Abstract
Tuberculosis (TB) is the leading global cause of death f rom an infectious bacterial agent. Therefore, limiting its epidemic spread is a pressing global health priority. The chaperone-like protein HtpG of M. tuberculosis (Mtb) is a large dimeric and multi-domain protein with a key role in Mtb pathogenesis and promising antigenic properties. This dual role, likely associated with the ability of Heat Shock proteins to act both intra- and extra-cellularly, makes HtpG highly exploitable both for drug and vaccine development. This review aims to gather the latest updates in HtpG structure and biological function, with HtpG operating in conjunction with a large number of chaperone molecules of Mtb. Altogether, these molecules help Mtb recovery after exposure to host-like stress by assisting the whole path of protein folding rescue, from the solubilisation of aggregated proteins to their refolding. Also, we highlight the role of structural biology in the development of safer and more effective subunit antigens. The larger availability of structural information on Mtb antigens and a better understanding of the host immune response to TB infection will aid the acceleration of TB vaccine development.
1. Introduction
Tuberculosis (TB) is the world’s second-deadliest infectious disease after the coronavirus disease 2019 (COVID-19) pandemic, causing 1.3 million deaths and 10.6 million new cases in 2022 and is the leading killer among people with HIV. During the 2020–2022 period, COVID-19 disruptions resulted in nearly half a million more deaths from TB (https://www.who.int/publications/i/item/9789240083851, accessed on 15 March 2024). TB is caused by the ancient pathogen Mycobacterium tuberculosis (Mtb), which is transmitted through inhalation of aerosol particles and transported to the lungs, where it infects alveolar macrophages and can produce cavitation, a hallmark of TB infection due to collagen degradation resulting from strong immune response [1,2]. Infected cells are sequestered at the core of the TB granuloma, a confined niche where Mtb can persist in a dormant state for decades [3,4]. During dormancy, cells are not able to replicate and be detected in biological samples, but this non-culturable state is transient [5]. The reactivation from dormancy is the secret weapon of Mtb, as it can also occur after decades in about 10% of infected individuals. This complex phenomenon has been related to the catalytic action of resuscitation-promoting factors, a set of peptidoglycan-degrading proteins [6,7,8,9,10].
In activated macrophages and in other host microenvironments, Mtb can be exposed to multiple chemical stresses, including acidic pH, reactive oxygen, nitrogen species, antibiotics, and high temperature [11,12,13,14]. All of these conditions can be deleterious to cellular proteins, which can undergo unfolding and aggregation [15,16]. Despite this, Mtb is able to survive under stressful conditions for years [17].
In dormant cells, which can be reckoned as an extreme case of survival in stressful conditions, proteomic characterisation has shown significant differences in the number of enzymes and proteins which normally participate in the metabolic pathways [18,19,20,21]. Indeed, despite the negligible metabolic activity of dormant Mtb, several consensus proteins with a stabilising and protective effect, like chaperones DnaJ1 (Rv0352), HtpG (Rv2299), DnaK (Rv0350), GroEL2 (Rv0440), GroES (Rv3418), GroEL1 (Rv3417), have been found in different dormancy models [20,22]. Consistently, nonreplicating bacterial cells contain greater amounts of protein aggregates than those in an actively replicating state [16,23,24,25,26] due to the suboptimal function of protein quality control machinery, lack of nutrients to support protein synthesis, and an increase in cellular oxidants [27]. Therefore, a finely tuned chaperone expression level is crucial for Mtb’s stress response [14].
Due to its importance in Mtb virulence, the chaperone machinery is an attractive target for therapeutical applications against TB. Among chaperones, HtpG, which belongs to the highly conserved Hsp90 family proteins, was shown to induce strong activation of dendritic cells (DCs), enhancing the protective immune response when fused to other Mtb antigens (e.g., ESAT6) [28,29,30,31]. This protein acts as an actor in a complex plot, which requires the cooperation of several chaperone molecules [28]. A large number of structural studies of Mtb chaperones have been conducted in the last few years [30,32,33,34]. In this review, we provide an overall picture of the contribution of these structural studies to the understanding of the complex molecular machinery devoted to protein refolding and to the role of HtpG in this scenario.
2. HtpG of M. tuberculosis, a Mycobacterial Virulence Factor
HtpG of Mtb is a metal-dependent ATPase, fully conserved among Mtb strains and highly conserved in pathogenic mycobacteria like M. leprae (Table 1). Importantly, it is not encoded in avirulent species like M. smegmatis. HtpG was recently shown to be essential for maintaining the function of the CRISPR/Cas system of Mtb, which is involved in the defence against invading nucleic acids [35].

Table 1.
Sequence conservation of HtpG in mycobacterial strains.
