stability upon Single Point Mutation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1762–1772. [Google Scholar] [CrossRef]
- Repecka, D.; Jauniskis, V.; Karpus, L.; Rembeza, E.; Rokaitis, I.; Zrimec, J.; Poviloniene, S.; Laurynenas, A.; Viknander, S.; Abuajwa, W.; et al. Expanding functional protein sequence spaces using generative adversarial networks. Nat. Mach. Intell. 2021, 3, 324–333. [Google Scholar] [CrossRef]
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust deep learning based protein sequence design using ProteinMPNN. bioRxiv 2022. [Google Scholar] [CrossRef]
- Reed, S.; Zolna, K.; Parisotto, E.; Colmenarejo, S.G.; Novikov, A.; Barth-Maron, G.; Gimenez, M.; Sulsky, Y.; Kay, J.; Springenberg, J.T.; et al. A Generalist Agent. arXiv 2022, arXiv:2205.06175. [Google Scholar]
- Chen, J.; Zheng, S.; Zhao, H.; Yang, Y. Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map. J. Cheminform. 2021, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Ning, W.; Ma, X.; Wang, X.; Zhou, K. Improving protein solubility and activity by introducing small peptide tags designed with machine learning models. Metab. Eng. Commun. 2020, 11, e00138. [Google Scholar] [CrossRef]
- Chen, L.; Oughtred, R.; Berman, H.M.; Westbrook, J. TargetDB: A target registration database for structural genomics projects. Bioinformatics 2004, 20, 2860–2862. [Google Scholar] [CrossRef]
- Madani, M.; Lin, K.; Tarakanova, A. DSResSol: A Sequence-Based Solubility Predictor Created with Dilated Squeeze Excitation Residual Networks. Int. J. Mol. Sci. 2021, 22, 3555. [Google Scholar] [CrossRef]
- Cai, Z.; Luo, F.; Wang, Y.; Li, E.; Huang, Y. Protein pK (a) Prediction with Machine Learning. ACS Omega 2021, 6, 34823–34831. [Google Scholar] [CrossRef]
- Ko, T.W.; Finkler, J.A.; Goedecker, S.; Behler, J. A fourth-generation high-dimensional neural network potential with accurate electrostatics including non-local charge transfer. Nat. Commun. 2021, 12, 398. [Google Scholar] [CrossRef]
- Chatzigoulas, A.; Cournia, Z. Predicting protein-membrane interfaces of peripheral membrane proteins using ensemble machine learning. Briefings Bioinform. 2022, 23, bbab518. [Google Scholar] [CrossRef]
- Lai, P.K.; Fernando, A.; Cloutier, T.K.; Kingsbury, J.S.; Gokarn, Y.; Halloran, K.T.; Calero-Rubio, C.; Trout, B.L. Machine Learning Feature Selection for Predicting High Concentration Therapeutic Antibody Aggregation. J. Pharm. Sci. 2021, 110, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Qin, Y.; Fontaine, N.T.; Ng Fuk Chong, M.; Maria-Solano, M.A.; Feixas, F.; Cadet, X.F.; Pandjaitan, R.; Garcia-Borràs, M.; Cadet, F.; et al. Machine Learning Enables Selection of Epistatic Enzyme Mutants for Stability Against Unfolding and Detrimental Aggregation. ChemBioChem 2021, 22, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, C.; Wang, M.; Webb, G.I.; Zhang, Y.; Whisstock, J.C.; Song, J. GlycoMine: A machine learning-based approach for predicting N-, C- and O-linked glycosylation in the human proteome. Bioinformatics 2015, 31, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Hassan, A.; Mitra, P. Boosting phosphorylation site prediction with sequence feature-based machine learning. Proteins Struct. Funct. Bioinform. 2020, 88, 284–291. [Google Scholar] [CrossRef]
- Arnold, F.H. Protein engineering for unusual environments. Curr. Opin. Biotechnol. 1993, 4, 450–455. [Google Scholar] [CrossRef]
- Prokop, M.; Damborský, J.; Koča, J. TRITON: In silico construction of protein mutants and prediction of their activities *. Bioinformatics 2000, 16, 845–846. [Google Scholar] [CrossRef]
- Gilis, D.; Rooman, M. PoPMuSiC, an algorithm for predicting protein mutant stability changes. Application to prion proteins. Protein Eng. Des. Sel. 2000, 13, 849–856. [Google Scholar] [CrossRef]
- Pasquier, C.; Hamodrakas, S. An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng. Des. Sel. 1999, 12, 631–634. [Google Scholar] [CrossRef]
- Marvin, J.S.; Corcoran, E.E.; Hattangadi, N.A.; Zhang, J.V.; Gere, S.A.; Hellinga, H.W. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. USA 1997, 94, 4366–4371. [Google Scholar] [CrossRef]
- Barany, F. Single-stranded hexameric linkers: A system for in-phase insertion mutagenesis and protein engineering. Gene 1985, 37, 111–123. [Google Scholar] [CrossRef]
- Kawai, F.; Nakamura, A.; Visootsat, A.; Iino, R. Plasmid-Based One-Pot Saturation Mutagenesis and Robot-Based Automated Screening for Protein Engineering. ACS Omega 2018, 3, 7715–7726. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.H.; Tsai, C.J.; Ma, B.; Nussinov, R. In silico protein design by combinatorial assembly of protein building blocks. Protein Sci. 2004, 13, 2753–2765. [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.J.; Kortemme, T. Backbone flexibility in computational protein design. Curr. Opin. Biotechnol. 2009, 20, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Lise, S.; Archambeau, C.; Pontil, M.; Jones, D.T. Prediction of hot spot residues at protein-protein interfaces by combining machine learning and energy-based methods. BMC Bioinform. 2009, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Nikam, R.; Kulandaisamy, A.; Harini, K.; Sharma, D.; Gromiha, M.M. ProThermDB: Thermodynamic database for proteins and mutants revisited after 15 years. Nucleic Acids Res. 2021, 49, D420–D424. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Yarlagadda, R.; Reed, C.C. Structure Based Thermostability Prediction Models for Protein Single Point Mutations with Machine Learning Tools. PLoS ONE 2015, 10, e0138022. [Google Scholar] [CrossRef]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J. Chem. Inf. Model 2019, 59, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Vangone, A.; Folkers, G.E.; Xue, L.C.; Bonvin, A. iSEE: Interface structure, evolution, and energy-based machine learning predictor of binding affinity changes upon mutations. Proteins 2019, 87, 110–119. [Google Scholar] [CrossRef]
- Wang, J.; Lisanza, S.; Juergens, D.; Tischer, D.; Anishchenko, I.; Baek, M.; Watson, J.L.; Chun, J.H.; Milles, L.F.; Dauparas, J.; et al. Deep learning methods for designing proteins scaffolding functional sites. bioRxiv 2021. [Google Scholar] [CrossRef]
- Harteveld, Z.; Bonet, J.; Rosset, S.; Yang, C.; Sesterhenn, F.; Correia, B.E. A generic framework for hierarchical de novo protein design. bioRxiv 2022. [Google Scholar] [CrossRef]
- Cang, Z.; Wei, G.W. TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput. Biol. 2017, 13, e1005690. [Google Scholar] [CrossRef] [PubMed]
- Moffat, L.; Kandathil, S.M.; Jones, D.T. Design in the DARK: Learning Deep Generative Models for De Novo Protein Design. bioRxiv 2022. [Google Scholar] [CrossRef]
- Keskin, O.; Gursoy, A.; Ma, B.; Nussinov, R. Principles of protein- protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 2008, 108, 1225–1244. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, P.; Li, F.; Marquez-Lago, T.T.; Leier, A.; Revote, J.; Zhu, Y.; Powell, D.R.; Akutsu, T.; Webb, G.I.; et al. iLearn: An integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Briefings Bioinform. 2020, 21, 1047–1057. [Google Scholar] [CrossRef]
