TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles
Abstract
:1. Introduction
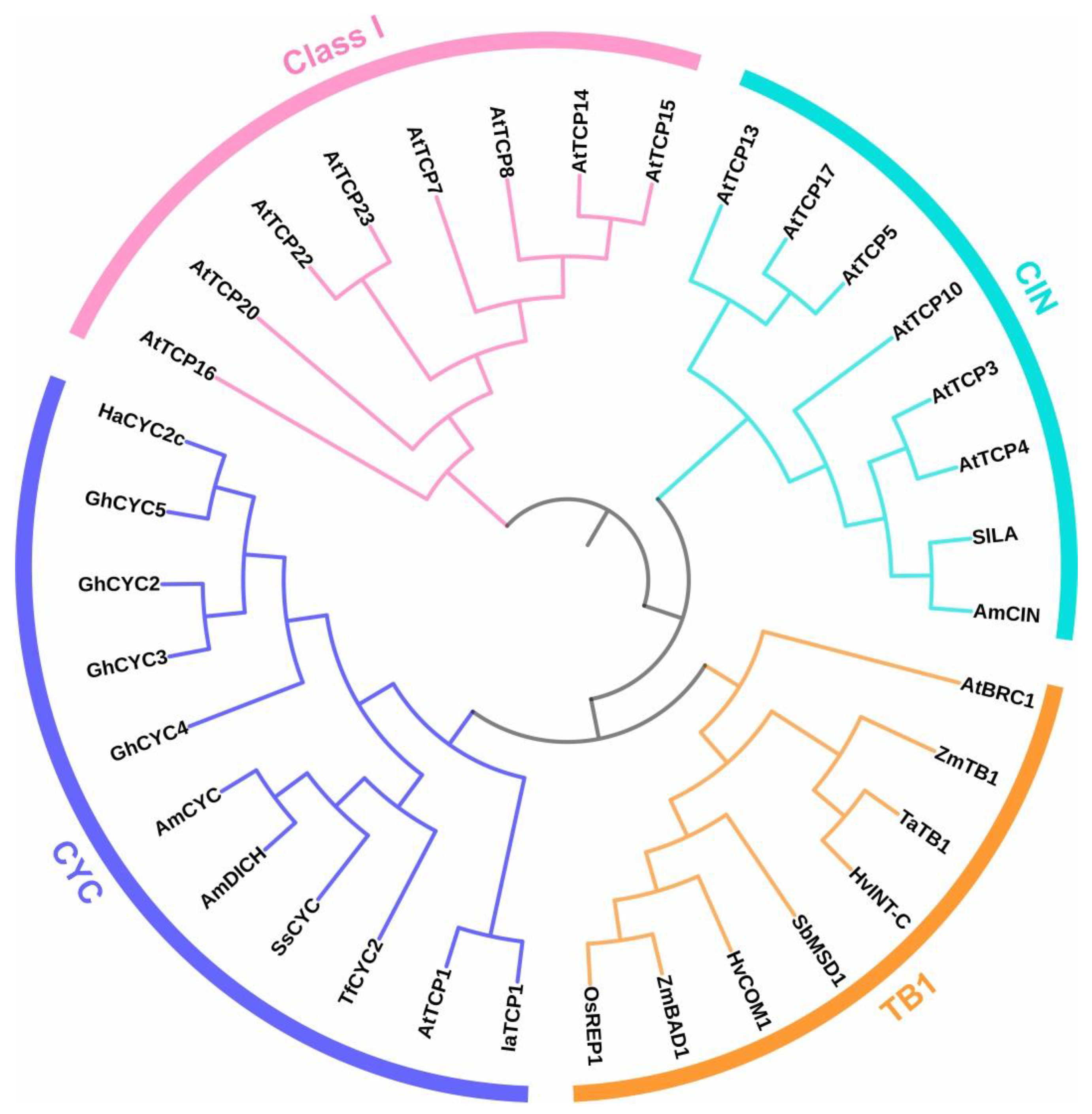
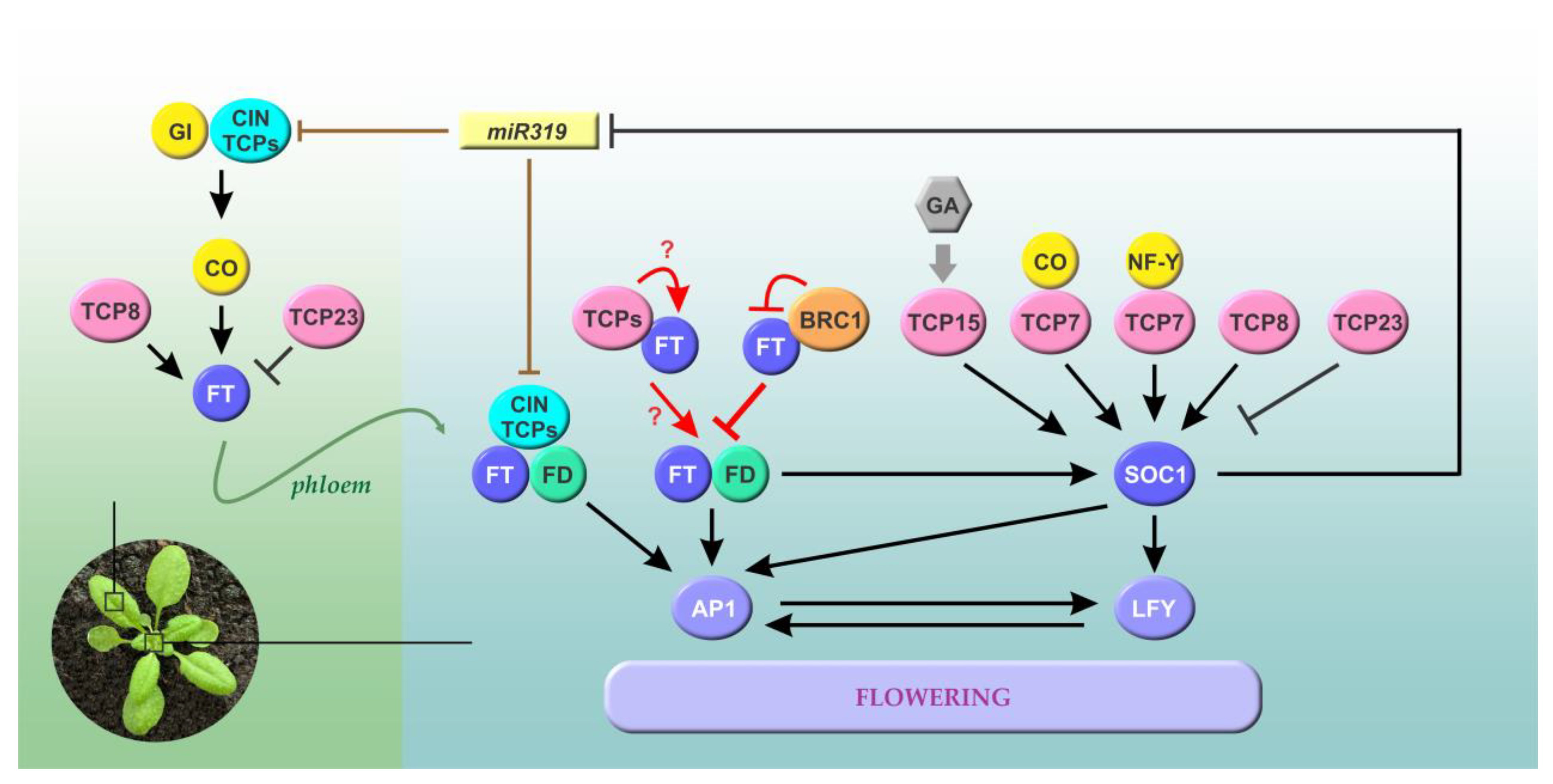
2. Class I and Class II TCPs Regulate Flowering Time Acting at Different Levels
3. CYCLOIDEA-like TCPs and the Determination of Flower Symmetry
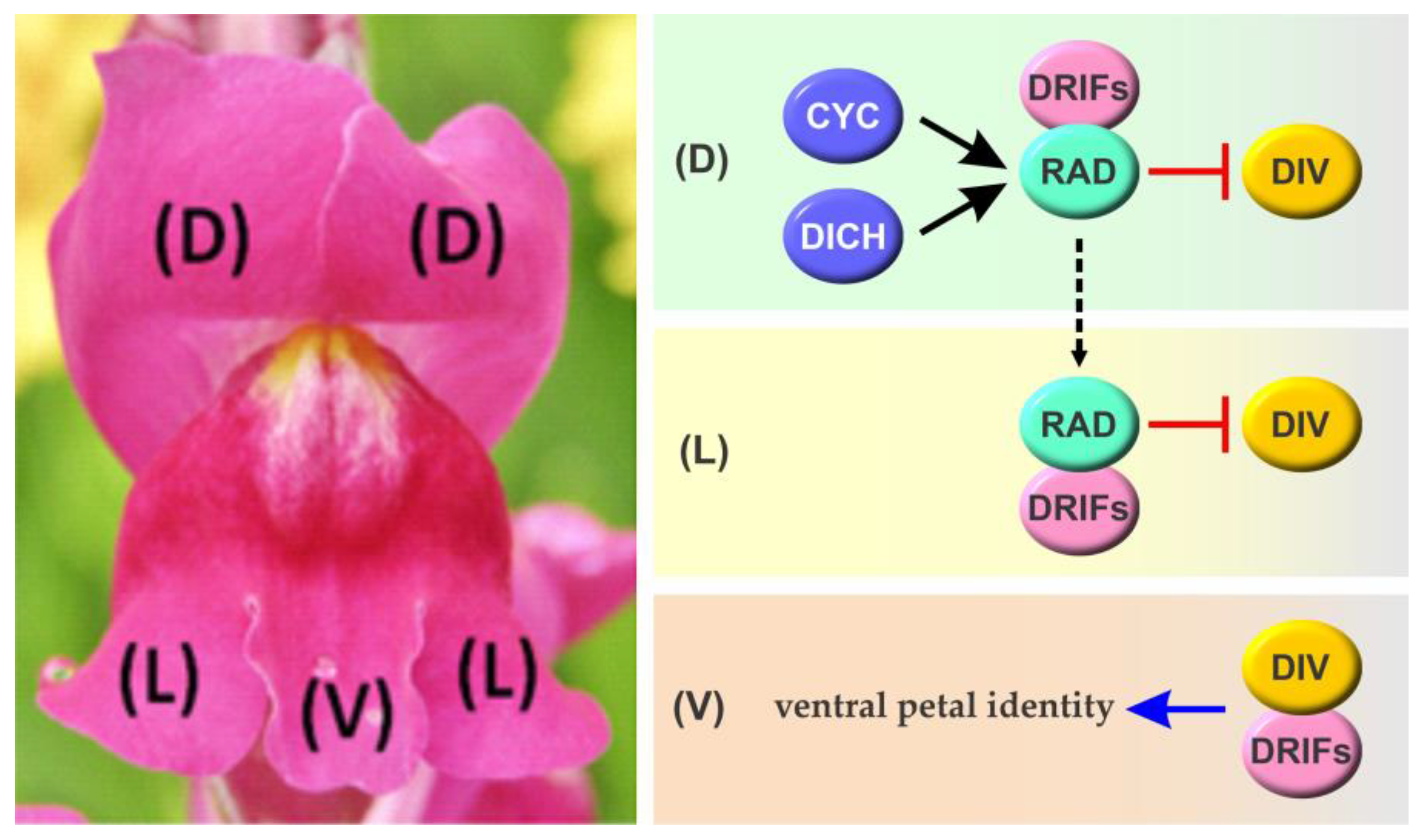
3.1. Molecular Mechanisms Involved in the Determination of Flower Symmetry in Antirrhinum majus
3.2. CYC-like TCPs in Other Eudicots
3.3. CYC-like TCPs and the Specification of Flower Identity in Asteraceae
3.4. CYC-like TCPs and the Specification of Other Flower Characteristics Associated with Symmetry
3.5. CYC/TB1 TCPs and the Specification of Flower Characteristics in Basal Eudicots and Monocots
4. CIN TCPs and the Regulation of Petal Differentiation and Growth
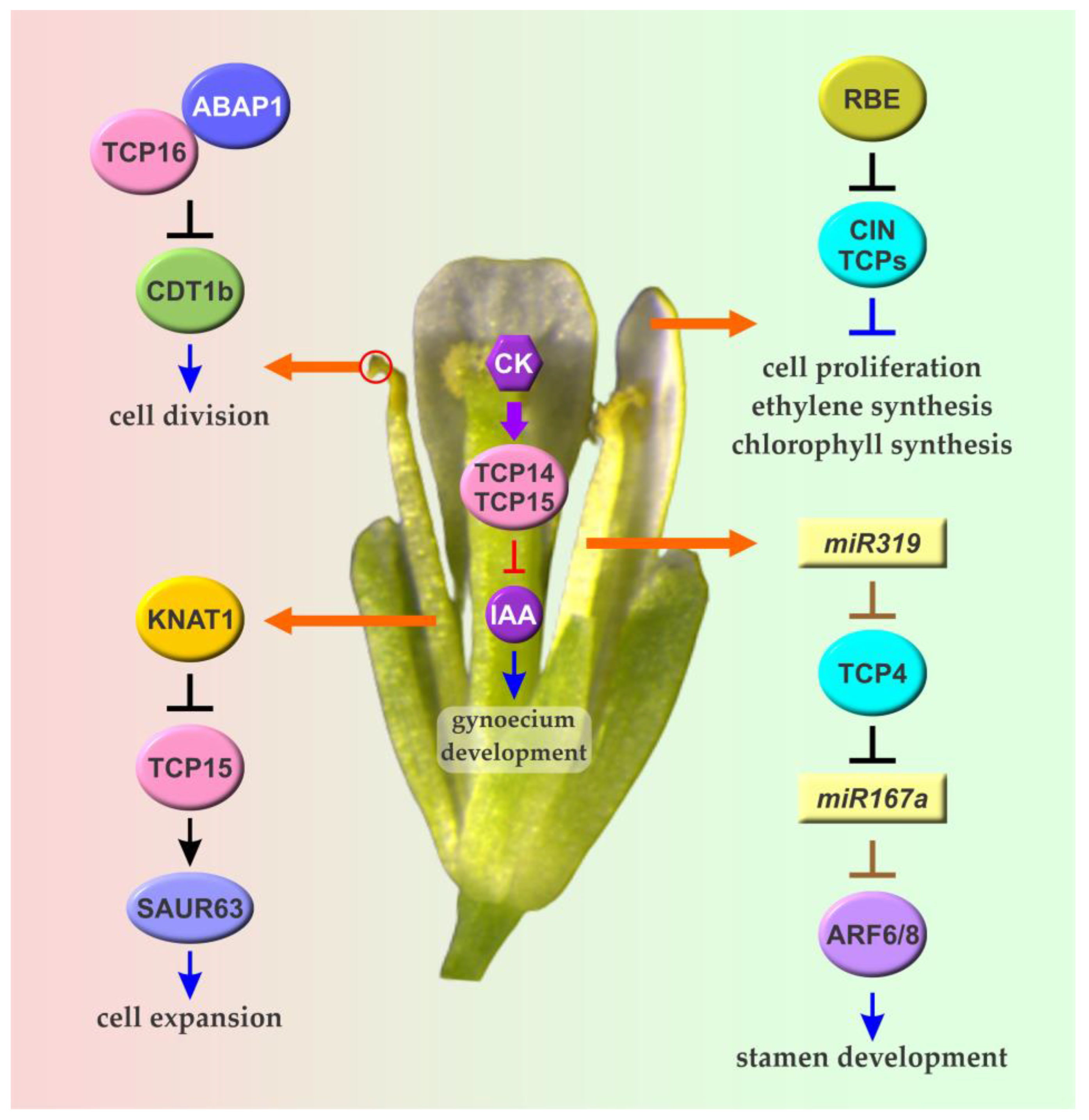
5. TCPs and the Modulation of Reproductive Organ Development and Growth
5.1. TCPs and the Regulation of Stamen Growth and Development
5.2. TCPs and the Regulation of Gynoecium Development
6. TB1-like TCPs and the Regulation of Inflorescence Architecture in Grasses
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef]
- Zhou, H.; Hwarari, D.; Ma, H.; Xu, H.; Yang, L.; Luo, Y. Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front. Genet. 2022, 13, 1060546. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2022, 13, 994567. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef]
- Aggarwal, P.; Das Gupta, M.; Joseph, A.P.; Chatterjee, N.; Srinivasan, N.; Nath, U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 2010, 22, 1174–1189. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Viola, I.L.; Uberti Manassero, N.G.; Ripoll, R.; Gonzalez, D.H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011, 435, 143–155. [Google Scholar] [CrossRef]
- Viola, I.L.; Reinheimer, R.; Ripoll, R.; Manassero, N.G.U.; Gonzalez, D.H. Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 2012, 287, 347–356. [Google Scholar] [CrossRef]
- Sun, L.; Zou, X.; Jiang, M.; Wu, X.; Chen, Y.; Wang, Q.; Wang, Q.; Chen, L.; Wu, Y. The crystal structure of the TCP domain of PCF6 in Oryza sativa L. reveals an RHH-like fold. FEBS Lett. 2020, 594, 1296–1306. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.P.; Nie, J.K.; Chen, H.; Qin, G.; Wang, B.; Su, X.D. DNA-TCP complex structures reveal a unique recognition mechanism for TCP transcription factor families. Nucleic Acids Res. 2023, 51, 434448. [Google Scholar] [CrossRef]
- Uberti-Manassero, N.G.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts 2013, 4, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Nath, U. CIN-TCP transcription factors: Transiting cell proliferation in plants. IUBMB Life 2018, 70, 718–731. [Google Scholar] [CrossRef]