Although not essential, HtpG is important for maintaining proteostasis in stressed Mtb cells. Indeed, transposon mutagenesis has shown that the loss of neither the gene encoding for HtpG nor for ClpB (another Mtb chaperone) is lethal to Mtb. However, the depletion of genes encoding for both chaperones impairs Mtb recovery after exposure to host-like stress [14]. Sequence analyses show that HtpG shares 46% sequence identity with the Hsp90 chaperone of Escherichia coli (PDB code 2IOP), a finding that suggests a similar function for HtpG. Consistently, like Hsp90 of E. coli, HtpG was shown to bind both ADP and AMP-PNP with a µM binding affinity [31].
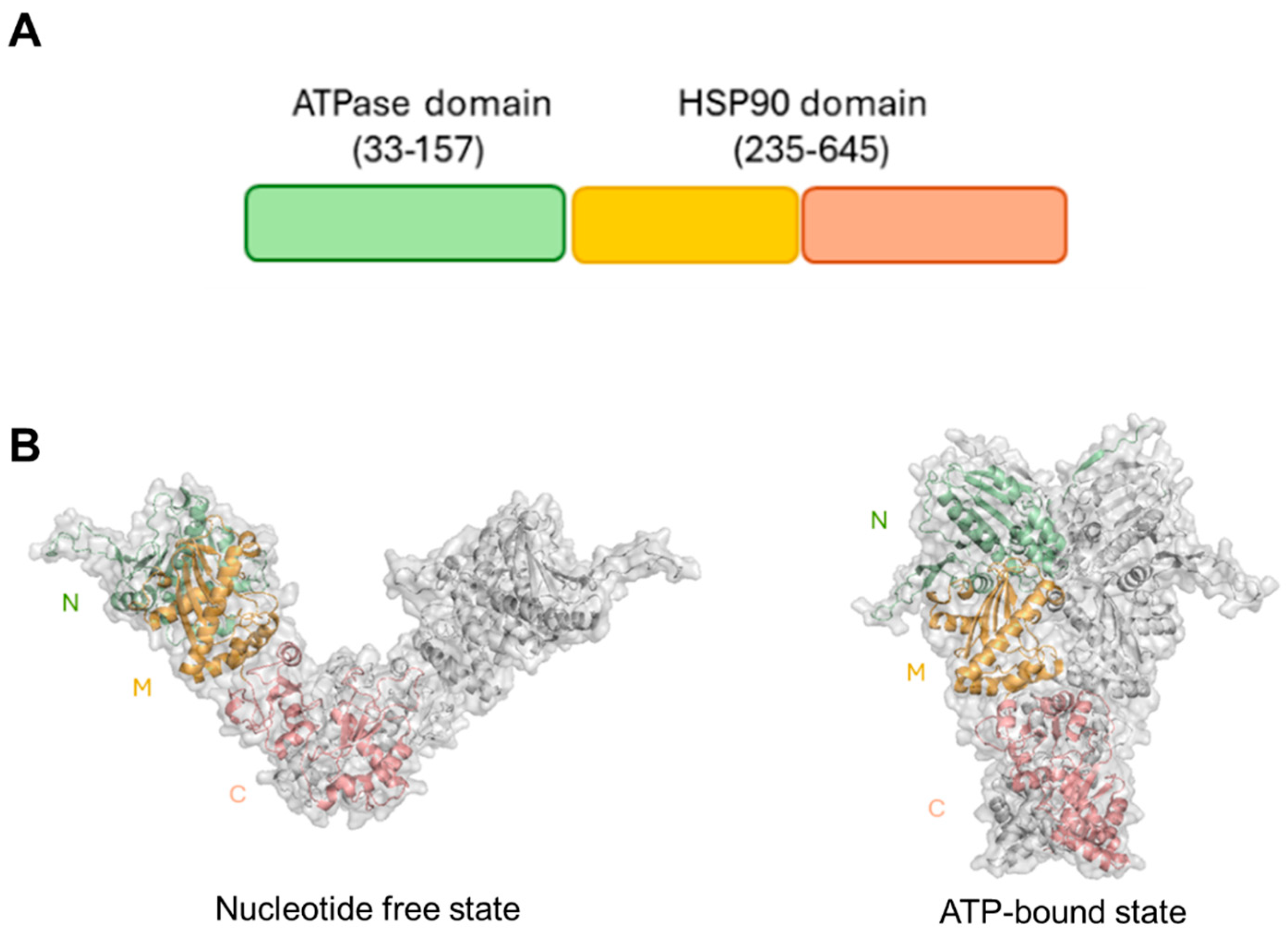
Although an experimental structure of HtpG is not yet available, computational studies have shown that HtpG adopts a highly stable dimeric structure, a property that is crucial for its functional role as a molecular chaperone [30]. HtpG is formed by three domains, which were shown to undergo large conformational variations from an open nucleotide-free to a compact ATP-bound state through a clamping mechanism (Figure 1).

Figure 1.
(A) Pfam organisation of HtpG. (B) Cartoon and surface representations of structural models of HtpG in its free state (on the left) and its ATPbound state (on the right). The two structures were derived by homology modelling, as previously reported [30]. Catalytic N–terminal, middle, and C–terminal domains are drawn in green, yellow, and salmon, respectively. Figures were generated with PyMol.