- Wang, J.T.L.; Ma, Q.; Shasha, D.; Wu, C.H. New techniques for extracting features from protein sequences. IBM Syst. J. 2001, 40, 426–441. [Google Scholar] [CrossRef]
- Singh, R.; Park, D.; Xu, J.; Hosur, R.; Berger, B. Struct2Net: A web service to predict protein–protein interactions using a structure-based approach. Nucleic Acids Res. 2010, 38, W508–W515. [Google Scholar] [CrossRef] [PubMed]
- Hashemifar, S.; Neyshabur, B.; Khan, A.A.; Xu, J. Predicting protein–protein interactions through sequence-based deep learning. Bioinformatics 2018, 34, i802–i810. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, G.; Xia, D.; Wang, J. Protein–protein interactions prediction based on ensemble deep neural networks. Neurocomputing 2019, 324, 10–19. [Google Scholar] [CrossRef]
- Lei, H.; Wen, Y.; You, Z.; Elazab, A.; Tan, E.L.; Zhao, Y.; Lei, B. Protein–protein interactions prediction via multimodal deep polynomial network and regularized extreme learning machine. IEEE J. Biomed. Health Inform. 2018, 23, 1290–1303. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.F.; Liu, S.R.; Yan, X.; Song, K.J. Predicting protein-protein interactions from matrix-based protein sequence using convolution neural network and feature-selective rotation forest. Sci. Rep. 2019, 9, 9848. [Google Scholar] [CrossRef]
- Yang, F.; Fan, K.; Song, D.; Lin, H. Graph-based prediction of Protein-protein interactions with attributed signed graph embedding. BMC Bioinform. 2020, 21, 323. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhu, F.; Ling, X.; Liu, Q. Protein interaction network reconstruction through ensemble deep learning with attention mechanism. Front. Bioeng. Biotechnol. 2020, 8, 390. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Chakrabarti, S. Classification and prediction of protein–protein interaction interface using machine learning algorithm. Sci. Rep. 2021, 11, 1761. [Google Scholar] [CrossRef]
- Lei, Y.; Li, S.; Liu, Z.; Wan, F.; Tian, T.; Li, S.; Zhao, D.; Zeng, J. A deep-learning framework for multi-level peptide–protein interaction prediction. Nat. Commun. 2021, 12, 5465. [Google Scholar] [CrossRef]
- Balogh, O.M.; Benczik, B.; Horváth, A.; Pétervári, M.; Csermely, P.; Ferdinandy, P.; Ágg, B. Efficient link prediction in the protein–protein interaction network using topological information in a generative adversarial network machine learning model. BMC Bioinform. 2022, 23, 78. [Google Scholar] [CrossRef]
- Song, B.; Luo, X.; Luo, X.; Liu, Y.; Niu, Z.; Zeng, X. Learning spatial structures of proteins improves protein–protein interaction prediction. Briefings Bioinform. 2022, 23, bbab558. [Google Scholar] [CrossRef]
- Daberdaku, S.; Ferrari, C. Exploring the potential of 3D Zernike descriptors and SVM for protein–protein interface prediction. BMC Bioinform. 2018, 19, 35. [Google Scholar] [CrossRef]
- Sanchez-Garcia, R.; Sorzano, C.O.S.; Carazo, J.M.; Segura, J. BIPSPI: A method for the prediction of partner-specific protein–protein interfaces. Bioinformatics 2019, 35, 470–477. [Google Scholar] [CrossRef]
- Northey, T.C.; Barešić, A.; Martin, A.C. IntPred: A structure-based predictor of protein–protein interaction sites. Bioinformatics 2018, 34, 223–229. [Google Scholar] [CrossRef]
- Gainza, P.; Sverrisson, F.; Monti, F.; Rodola, E.; Boscaini, D.; Bronstein, M.; Correia, B. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 2020, 17, 184–192. [Google Scholar] [CrossRef]
- Yuan, Q.; Chen, J.; Zhao, H.; Zhou, Y.; Yang, Y. Structure-aware protein–protein interaction site prediction using deep graph convolutional network. Bioinformatics 2022, 38, 125–132. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins and their “mysterious”(meta) physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef]
- Katuwawala, A.; Peng, Z.; Yang, J.; Kurgan, L. Computational Prediction of MoRFs, Short Disorder-to-order Transitioning Protein Binding Regions. Comput. Struct. Biotechnol. J. 2019, 17, 454–462. [Google Scholar] [CrossRef]
- Zhao, B.; Kurgan, L. Surveying over 100 predictors of intrinsic disorder in proteins. Expert Rev. Proteom. 2021, 18, 1019–1029. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Tosatto, S.C. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef]
- Hatos, A.; Hajdu-Soltész, B.; Monzon, A.M.; Palopoli, N.; Álvarez, L.; Aykac-Fas, B.; Bassot, C.; Benítez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276. [Google Scholar] [CrossRef]
- Malhis, N.; Jacobson, M.; Gsponer, J. MoRFchibi SYSTEM: Software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 2016, 44, W488–W493. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, J.; Xu, J. AUCpreD: Proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. Bioinformatics 2016, 32, i672–i679. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Kumar, S.; Tsunoda, T.; Patil, A.; Sharma, A. Predicting MoRFs in protein sequences using HMM profiles. BMC Bioinform. 2016, 17, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Yang, Y.; Paliwal, K.; Zhou, Y. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics 2017, 33, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Bayarjargal, M.; Tsunoda, T.; Patil, A.; Sharma, A. MoRFPred-plus: Computational identification of MoRFs in protein sequences using physicochemical properties and HMM profiles. J. Theor. Biol. 2018, 437, 9–16. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, A.; Raicar, G.; Tsunoda, T.; Patil, A. OPAL+: Length-specific MoRF prediction in intrinsically disordered protein sequences. Proteomics 2019, 19, 1800058. [Google Scholar] [CrossRef]
- Mirabello, C.; Wallner, B. RAWMSA: End-to-end deep learning using raw multiple sequence alignments. PLoS ONE 2019, 14, e0220182. [Google Scholar]
- Hanson, J.; Paliwal, K.K.; Litfin, T.; Zhou, Y. SPOT-Disorder2: Improved protein intrinsic disorder prediction by ensembled deep learning. Genom. Proteom. Bioinform. 2019, 17, 645–656. [Google Scholar] [CrossRef]
- Dass, R.; Mulder, F.A.; Nielsen, J.T. ODiNPred: Comprehensive prediction of protein order and disorder. Sci. Rep. 2020, 10, 14780. [Google Scholar] [CrossRef]
- Tang, Y.J.; Pang, Y.H.; Liu, B. IDP-Seq2Seq: Identification of intrinsically disordered regions based on sequence to sequence learning. Bioinformatics 2020, 36, 5177–5186. [Google Scholar] [CrossRef]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Liu, B. RFPR-IDP: Reduce the false positive rates for intrinsically disordered protein and region prediction by incorporating both fully ordered proteins and disordered proteins. Briefings Bioinform. 2021, 22, 2000–2011. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Griffith, D.; Holehouse, A.S. Metapredict: A fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys. J. 2021, 120, 4312–4319. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, B.; Shi, W.; Li, M.; Kurgan, L. DeepDISOBind: Accurate prediction of RNA-, DNA-and protein-binding intrinsically disordered residues with deep multi-task learning. Briefings Bioinform. 2022, 23, bbab521. [Google Scholar] [CrossRef]