- Viola, I.L.; Alem, A.L.; Jure, R.M.; Gonzalez, D.H. Physiological Roles and Mechanisms of Action of Class I TCP Transcription Factors. Int. J. Mol. Sci. 2023, 24, 5437. [Google Scholar] [CrossRef]
- Blázquez, M.A.; Weigel, D. Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Balsemão-Pires, E.; Andrade, L.R.; Sachetto-Martins, G. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 2013, 67, 120–125. [Google Scholar] [CrossRef]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef]
- Kubota, A.; Ito, S.; Shim, J.S.; Johnson, R.S.; Song, Y.H.; Breton, G.; Goralogia, G.S.; Kwon, M.S.; Laboy Cintrón, D.; Koyama, T.; et al. TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 2017, 13, e1006856. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, X.; Liu, P.; Li, D.; Chen, T.; Gu, X.; Sun, J. MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PLoS Genet. 2017, 13, e1006833. [Google Scholar] [CrossRef]
- Lucero, L.E.; Manavella, P.A.; Gras, D.E.; Ariel, F.D.; Gonzalez, D.H. Class I and Class II TCP Transcription Factors Modulate SOC1-Dependent Flowering at Multiple Levels. Mol. Plant 2017, 10, 1571–1574. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis Class II TCP Transcription Factors Integrate with the FT-FD Module to Control Flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef]
- Li, X.; Zhang, G.; Liang, Y.; Hu, L.; Zhu, B.; Qi, D.; Cui, S.; Zhao, H. TCP7 interacts with Nuclear Factor-Ys to promote flowering by directly regulating SOC1 in Arabidopsis. Plant J. 2021, 108, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Camoirano, A.; Alem, A.L.; Gonzalez, D.H.; Viola, I.L. The N-terminal region located upstream of the TCP domain is responsible for the antagonistic action of the Arabidopsis thaliana TCP8 and TCP23 transcription factors on flowering time. Plant Sci. 2023, 328, 111571. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.W.; Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 2014, 26, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Deng, Q.; Lu, H.; Wei, D.; Wang, Z.; Tang, Q. Brassica juncea BRC1-1 induced by SD negatively regulates flowering by directly interacting with BjuFT and BjuFUL promoter. Front. Plant Sci. 2022, 13, 986811. [Google Scholar] [CrossRef]
- Danisman, S.; van Dijk, A.D.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef]
- Burko, Y.; Shleizer-Burko, S.; Yanai, O.; Shwartz, I.; Zelnik, I.D.; Jacob-Hirsch, J.; Kela, I.; Eshed-Williams, L.; Ori, N. A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell 2013, 25, 2070–2083. [Google Scholar] [CrossRef]
- Silva, G.F.F.; Silva, E.M.; Correa, J.P.O.; Vicente, M.H.; Jiang, N.; Notini, M.M.; Junior, A.C.; De Jesus, F.A.; Castilho, P.; Carrera, E.; et al. Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 2019, 221, 1328–1344. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of organ asymmetry in flowers of Antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef]