- Li, H.; Pang, Y.; Liu, B.; Yu, L. MoRF-FUNCpred: Molecular Recognition Feature Function Prediction Based on Multi-Label Learning and Ensemble Learning. Front. Pharmacol. 2022, 13, 856417. [Google Scholar] [CrossRef] [PubMed]
- Orlando, G.; Raimondi, D.; Codice, F.; Tabaro, F.; Vranken, W. Prediction of disordered regions in proteins with recurrent neural networks and protein dynamics. J. Mol. Biol. 2022, 434, 167579. [Google Scholar] [CrossRef]
- Wilson, C.J.; Choy, W.Y.; Karttunen, M. AlphaFold2: A Role for Disordered Protein/Region Prediction? Int. J. Mol. Sci. 2022, 23, 4591. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-Factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Naturwissenschaften 2005, 72, 212–213. [Google Scholar] [CrossRef]
- Kuboniwa, H.; Tjandra, N.; Grzesiek, S.; Ren, H.; Klee, C.B.; Bax, A. Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 1995, 2, 768–776. [Google Scholar] [CrossRef]
- Yun, C.H.; Bai, J.; Sun, D.Y.; Cui, D.F.; Chang, W.R.; Liang, D.C. Structure of potato calmodulin PCM6: The first report of the three-dimensional structure of a plant calmodulin. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Vertessy, B.G.; Harmat, V.; Bocskei, Z.; Naray-Szabo, G.; Orosz, F.; Ovadi, J. Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: Crystallographic and spectroscopic studies. Biochemistry 1998, 37, 15300–15310. [Google Scholar] [CrossRef]
- Komeiji, Y.; Ueno, Y.; Uebayasi, M. Molecular dynamics simulations revealed Ca(2+)-dependent conformational change of Calmodulin. FEBS Lett. 2002, 521, 133–139. [Google Scholar] [CrossRef]
- Fonze, E.; Charlier, P.; To’th, Y.; Vermeire, M.; Raquet, X.; Dubus, A.; Frere, J.M. TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr. D Biol. Crystallogr. 1995, 51, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Avery, C.; Baker, L.; Jacobs, D.J. Functional Dynamics of Substrate Recognition in TEM Beta-Lactamase. Entropy 2022, 24, 729. [Google Scholar] [CrossRef]
- Hsiao, C.D.; Sun, Y.J.; Rose, J.; Wang, B.C. The crystal structure of glutamine-binding protein from Escherichia coli. J. Mol. Biol. 1996, 262, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.J. Do Dynamic Allosteric Effects Occur in IGG4 Antibodies? Ph.D. Thesis, The University of North Carolina at Charlotte, Charlotte, NC, USA, 2020. [Google Scholar]
- Carugo, O.; Argos, P. Protein—Protein crystal-packing contacts. Protein Sci. 1997, 6, 2261–2263. [Google Scholar] [CrossRef] [PubMed]
- Berjanskii, M.V.; Wishart, D.S. Application of the random coil index to studying protein flexibility. J. Biomol. NMR 2008, 40, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Livesay, D.R.; Huynh, D.H.; Dallakyan, S.; Jacobs, D.J. Hydrogen bond networks determine emergent mechanical and thermodynamic properties across a protein family. Chem. Cent. J. 2008, 2, 17. [Google Scholar] [CrossRef]
- Li, T.; Tracka, M.B.; Uddin, S.; Casas-Finet, J.; Jacobs, D.J.; Livesay, D.R. Redistribution of flexibility in stabilizing antibody fragment mutants follows Le Châtelier’s principle. PLoS ONE 2014, 9, e92870. [Google Scholar] [CrossRef]
- Atilgan, A.R.; Durell, S.R.; Jernigan, R.L.; Demirel, M.C.; Keskin, O.; Bahar, I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001, 80, 505–515. [Google Scholar] [CrossRef]
- Xia, K.; Opron, K.; Wei, G.W. Multiscale multiphysics and multidomain models–flexibility and rigidity. J. Chem. Phys. 2013, 139, 194109. [Google Scholar] [CrossRef]
- Opron, K.; Xia, K.; Wei, G.W. Fast and anisotropic flexibility-rigidity index for protein flexibility and fluctuation analysis. J. Chem. Phys. 2014, 140, 234105. [Google Scholar] [CrossRef]
- Bramer, D.; Wei, G.W. Blind prediction of protein B-factor and flexibility. J. Chem. Phys. 2018, 149, 134107. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Siggers, K.; Rost, B.; Palmer, A.G., 3rd. Protein conformational flexibility prediction using machine learning. J. Magn. Reson. 2008, 192, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ludtke, S.J. Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM. Nat. Methods 2021, 18, 930–936. [Google Scholar] [CrossRef]
- Nembrini, S.; König, I.R.; Wright, M.N. The revival of the Gini importance? Bioinformatics 2018, 34, 3711–3718. [Google Scholar] [CrossRef]
- Grisci, B.; Dorn, M. NEAT-FLEX: Predicting the conformational flexibility of amino acids using neuroevolution of augmenting topologies. J. Bioinform. Comput. Biol. 2017, 15, 1750009. [Google Scholar] [CrossRef]
- Spiwok, V.; Kriz, P. Time-Lagged t-Distributed Stochastic Neighbor Embedding (t-SNE) of Molecular Simulation Trajectories. Front. Mol. Biosci. 2020, 7, 132. [Google Scholar] [CrossRef]
- Grear, T.; Avery, C.; Patterson, J.; Jacobs, D.J. Molecular function recognition by supervised projection pursuit machine learning. Sci. Rep. 2021, 11, 4247. [Google Scholar] [CrossRef]
- Patterson, J.; Grear, T.; Jacobs, D.J. Biased Hypothesis Formation From Projection Pursuit 2021. Adv. Artif. Intell. Mach. Learn. 2021, 3. [Google Scholar]
- Zheng, W. Predicting cryptic ligand binding sites based on normal modes guided conformational sampling. Proteins 2021, 89, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Degiacomi, M.T. Coupling Molecular Dynamics and Deep Learning to Mine Protein Conformational Space. Structure 2019, 27, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Jiang, X.; Trozzi, F.; Xiao, S.; Larson, E.C.; Tao, P. Explore Protein Conformational Space With Variational Autoencoder. Front. Mol. Biosci. 2021, 8, 781635. [Google Scholar] [CrossRef] [PubMed]
- Romero, R.; Ramanathan, A.; Yuen, T.; Bhowmik, D.; Mathew, M.; Munshi, L.B.; Javaid, S.; Bloch, M.; Lizneva, D.; Rahimova, A.; et al. Mechanism of glucocerebrosidase activation and dysfunction in Gaucher disease unraveled by molecular dynamics and deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 5086–5095. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.G.F.; Kim, P.M. Data driven flexible backbone protein design. PLoS Comput. Biol. 2017, 13, e1005722. [Google Scholar] [CrossRef]
- Monzon, A.M.; Rohr, C.O.; Fornasari, M.S.; Parisi, G. CoDNaS 2.0: A comprehensive database of protein conformational diversity in the native state. Database 2016, 2016, baw038. [Google Scholar] [CrossRef]
- Srivastava, A.; Tracka, M.B.; Uddin, S.; Casas-Finet, J.; Livesay, D.R.; Jacobs, D.J. Mutations in Antibody Fragments Modulate Allosteric Response Via Hydrogen-Bond Network Fluctuations. Biophys. J. 2016, 110, 1933–1942. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, H.X. Protein Allostery and Conformational Dynamics. Chem. Rev. 2016, 116, 6503–6515. [Google Scholar] [CrossRef]
- Liu, J.; Nussinov, R. Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput. Biol. 2016, 12, e1004966. [Google Scholar] [CrossRef]
- Perutz, M.F.; Fermi, G.; Luisi, B.; Shaanan, B.; Liddington, R.C. Stereochemistry of cooperative mechanisms in hemoglobin. Accounts Chem. Res. 1987, 20, 309–321. [Google Scholar] [CrossRef]
- Nussinov, R. Introduction to Protein Ensembles and Allostery. Chem. Rev. 2016, 116, 6263–6266. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Ma, B.; Nussinov, R. Is allostery an intrinsic property of all dynamic proteins? Proteins Struct. Funct. Bioinform. 2004, 57, 433–443. [Google Scholar] [CrossRef]
- Istomin, A.Y.; Gromiha, M.M.; Vorov, O.K.; Jacobs, D.J.; Livesay, D.R. New insight into long-range nonadditivity within protein double-mutant cycles. Proteins Struct. Funct. Bioinform. 2008, 70, 915–924. [Google Scholar] [CrossRef]
- Skjaerven, L.; Hollup, S.M.; Reuter, N. Normal mode analysis for proteins. J. Mol. Struct. THEOCHEM 2009, 898, 42–48. [Google Scholar] [CrossRef]
- Tama, F.; Sanejouand, Y.H. Conformational change of proteins arising from normal mode calculations. Protein Eng. Des. Sel. 2001, 14, 1–6. [Google Scholar] [CrossRef]
- Hayward, S.; Kitao, A.; Berendsen, H.J. Model-free methods of analyzing domain motions in proteins from simulation: A comparison of normal mode analysis and molecular dynamics simulation of lysozyme. Proteins Struct. Funct. Bioinform. 1997, 27, 425–437. [Google Scholar] [CrossRef]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef]
- Wells, S.; Menor, S.; Hespenheide, B.; Thorpe, M.F. Constrained geometric simulation of diffusive motion in proteins. Phys. Biol. 2005, 2, S127–S136. [Google Scholar] [CrossRef]
- Ma, B.; Tsai, C.J.; Haliloğlu, T.; Nussinov, R. Dynamic Allostery: Linkers Are Not Merely Flexible. Structure 2011, 19, 907–917. [Google Scholar] [CrossRef]
- Pandey, R.B.; Jacobs, D.J.; Farmer, B.L. Preferential binding effects on protein structure and dynamics revealed by coarse-grained Monte Carlo simulation. J. Chem. Phys. 2017, 146, 195101. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, M.; Moroni, E.; Ippoliti, E.; Rinaldi, S.; Sanchez-Martin, C.; Rasola, A.; Pavarino, L.F.; Colombo, G. Machine Learning of Allosteric Effects: The Analysis of Ligand-Induced Dynamics to Predict Functional Effects in TRAP1. J. Phys. Chem. B 2021, 125, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, F.; Moroni, E.; Pandini, A.; Colombo, G. Machine Learning Prediction of Allosteric Drug Activity from Molecular Dynamics. J. Phys. Chem. Lett. 2021, 12, 3724–3732. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, J.; Han, W.; Xu, D. Neural relational inference to learn long-range allosteric interactions in proteins from molecular dynamics simulations. Nat. Commun. 2022, 13, 1661. [Google Scholar] [CrossRef]
- Tian, H.; Jiang, X.; Tao, P. PASSer: Prediction of allosteric sites server. Mach. Learn. Sci. Technol. 2021, 2, 035015. [Google Scholar] [CrossRef]
- Vishweshwaraiah, Y.L.; Chen, J.; Dokholyan, N.V. Engineering an Allosteric Control of Protein Function. J. Phys. Chem. B 2021, 125, 1806–1814. [Google Scholar] [CrossRef]
- Gorman, S.D.; D’Amico, R.N.; Winston, D.S.; Boehr, D.D. Engineering Allostery into Proteins. Adv. Exp. Med. Biol. 2019, 1163, 359–384. [Google Scholar] [CrossRef]
- Quijano-Rubio, A.; Yeh, H.W.; Park, J.; Lee, H.; Langan, R.A.; Boyken, S.E.; Lajoie, M.J.; Cao, L.; Chow, C.M.; Miranda, M.C.; et al. De novo design of modular and tunable protein biosensors. Nature 2021, 591, 482–487. [Google Scholar] [CrossRef]
- Unke, O.T.; Chmiela, S.; Sauceda, H.E.; Gastegger, M.; Poltavsky, I.; Schutt, K.T.; Tkatchenko, A.; Muller, K.R. Machine Learning Force Fields. Chem. Rev. 2021, 121, 10142–10186. [Google Scholar] [CrossRef]
- Behler, J. Perspective: Machine learning potentials for atomistic simulations. J. Chem. Phys. 2016, 145, 170901. [Google Scholar] [CrossRef]
- Behler, J. Atom-centered symmetry functions for constructing high-dimensional neural network potentials. J. Chem. Phys. 2011, 134, 074106. [Google Scholar] [CrossRef] [PubMed]
- Gastegger, M.; Schwiedrzik, L.; Bittermann, M.; Berzsenyi, F.; Marquetand, P. wACSF-Weighted atom-centered symmetry functions as descriptors in machine learning potentials. J. Chem. Phys. 2018, 148, 241709. [Google Scholar] [CrossRef] [PubMed]
- Bartok, A.P.; Kondor, R.; Csányi, G. On representing chemical environments. Phys. Rev. B 2013, 87, 184115. [Google Scholar] [CrossRef]
- Bartok, A.P.; Payne, M.C.; Kondor, R.; Csanyi, G. Gaussian approximation potentials: The accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 2010, 104, 136403. [Google Scholar] [CrossRef]
- Csányi, G.; Winfield, S.; Kermode, J.R.; De Vita, A.; Comisso, A.; Bernstein, N.; Payne, M.C. Expressive Programming for Computational Physics in Fortran 95+. In IoP Computational Physics Group Newsletter; Spring: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Sumpter, B.G.; Noid, D.W. Potential energy surfaces for macromolecules. A neural network technique. Chem. Phys. Lett. 1992, 192, 455–462. [Google Scholar] [CrossRef]
- Blank, T.B.; Brown, S.D.; Calhoun, A.W.; Doren, D.J. Neural network models of potential energy surfaces. J. Chem. Phys. 1995, 103, 4129–4137. [Google Scholar] [CrossRef]
- Prudente, F.V.; Acioli, P.H.; Neto, J.S. The fitting of potential energy surfaces using neural networks: Application to the study of vibrational levels of H3+. J. Chem. Phys. 1998, 109, 8801–8808. [Google Scholar] [CrossRef]
- Hunger, J.; Huttner, G. Optimization and analysis of force field parameters by combination of genetic algorithms and neural networks. J. Comput. Chem. 1999, 20, 455–471. [Google Scholar] [CrossRef]
- Lorenz, S.; Groß, A.; Scheffler, M. Representing high-dimensional potential-energy surfaces for reactions at surfaces by neural networks. Chem. Phys. Lett. 2004, 395, 210–215. [Google Scholar] [CrossRef]
- Behler, J.; Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 2007, 98, 146401. [Google Scholar] [CrossRef]
- Behler, J. Constructing high-dimensional neural network potentials: A tutorial review. Int. J. Quantum Chem. 2015, 115, 1032–1050. [Google Scholar] [CrossRef]
- Unke, O.T.; Meuwly, M. PhysNet: A Neural Network for Predicting Energies, Forces, Dipole Moments, and Partial Charges. J. Chem. Theory Comput. 2019, 15, 3678–3693. [Google Scholar] [CrossRef] [PubMed]
- Schütt, K.; Kindermans, P.J.; Sauceda Felix, H.E.; Chmiela, S.; Tkatchenko, A.; Müller, K.R. SchNet: A continuous-filter convolutional neural network for modeling quantum interactions. In Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Schütt, K.T.; Sauceda, H.E.; Kindermans, P.J.; Tkatchenko, A.; Muller, K.R. SchNet—A deep learning architecture for molecules and materials. J. Chem. Phys. 2018, 148, 241722. [Google Scholar] [CrossRef]
- Schütt, K.T.; Kessel, P.; Gastegger, M.; Nicoli, K.A.; Tkatchenko, A.; Muller, K.R. SchNetPack: A Deep Learning Toolbox For Atomistic Systems. J. Chem. Theory Comput. 2019, 15, 448–455. [Google Scholar] [CrossRef]
- Gasteiger, J.; Groß, J.; Günnemann, S. Directional Message Passing for Molecular Graphs. In Proceedings of the International Conference on Learning Representations, Addis Ababa, Ethiopia, 26 April–1 May 2020. [Google Scholar]
- Park, C.W.; Kornbluth, M.; Vandermause, J.; Wolverton, C.; Kozinsky, B.; Mailoa, J.P. Accurate and scalable graph neural network force field and molecular dynamics with direct force architecture. NPJ Comput. Mater. 2021, 7, 73. [Google Scholar] [CrossRef]