- Corley, S.B.; Carpenter, R.; Copsey, L.; Coen, E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl. Acad. Sci. USA 2005, 102, 5068–5073. [Google Scholar] [CrossRef]
- Costa, M.M.; Fox, S.; Hanna, A.I.; Baxter, C.; Coen, E. Evolution of regulatory interactions controlling floral asymmetry. Development 2005, 132, 5093–5101. [Google Scholar] [CrossRef]
- Galego, L.; Almeida, J. Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002, 16, 880–891. [Google Scholar] [CrossRef]
- Raimundo, J.; Sobral, R.; Bailey, P.; Azevedo, H.; Galego, L.; Almeida, J.; Coen, E.; Costa, M.M. A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J. 2013, 75, 527–538. [Google Scholar] [CrossRef]
- Machemer, K.; Shaiman, O.; Salts, Y.; Shabtai, S.; Sobolev, I.; Belausov, E.; Grotewold, E.; Barg, R. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant J. 2011, 68, 337–350. [Google Scholar] [CrossRef]
- Sengupta, A.; Hileman, L.C. A CYC-RAD-DIV-DRIF interaction likely pre-dates the origin of floral monosymmetry in Lamiales. Evodevo 2022, 13, 3. [Google Scholar] [CrossRef]
- Busch, A.; Zachgo, S. Flower symmetry evolution: Towards understanding the abominable mystery of angiosperm radiation. Bioessays 2009, 31, 1181–1190. [Google Scholar] [CrossRef]
- Li, M.; Zhang, D.; Gao, Q.; Luo, Y.; Zhang, H.; Ma, B.; Chen, C.; Whibley, A.; Zhang, Y.; Cao, Y.; et al. Genome structure and evolution of Antirrhinum majus L. Nat. Plants 2019, 5, 174–183. [Google Scholar] [CrossRef]
- Guo, Z.; Fujioka, S.; Blancaflor, E.B.; Miao, S.; Gou, X.; Li, J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 2010, 22, 1161–1173. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G.; et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef]
- Cubas, P.; Coen, E.; Zapater, J.M. Ancient asymmetries in the evolution of flowers. Curr. Biol. 2001, 11, 1050–1052. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Zachgo, S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. USA 2007, 104, 16714–16719. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Developmental genetics of floral symmetry evolution. Trends Plant Sci. 2009, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Reyes, E.; Sauquet, H.; Nadot, S. Perianth symmetry changed at least 199 times in angiosperm evolution. Taxon 2016, 65, 945–964. [Google Scholar] [CrossRef]
- Spencer, V.; Kim, M. Re“CYC”ling molecular regulators in the evolution and development of flower symmetry. Semin. Cell Dev. Biol. 2018, 79, 16–26. [Google Scholar] [CrossRef]