- Haghighatlari, M.; Li, J.; Guan, X.; Zhang, O.; Das, A.; Stein, C.J.; Heidar-Zadeh, F.; Liu, M.; Head-Gordon, M.; Bertels, L.; et al. NewtonNet: A Newtonian message passing network for deep learning of interatomic potentials and forces. Digit Discov. 2022, 1, 333–343. [Google Scholar] [CrossRef]
- Doerr, S.; Majewski, M.; Pérez, A.; Kramer, A.; Clementi, C.; Noe, F.; Giorgino, T.; De Fabritiis, G. Torchmd: A deep learning framework for molecular simulations. J. Chem. Theory Comput. 2021, 17, 2355–2363. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Han, J.; E, W. DeePMD-kit: A deep learning package for many-body potential energy representation and molecular dynamics. Comput. Phys. Commun. 2018, 228, 178–184. [Google Scholar] [CrossRef]
- Sinha, S.; Vohora, D. Drug discovery and development: An overview. Pharm. Med. Transl. Clin. Res. 2018, 19–32. [Google Scholar] [CrossRef]
- Shen, C.; Ding, J.; Wang, Z.; Cao, D.; Ding, X.; Hou, T. From machine learning to deep learning: Advances in scoring functions for protein–ligand docking. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2020, 10, e1429. [Google Scholar] [CrossRef]
- Lavecchia, A. Machine-learning approaches in drug discovery: Methods and applications. Drug Discov. Today 2015, 20, 318–331. [Google Scholar] [CrossRef]
- Lo, Y.C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Ghasemi, F.; Mehridehnavi, A.; Pérez-Garrido, A.; Pérez-Sánchez, H. Neural network and deep-learning algorithms used in QSAR studies: Merits and drawbacks. Drug Discov. Today 2018, 23, 1784–1790. [Google Scholar] [CrossRef]
- Rifaioglu, A.S.; Atas, H.; Martin, M.J.; Cetin-Atalay, R.; Atalay, V.; Doğan, T. Recent applications of deep learning and machine intelligence on in silico drug discovery: Methods, tools and databases. Briefings Bioinform. 2019, 20, 1878–1912. [Google Scholar] [CrossRef]
- Jing, Y.; Bian, Y.; Hu, Z.; Wang, L.; Xie, X.Q.S. Deep learning for drug design: An artificial intelligence paradigm for drug discovery in the big data era. AAPS J. 2018, 20, 58. [Google Scholar] [CrossRef]
- Dana, D.; Gadhiya, S.V.; St. Surin, L.G.; Li, D.; Naaz, F.; Ali, Q.; Paka, L.; Yamin, M.A.; Narayan, M.; Goldberg, I.D.; et al. Deep learning in drug discovery and medicine; scratching the surface. Molecules 2018, 23, 2384. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in de novo drug design: From conventional to machine learning methods. Int. J. Mol. Sci. 2021, 22, 1676. [Google Scholar] [CrossRef]
- Peña-Guerrero, J.; Nguewa, P.A.; García-Sosa, A.T. Machine learning, artificial intelligence, and data science breaking into drug design and neglected diseases. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1513. [Google Scholar] [CrossRef]
- Maia, E.H.B.; Assis, L.C.; De Oliveira, T.A.; Da Silva, A.M.; Taranto, A.G. Structure-based virtual screening: From classical to artificial intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Sunny, S.; Jayaraj, P. Protein–protein docking: Past, present, and future. Protein J. 2022, 41, 1–26. [Google Scholar] [CrossRef]
- Crampon, K.; Giorkallos, A.; Deldossi, M.; Baud, S.; Steffenel, L.A. Machine-learning methods for ligand–protein molecular docking. Drug Discov. Today 2021, 27, 151–164. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Baum, B.; Muley, L.; Smolinski, M.; Heine, A.; Hangauer, D.; Klebe, G. Non-additivity of functional group contributions in protein–ligand binding: A comprehensive study by crystallography and isothermal titration calorimetry. J. Mol. Biol. 2010, 397, 1042–1054. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y. Improving scoring-docking-screening powers of protein–ligand scoring functions using random forest. J. Comput. Chem. 2017, 38, 169–177. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New machine learning and physics-based scoring functions for drug discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar] [CrossRef]
- Wang, X.; Terashi, G.; Christoffer, C.W.; Zhu, M.; Kihara, D. Protein docking model evaluation by 3D deep convolutional neural networks. Bioinformatics 2020, 36, 2113–2118. [Google Scholar] [CrossRef]
- Yang, L.; Yang, G.; Chen, X.; Yang, Q.; Yao, X.; Bing, Z.; Niu, Y.; Huang, L.; Yang, L. Deep scoring neural network replacing the scoring function components to improve the performance of structure-based molecular docking. ACS Chem. Neurosci. 2021, 12, 2133–2142. [Google Scholar] [CrossRef]
- Xie, Z.; Deng, X.; Shu, K. Prediction of protein–protein interaction sites using convolutional neural network and improved data sets. Int. J. Mol. Sci. 2020, 21, 467. [Google Scholar] [CrossRef]
- Townshend, R.; Bedi, R.; Suriana, P.; Dror, R. End-to-end learning on 3d protein structure for interface prediction. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, CO, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Zhu, H.; Du, X.; Yao, Y. ConvsPPIS: Identifying protein-protein interaction sites by an ensemble convolutional neural network with feature graph. Curr. Bioinform. 2020, 15, 368–378. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, H.; Cai, L.; Ji, S. Deep learning of high-order interactions for protein interface prediction. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Event, 6–10 July 2020; pp. 679–687. [Google Scholar]
- Fout, A.; Byrd, J.; Shariat, B.; Ben-Hur, A. Protein interface prediction using graph convolutional networks. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Cao, Y.; Shen, Y. Energy-based graph convolutional networks for scoring protein docking models. Proteins Struct. Funct. Bioinform. 2020, 88, 1091–1099. [Google Scholar] [CrossRef]
- Wang, X.; Flannery, S.T.; Kihara, D. Protein docking model evaluation by graph neural networks. Front. Mol. Biosci. 2021, 8, 647915. [Google Scholar] [CrossRef]
- Ramaswamy, V.K.; Musson, S.C.; Willcocks, C.G.; Degiacomi, M.T. Deep learning protein conformational space with convolutions and latent interpolations. Phys. Rev. X 2021, 11, 011052. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Gao, K.; Wang, M.; Wei, G.W. MathDL: Mathematical deep learning for D3R Grand Challenge 4. J. Comput.-Aided Mol. Des. 2020, 34, 131–147. [Google Scholar] [CrossRef]
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction tree variational autoencoder for molecular graph generation. In Proceedings of the International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; pp. 2323–2332. [Google Scholar]
- Adeshina, Y.O.; Deeds, E.J.; Karanicolas, J. Machine learning classification can reduce false positives in structure-based virtual screening. Proc. Natl. Acad. Sci. USA 2020, 117, 18477–18488. [Google Scholar] [CrossRef]
- Schreiber, G. Protein–Protein Interaction Interfaces and Their Functional Implications. Protein–Protein Interaction Regulators. 2020. Available online: https://pubs.rsc.org/en/content/chapterhtml/2020/bk9781788011877-00001?isbn=978-1-78801-187-7&sercode=bk (accessed on 15 July 2022).
- Fan, J.; Fu, A.; Zhang, L. Progress in molecular docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. BioLiP: A semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013, 41, D1096–D1103. [Google Scholar] [CrossRef]
- Smith, R.D.; Clark, J.J.; Ahmed, A.; Orban, Z.J.; Dunbar, J.B., Jr.; Carlson, H.A. Updates to Binding MOAD (Mother of All Databases): Polypharmacology Tools and Their Utility in Drug Repurposing. J. Mol. Biol. 2019, 431, 2423–2433. [Google Scholar] [CrossRef]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2015, 44, D1045–D1053. [Google Scholar] [CrossRef]
- Su, M.; Yang, Q.; Du, Y.; Feng, G.; Liu, Z.; Li, Y.; Wang, R. Comparative Assessment of Scoring Functions: The CASF-2016 Update. J. Chem. Inf. Model 2019, 59, 895–913. [Google Scholar] [CrossRef]
- Liu, Z.; Su, M.; Han, L.; Liu, J.; Yang, Q.; Li, Y.; Wang, R. Forging the Basis for Developing Protein-Ligand Interaction Scoring Functions. Acc. Chem. Res. 2017, 50, 302–309. [Google Scholar] [CrossRef]
- Ballester, P.J.; Mitchell, J.B. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 2010, 26, 1169–1175. [Google Scholar] [CrossRef]
- Li, H.; Leung, K.S.; Wong, M.H.; Ballester, P.J. Improving AutoDock Vina Using Random Forest: The Growing Accuracy of Binding Affinity Prediction by the Effective Exploitation of Larger Data Sets. Mol. Inform. 2015, 34, 115–126. [Google Scholar] [CrossRef]
- Li, H.; Leung, K.S.; Wong, M.H.; Ballester, P.J. Substituting random forest for multiple linear regression improves binding affinity prediction of scoring functions: Cyscore as a case study. BMC Bioinform. 2014, 15, 291. [Google Scholar] [CrossRef]
- Ashtawy, H.M.; Mahapatra, N.R. A Comparative Assessment of Predictive Accuracies of Conventional and Machine Learning Scoring Functions for Protein-Ligand Binding Affinity Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 12, 335–347. [Google Scholar] [CrossRef]
- Shar, P.A.; Tao, W.; Gao, S.; Huang, C.; Li, B.; Zhang, W.; Shahen, M.; Zheng, C.; Bai, Y.; Wang, Y. Pred-binding: Large-scale protein-ligand binding affinity prediction. J. Enzym. Inhib. Med. Chem. 2016, 31, 1443–1450. [Google Scholar] [CrossRef]
- Jover, J.; Bosque, R.; Sales, J. Quantitative structure-property relationship estimation of cation binding affinity of the common amino acids. J. Phys. Chem. A 2009, 113, 3703–3708. [Google Scholar] [CrossRef]
- Ragoza, M.; Hochuli, J.; Idrobo, E.; Sunseri, J.; Koes, D.R. Protein-Ligand Scoring with Convolutional Neural Networks. J. Chem. Inf. Model 2017, 57, 942–957. [Google Scholar] [CrossRef]
- Jimenez, J.; Skalic, M.; Martinez-Rosell, G.; De Fabritiis, G. KDEEP: Protein-Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J. Chem. Inf. Model 2018, 58, 287–296. [Google Scholar] [CrossRef]
- Stepniewska-Dziubinska, M.M.; Zielenkiewicz, P.; Siedlecki, P. Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 2018, 34, 3666–3674. [Google Scholar] [CrossRef]
- Li, Y.; Rezaei, M.A.; Li, C.; Li, X. DeepAtom: A Framework for Protein-Ligand Binding Affinity Prediction. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 303–310. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, L.; Saravanan, K.M.; Yin, P.; Wei, Y. DeepBindRG: A deep learning based method for estimating effective protein-ligand affinity. PeerJ 2019, 7, e7362. [Google Scholar] [CrossRef]