- Citerne, H.L.; Pennington, R.T.; Cronk, Q.C. An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc. Natl. Acad. Sci. USA 2006, 103, 12017–12020. [Google Scholar] [CrossRef]
- Preston, J.C.; Martinez, C.C.; Hileman, L.C. Gradual disintegration of the floral symmetry gene network is implicated in the evolution of a wind-pollination syndrome. Proc. Natl. Acad. Sci. USA 2011, 108, 2343–2348. [Google Scholar] [CrossRef]
- Zhang, W.; Steinmann, V.W.; Nikolov, L.; Kramer, E.M.; Davis, C.C. Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front. Plant Sci. 2013, 4, 302. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, J.; Li, P.W.; Li, C.Q.; Lü, T.F.; Yang, X.; Wang, Y.Z. Evolution of Darwin’s Peloric Gloxinia (Sinningia speciosa) Is Caused by a Null Mutation in a Pleiotropic TCP Gene. Mol. Biol. Evol. 2018, 35, 1901–1915. [Google Scholar] [CrossRef]
- Gómez, J.M.; Perfectti, F.; Camacho, J.P. Natural selection on Erysimum mediohispanicum flower shape: Insights into the evolution of zygomorphy. Am. Nat. 2006, 168, 531–545. [Google Scholar] [CrossRef]
- Zhang, W.; Kramer, E.M.; Davis, C.C. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. USA 2010, 107, 6388–6393. [Google Scholar] [CrossRef]
- van der Niet, T.; Johnson, S.D. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends Ecol. Evol. 2012, 27, 353–361. [Google Scholar] [CrossRef]
- Gaudin, V.; Lunness, P.A.; Fobert, P.R.; Towers, M.; Riou-Khamlichi, C.; Murray, J.A.; Coen, E.; Doonan, J.H. The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 2000, 122, 1137–1148. [Google Scholar] [CrossRef]
- Elomaa, P.; Zhao, Y.; Zhang, T. Flower heads in Asteraceae-recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Hortic. Res. 2018, 5, 36. [Google Scholar] [CrossRef]
- Broholm, S.K.; Tähtiharju, S.; Laitinen, R.A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef]
- Chapman, M.A.; Leebens-Mack, J.H.; Burke, J.M. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 2008, 25, 1260–1273. [Google Scholar] [CrossRef]
- Kim, M.; Cui, M.L.; Cubas, P.; Gillies, A.; Lee, K.; Chapman, M.A.; Abbott, R.J.; Coen, E. Regulatory genes control a key morphological and ecological trait transferred between species. Science 2008, 322, 1116–1119. [Google Scholar] [CrossRef]
- Fambrini, M.; Salvini, M.; Pugliesi, C. A transposon-mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus annuus. Genetica 2011, 139, 1521–1529. [Google Scholar] [CrossRef]
- Chapman, M.A.; Tang, S.; Draeger, D.; Nambeesan, S.; Shaffer, H.; Barb, J.G.; Knapp, S.J.; Burke, J.M. Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet. 2012, 8, e1002628. [Google Scholar] [CrossRef]
- Shen, C.Z.; Chen, J.; Zhang, C.J.; Rao, G.Y.; Guo, Y.P. Dysfunction of CYC2g is responsible for the evolutionary shift from radiate to disciform flowerheads in the Chrysanthemum group (Asteraceae: Anthemideae). Plant J. 2021, 106, 1024–1038. [Google Scholar] [CrossRef]