- Zheng, L.; Fan, J.; Mu, Y. OnionNet: A Multiple-Layer Intermolecular-Contact-Based Convolutional Neural Network for Protein-Ligand Binding Affinity Prediction. ACS Omega 2019, 4, 15956–15965. [Google Scholar] [CrossRef]
- Wang, S.; Liu, D.; Ding, M.; Du, Z.; Zhong, Y.; Song, T.; Zhu, J.; Zhao, R. SE-OnionNet: A Convolution Neural Network for Protein-Ligand Binding Affinity Prediction. Front. Genet. 2020, 11, 607824. [Google Scholar] [CrossRef]
- Ozturk, H.; Ozgur, A.; Ozkirimli, E. DeepDTA: Deep drug-target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Pang, L.; Liu, Y.; Zhang, J. GANsDTA: Predicting Drug-Target Binding Affinity Using GANs. Front. Genet. 2019, 10, 1243. [Google Scholar] [CrossRef]
- Zhao, Q.; Duan, G.; Yang, M.; Cheng, Z.; Li, Y.; Wang, J. AttentionDTA: Drug-target binding affinity prediction by sequence-based deep learning with attention mechanism. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022. [Google Scholar] [CrossRef]
- Nguyen, T.; Le, H.; Quinn, T.P.; Nguyen, T.; Le, T.D.; Venkatesh, S. GraphDTA: Predicting drug-target binding affinity with graph neural networks. Bioinformatics 2021, 37, 1140–1147. [Google Scholar] [CrossRef]
- Son, J.; Kim, D. Development of a graph convolutional neural network model for efficient prediction of protein-ligand binding affinities. PLoS ONE 2021, 16, e0249404. [Google Scholar] [CrossRef]
- Jankauskaite, J.; Jimenez-Garcia, B.; Dapkunas, J.; Fernandez-Recio, J.; Moal, I.H. SKEMPI 2.0: An updated benchmark of changes in protein-protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics 2019, 35, 462–469. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Benedix, A.; Becker, C.M.; de Groot, B.L.; Caflisch, A.; Bockmann, R.A. Predicting free energy changes using structural ensembles. Nat. Methods 2009, 6, 3–4. [Google Scholar] [CrossRef]
- Dehouck, Y.; Kwasigroch, J.M.; Rooman, M.; Gilis, D. BeAtMuSiC: Prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 2013, 41, W333–W339. [Google Scholar] [CrossRef]
- Xiong, P.; Zhang, C.; Zheng, W.; Zhang, Y. BindProfX: Assessing Mutation-Induced Binding Affinity Change by Protein Interface Profiles with Pseudo-Counts. J. Mol. Biol. 2017, 429, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef]
- Rodrigues, C.H.M.; Myung, Y.; Pires, D.E.V.; Ascher, D.B. mCSM-PPI2: Predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019, 47, W338–W344. [Google Scholar] [CrossRef] [PubMed]
- Timasheff, S.N. Protein-solvent preferential interactions, protein hydration, and the modulation of biochemical reactions by solvent components. Proc. Natl. Acad. Sci. USA 2002, 99, 9721–9726. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.M.; Ferreon, J.C.; Bolen, D.W.; Rösgen, J. Protein Phase Diagrams II: Nonideal Behavior of Biochemical Reactions in the Presence of Osmolytes. Biophys. J. 2007, 92, 245–256. [Google Scholar] [CrossRef][Green Version]
- Duff, M.R.; Howell, E.E. Thermodynamics and solvent linkage of macromolecule–ligand interactions. Methods 2015, 76, 51–60. [Google Scholar] [CrossRef]
- Völker, J.; Breslauer, K.J. Communication between noncontacting macromolecules. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 21–42. [Google Scholar] [CrossRef][Green Version]
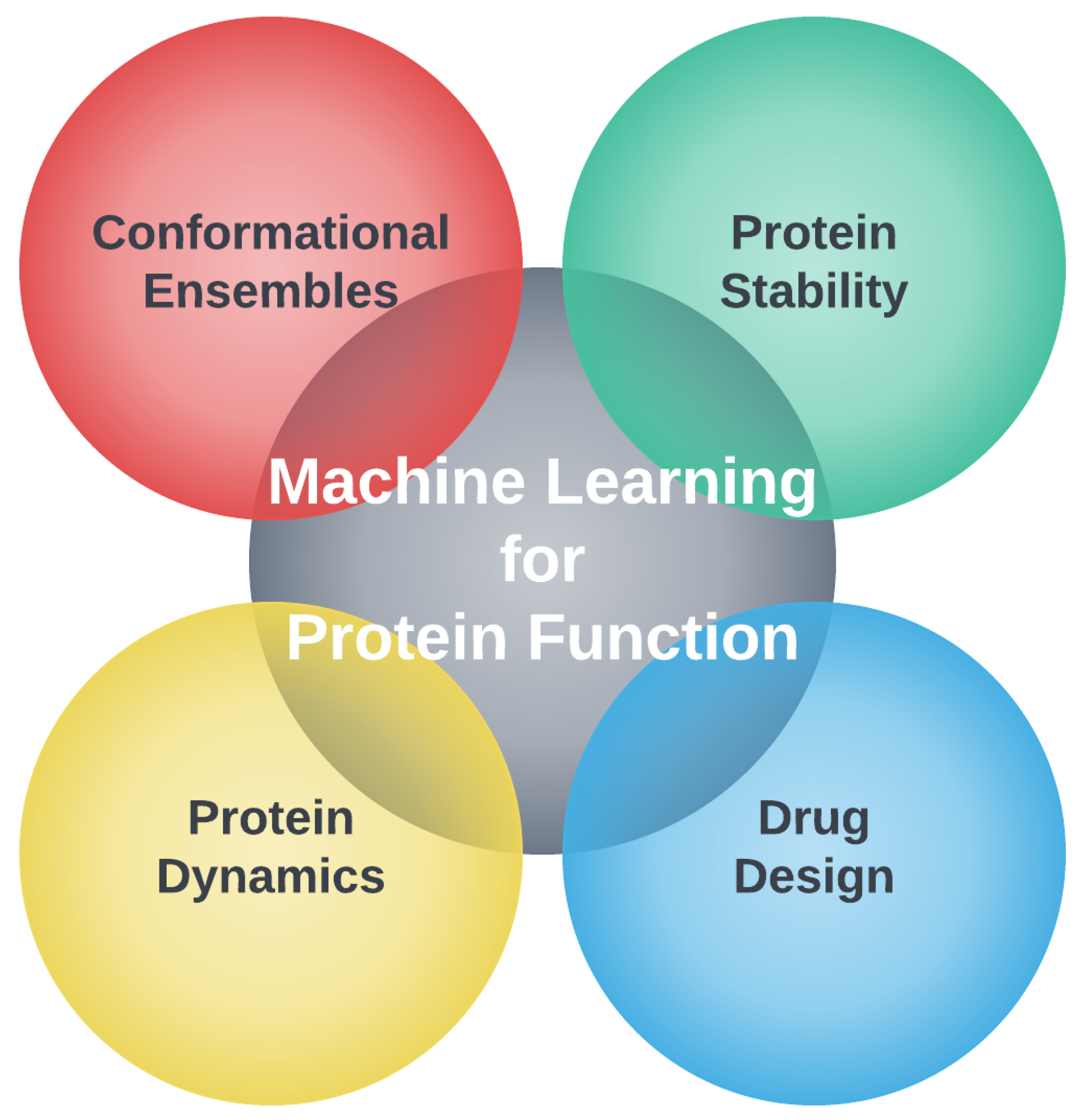
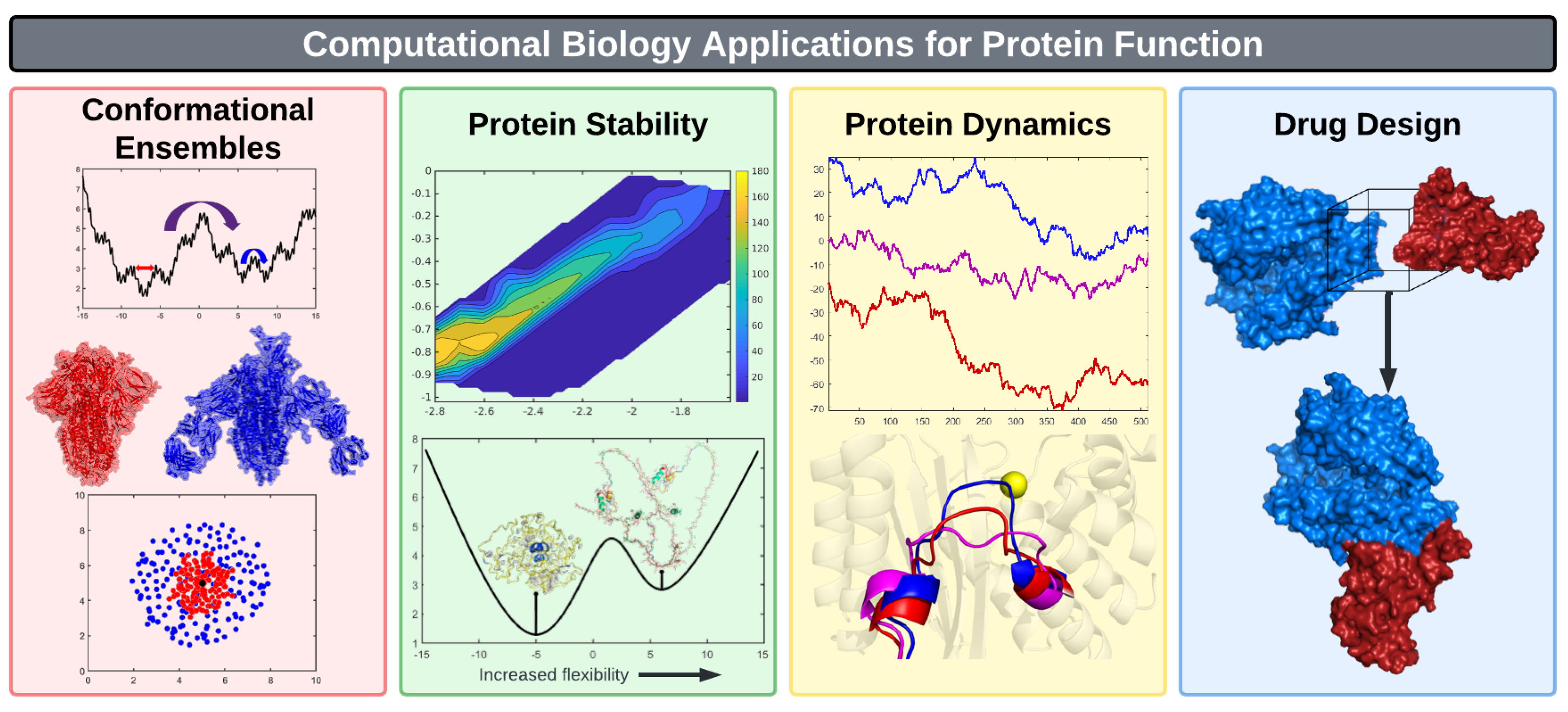




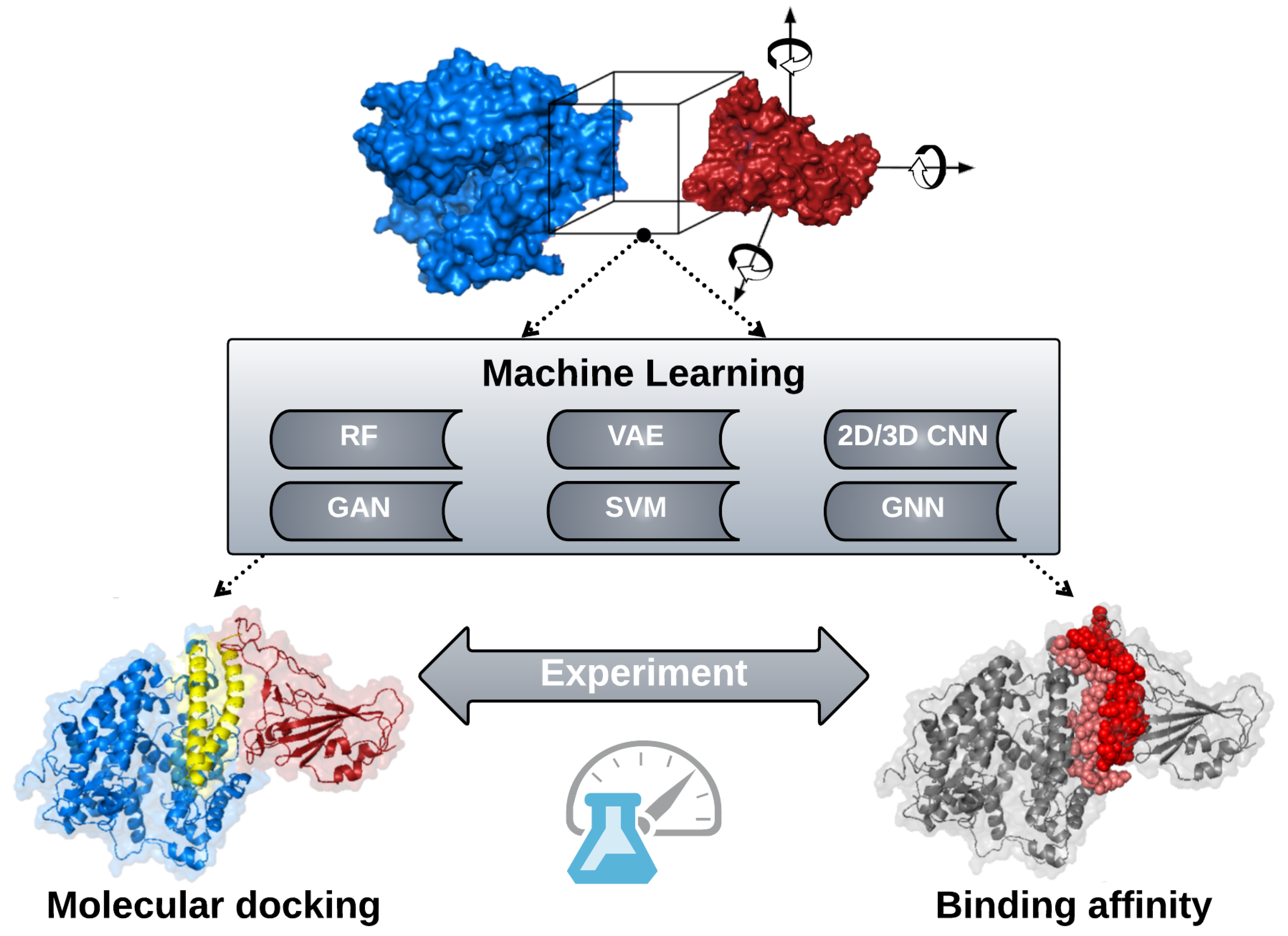
| Method | Task | Paradigm | Abbreviation |
|---|---|---|---|
| k-Means | clustering | Unsupervised | - |
| Agglomerative Clustering | clustering | Unsupervised | - |