- Juntheikki-Palovaara, I.; Tähtiharju, S.; Lan, T.; Broholm, S.K.; Rijpkema, A.S.; Ruonala, R.; Kale, L.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 2014, 79, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Broholm, S.K.; Wang, F.; Rijpkema, A.S.; Lan, T.; Albert, V.A.; Teeri, T.H.; Elomaa, P. TCP and MADS-Box Transcription Factor Networks Regulate Heteromorphic Flower Type Identity in Gerbera hybrida. Plant Physiol. 2020, 184, 1455–1468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.J.; Rong, Y.L.; Jiang, C.K.; Guo, Y.P.; Rao, G.Y. Co-option of a carotenoid cleavage dioxygenase gene (CCD4a) into the floral symmetry gene regulatory network contributes to the polymorphic floral shape-color combinations in Chrysanthemum sensu lato. New Phytol. 2022, 236, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Xiao, W.; Guo, W.; Yao, X.; Xiao, J.; Ye, Z.; Wang, N.; Jiao, K.; Lei, M.; Peng, Q.; et al. The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 2017, 215, 1582–1593. [Google Scholar] [CrossRef]
- Zhao, Y.; Pfannebecker, K.; Dommes, A.B.; Hidalgo, O.; Becker, A.; Elomaa, P. Evolutionary diversification of CYC/TB1-like TCP homologs and their recruitment for the control of branching and floral morphology in Papaveraceae (basal eudicots). New Phytol. 2018, 220, 317–331. [Google Scholar] [CrossRef]
- Yuan, Z.; Gao, S.; Xue, D.W.; Luo, D.; Li, L.T.; Ding, S.Y.; Yao, X.; Wilson, Z.A.; Qian, Q.; Zhang, D.B. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 2009, 149, 235–244. [Google Scholar] [CrossRef]
- Rudall, P.J.; Bateman, R.M. Evolution of zygomorphy in monocot flowers: Iterative patterns and developmental constraints. New Phytol. 2004, 162, 25–44. [Google Scholar] [CrossRef]
- Crawford, B.C.; Nath, U.; Carpenter, R.; Coen, E.S. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 2004, 135, 244–253. [Google Scholar] [CrossRef]
- Chen, H.W.; Lee, P.L.; Wang, C.N.; Hsu, H.J.; Chen, J.C. Silencing of PhLA, a CIN-TCP gene, causes defected petal conical epidermal cell formation and results in reflexed corolla lobes in petunia. Bot. Stud. 2020, 61, 24. [Google Scholar] [CrossRef]
- Koyama, T.; Ohme-Takagi, M.; Sato, F. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 697–699. [Google Scholar] [CrossRef]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Narumi, T.; Aida, R.; Koyama, T.; Yamaguchi, H.; Sasaki, K.; Shikata, M.; Nakayama, M.; Ohme-Takagi, M.; Ohtsubo, N. Arabidopsis chimeric TCP3 repressor produces novel floral traits in Torenia fournieri and Chrysanthemum morifolium. Plant Biotechnol. 2011, 28, 131–140. [Google Scholar] [CrossRef]