| Spectral Clustering | clustering | Unsupervised | - |
| Self-Organizing Maps | clustering | Unsupervised | SOM |
| Principal Component Analysis | DR | Unsupervised | PCA |
| Time-Lagged Independent Component Analysis | DR | Unsupervised | tICA |
| t-Distributed Stochastic Neighbor Embedding | DR | Unsupervised | t-SNE |
| Supervised Projection Learning for Orthogonal Completeness | clustering/classification/DR | Unsupervised | SPLOC |
| Naive Bayes | classification/regression | Supervised | NB |
| Support Vector Machines | classification | Supervised | SVM |
| Decision Trees | classification/regression | Supervised | DT |
| Random Forest | classification/regression | Supervised | RF |
| Gaussian Mixture Model | classification/regression | Supervised | GMM |
| Artificial Neural Network | classification/regression/NLP | Supervised | ANN |
| Shallow Neural Network | classification/regression/NLP | Supervised | SNN |
| Deep Neural Network | classification/regression/NLP | Supervised | DNN |
| Convolutional Neural Network | classification/regression/NLP | Supervised | CNN |
| 3D Convolutional Neural Network | classification/regression/NLP | Supervised | 3D-CNN |
| Recurrent Neural Network | classification/regression/NLP | Supervised | RNN |
| Autoencoder | DR/generative modeling | Supervised | - |
| Variational Autoencoder | DR/generative modeling | Supervised | VAE |
| Generative Adversarial Network | classification/generative modeling | Supervised | GAN |
| Message Passing Neural Network | classification/regression/DR | Supervised | MPNN |
| Graph Neural Network | classification/regression/DR | Supervised | GNN |
| Graph Convolutional Neural Network | classification/regression/DR | Supervised | GCNN |
| CASP | Method/Group | Year | Model | Stand Alone | Webserver |
|---|---|---|---|---|---|
| 12 | Baker [75] | 2016 | Rosetta | Yes | Yes |
| LEEab [76] | 2016 | GOAL | No | No | |
| Zhang [77] | 2016 | I-TASSER | Yes | Yes | |
| LEE [76] | 2016 | GOAL | No | No | |
| VoroMQA-select [78] | 2016 | VoroMQA | Yes | Yes | |
| 13 | A7D (AlphaFold) [79,80] | 2018 | FreeM + DNN | Yes | No |
| Zhang [81] | 2018 | TripletRes | Yes | Yes | |
| MULTICOM [82] | 2018 | CDNN + ab initio | Yes | Yes | |
| QUARK [83] | 2018 | C-QUARK | Yes | Yes | |
| Zhang-Server [83] | 2018 | C-I-TASSER | Yes | Yes | |
| 14 | AlphaFold2 [84] | 2020 | TR/ATT + ANN | Yes | Yes |
| Baker [85] | 2020 | ResNet + trRosetta | Yes | Yes | |
| Baker-exp. [86] | 2020 | ResNet + trRosetta | Yes | Yes | |
| FEIG-R2 [87] | 2020 | PREFMD2 | Yes | Yes | |
| Zhang [88] | 2020 | UnknownI | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avery, C.; Patterson, J.; Grear, T.; Frater, T.; Jacobs, D.J. Protein Function Analysis through Machine Learning. Biomolecules 2022, 12, 1246. https://doi.org/10.3390/biom12091246
Avery C, Patterson J, Grear T, Frater T, Jacobs DJ. Protein Function Analysis through Machine Learning. Biomolecules. 2022; 12(9):1246. https://doi.org/10.3390/biom12091246
Chicago/Turabian StyleAvery, Chris, John Patterson, Tyler Grear, Theodore Frater, and Donald J. Jacobs. 2022. "Protein Function Analysis through Machine Learning" Biomolecules 12, no. 9: 1246. https://doi.org/10.3390/biom12091246
APA StyleAvery, C., Patterson, J., Grear, T., Frater, T., & Jacobs, D. J. (2022). Protein Function Analysis through Machine Learning. Biomolecules, 12(9), 1246. https://doi.org/10.3390/biom12091246