- Tanaka, Y.L.; Yamamura, T.; Oshima, Y.; Mitsuda, N.; Koyama, T.; Ohme-Takagi, M.; Terakawa, T. Creating ruffled flower petals in Cyclamen persicum by expression of the chimeric cyclamen TCP repressor. Plant Biotechnol. 2011, 28, 141–147. [Google Scholar] [CrossRef]
- Huang, T.; Irish, V.F. Temporal Control of Plant Organ Growth by TCP Transcription Factors. Curr. Biol. 2015, 25, 1765–1770. [Google Scholar] [CrossRef]
- van Es, S.W.; Silveira, S.R.; Rocha, D.I.; Bimbo, A.; Martinelli, A.P.; Dornelas, M.C.; Angenent, G.C.; Immink, R.G.H. Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 2018, 94, 867–879. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zhang, Y.; Wang, W.; Irish, V.F.; Huang, T. RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 2016, 67, 6473–6480. [Google Scholar] [CrossRef]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Whitney, H.M.; Bennett, K.M.; Dorling, M.; Sandbach, L.; Prince, D.; Chittka, L.; Glover, B.J. Why do so many petals have conical epidermal cells? Ann. Bot. 2011, 108, 609–616. [Google Scholar] [CrossRef]
- Papiorek, S.; Junker, R.R.; Lunau, K. Gloss, colour and grip: Multifunctional epidermal cell shapes in bee- and bird-pollinated flowers. PLoS ONE 2014, 9, e112013. [Google Scholar] [CrossRef]
- Whitney, H.M.; Chittka, L.; Bruce, T.J.; Glover, B.J. Conical epidermal cells allow bees to grip flowers and increase foraging efficiency. Curr. Biol. 2009, 19, 948–953. [Google Scholar] [CrossRef]
- Zheng, X.; Lan, J.; Yu, H.; Zhang, J.; Zhang, Y.; Qin, Y.; Su, X.D.; Qin, G. Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening. Plant Commun. 2022, 3, 100309. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef] [PubMed]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP Transcription Factors Activate the SAUR63 Gene Subfamily in Gibberellin-Dependent Stamen Filament Elongation. Plant Physiol. 2020, 182, 2096–2110. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef]
- Gastaldi, V.; Alem, A.L.; Mansilla, N.; Ariel, F.D.; Viola, I.L.; Lucero, L.E.; Gonzalez, D.H. BREVIPEDICELLUS/KNAT1 targets TCP15 to modulate filament elongation during Arabidopsis late stamen development. Plant Physiol. 2023, 191, 29–34. [Google Scholar] [CrossRef]
- Takeda, T.; Amano, K.; Ohto, M.A.; Nakamura, K.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 2006, 61, 165–177. [Google Scholar] [CrossRef]
- Cabral, L.M.; Masuda, H.P.; Ballesteros, H.F.; de Almeida-Engler, J.; Alves-Ferreira, M.; De Toni, K.L.G.; Bizotto, F.M.; Ferreira, P.C.G.; Hemerly, A.S. ABAP1 Plays a Role in the Differentiation of Male and Female Gametes in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 642758. [Google Scholar] [CrossRef]
- Andrés-Colás, N.; Carrió-Seguí, A.; Abdel-Ghany, S.E.; Pilon, M.; Peñarrubia, L. Expression of the Intracellular COPT3-Mediated Cu Transport Is Temporally Regulated by the TCP16 Transcription Factor. Front. Plant Sci. 2018, 9, 910. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Weigel, D. Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 2013, 9, e1003374. [Google Scholar] [CrossRef]
- Wang, H.; Mao, Y.; Yang, J.; He, Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 2015, 6, 436. [Google Scholar] [CrossRef]
- Lucero, L.E.; Uberti-Manassero, N.G.; Arce, A.L.; Colombatti, F.; Alemano, S.G.; Gonzalez, D.H. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 2015, 84, 267–282. [Google Scholar] [CrossRef]
- Alonso-Cantabrana, H.; Ripoll, J.J.; Ochando, I.; Vera, A.; Ferrándiz, C.; Martínez-Laborda, A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 2007, 134, 2663–2671. [Google Scholar] [CrossRef]
- Herrera-Ubaldo, H.; Campos, S.E.; López-Gómez, P.; Luna-García, V.; Zúñiga-Mayo, V.M.; Armas-Caballero, G.E.; González-Aguilera, K.L.; DeLuna, A.; Marsch-Martínez, N.; Espinosa-Soto, C.; et al. The protein-protein interaction landscape of transcription factors during gynoecium development in Arabidopsis. Mol. Plant 2023, 16, 260–278. [Google Scholar] [CrossRef]
- Cao, B.; Wang, H.; Bai, J.; Wang, X.; Li, X.; Zhang, Y.; Yang, S.; He, Y.; Yu, X. miR319-Regulated TCP3 Modulates Silique Development Associated with Seed Shattering in Brassicaceae. Cells 2022, 11, 3096. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, J.; Pang, C.; Yu, H.; Guo, D.; Jiang, H.; Ding, M.; Chen, Z.; Tao, Q.; Gu, H.; et al. The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 2015, 25, 121–134. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar] [CrossRef]
- Braun, N.; de Saint Germain, A.; Pillot, J.P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, X.; Xi, L.; Li, J.; Zhao, R.; Ma, N.; Zhao, L. Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema ×grandiflora cv. Jinba). PLoS ONE 2013, 8, e61717. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Palomino, M.; Trontin, C. High time for a roll call: Gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 2011, 107, 1533–1544. [Google Scholar] [CrossRef] [PubMed]
- Whipple, C.J. Grass inflorescence architecture and evolution: The origin of novel signaling centers. New Phytol. 2017, 216, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Reinheimer, R.; Durantini, D.; Kellogg, E.A.; Schmidt, R.J. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 12225–12230. [Google Scholar] [CrossRef]
- Lewis, M.W.; Bolduc, N.; Hake, K.; Htike, Y.; Hay, A.; Candela, H.; Hake, S. Gene regulatory interactions at lateral organ boundaries in maize. Development 2014, 141, 4590–4597. [Google Scholar] [CrossRef]
- Poursarebani, N.; Trautewig, C.; Melzer, M.; Nussbaumer, T.; Lundqvist, U.; Rutten, T.; Schmutzer, T.; Brandt, R.; Himmelbach, A.; Altschmied, L.; et al. COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 2020, 11, 5138. [Google Scholar] [CrossRef]
- Shang, Y.; Yuan, L.; Di, Z.; Jia, Y.; Zhang, Z.; Li, S.; Xing, L.; Qi, Z.; Wang, X.; Zhu, J.; et al. A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 2020, 71, 7118–7131. [Google Scholar] [CrossRef]
- Ramsay, L.; Comadran, J.; Druka, A.; Marshall, D.F.; Thomas, W.T.; Macaulay, M.; MacKenzie, K.; Simpson, C.; Fuller, J.; Bonar, N.; et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 2011, 43, 169–172. [Google Scholar] [CrossRef]
- Koppolu, R.; Schnurbusch, T. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J. Integr. Plant Biol. 2019, 61, 278–295. [Google Scholar] [CrossRef]
- Gao, G.; Kan, J.; Jiang, C.; Ahmar, S.; Zhang, J.; Yang, P. Genome-wide diversity analysis of TCP transcription factors revealed cases of selection from wild to cultivated barley. Funct. Integr. Genom. 2021, 21, 31–42. [Google Scholar] [CrossRef]
- Jiao, Y.; Lee, Y.K.; Gladman, N.; Chopra, R.; Christensen, S.A.; Regulski, M.; Burow, G.; Hayes, C.; Burke, J.; Ware, D.; et al. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun. 2018, 9, 822. [Google Scholar] [CrossRef]
- Li, M.; Shao, M.R.; Zeng, D.; Ju, T.; Kellogg, E.A.; Topp, C.N. Comprehensive 3D phenotyping reveals continuous morphological variation across genetically diverse sorghum inflorescences. New Phytol. 2020, 226, 1873–1885. [Google Scholar] [CrossRef]
- Studer, A.J.; Wang, H.; Doebley, J.F. Selection During Maize Domestication Targeted a Gene Network Controlling Plant and Inflorescence Architecture. Genetics 2017, 207, 755–765. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Gustus, C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef]
- Dixon, L.E.; Greenwood, J.R.; Bencivenga, S.; Zhang, P.; Cockram, J.; Mellers, G.; Ramm, K.; Cavanagh, C.; Swain, S.M.; Boden, S.A. TEOSINTE BRANCHED1 Regulates Inflorescence Architecture and Development in Bread Wheat (Triticum aestivum). Plant Cell 2018, 30, 563–581. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viola, I.L.; Gonzalez, D.H. TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles. Biomolecules 2023, 13, 750. https://doi.org/10.3390/biom13050750
Viola IL, Gonzalez DH. TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles. Biomolecules. 2023; 13(5):750. https://doi.org/10.3390/biom13050750
Chicago/Turabian StyleViola, Ivana L., and Daniel H. Gonzalez. 2023. "TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles" Biomolecules 13, no. 5: 750. https://doi.org/10.3390/biom13050750
APA StyleViola, I. L., & Gonzalez, D. H. (2023). TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles. Biomolecules, 13(5), 750. https://doi.org/10.3390/biom13050750