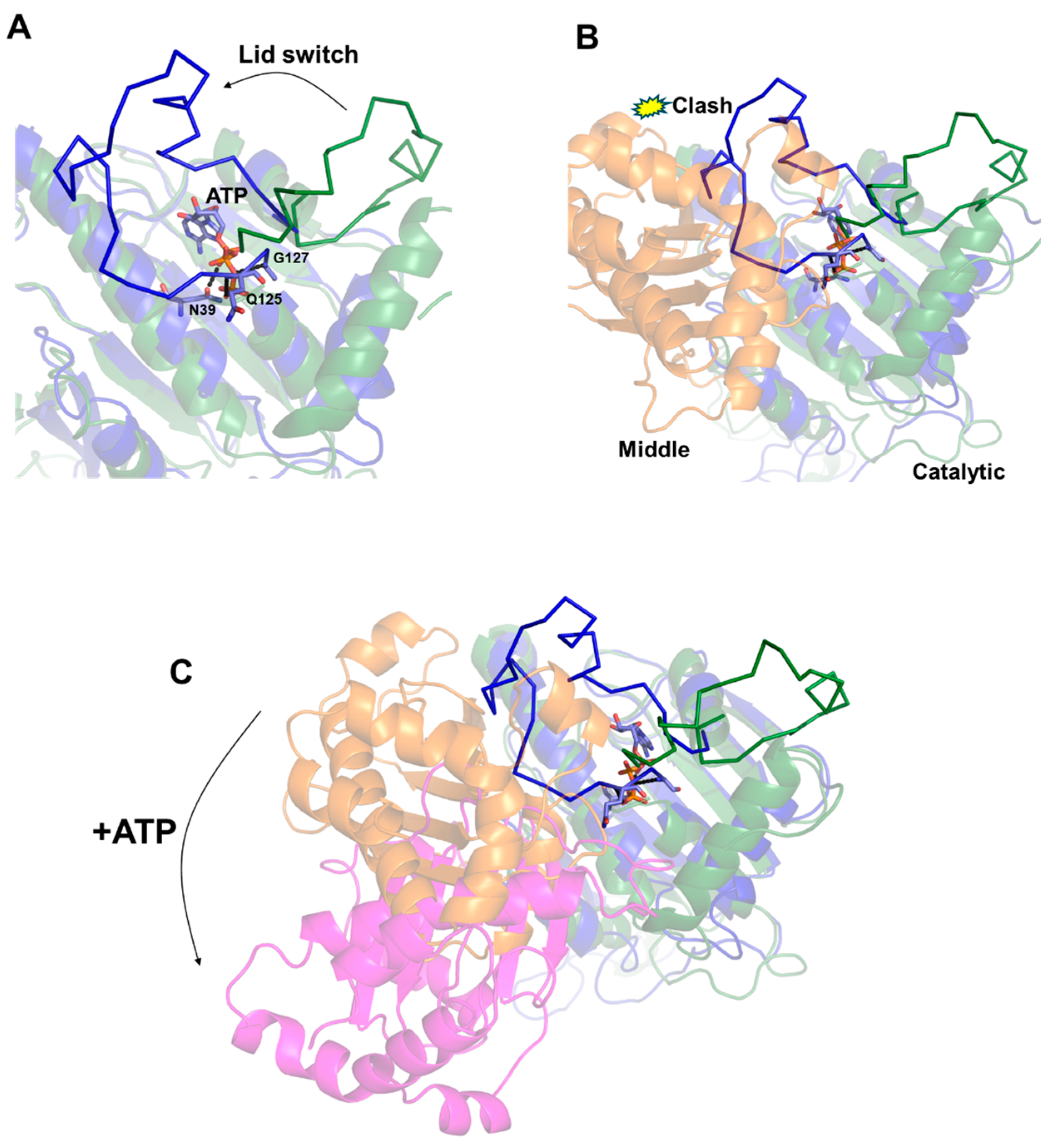
The structural determinants governing these large conformational changes were suggested by Molecular Dynamics (MDs) approaches [30]. These studies have shown that ATP rigidly anchors an ATP-binding loop of the N-terminal domain, denoted as the “lid”. The ATP-bound lid was shown to exhibit a low level of flexibility due to the strong interactions mediated by the phosphate groups of ATP with lid residues (e.g., Gln125, Gly127) (Figure 2A). This conformation is the result of a conformational “switch” of the lid region from the structure of the free to the ATP-bound enzyme [30] (Figure 2A). Importantly, the conformational switch of the “lid” is not compatible with the structure of HtpG in its free state due to collisions of the “lid” with the middle domain (Figure 2B). Indeed, it was proposed that the low flexibility of the ATP-bound “lid” makes it unable to accommodate its structure compatibly with the orientation between middle and catalytic domains in the free enzyme [30].

Figure 2.
A cartoon representation of the structural modifications in the “lid” upon ATP binding in the computed structures of HtpG [30]. (A,B). The catalytic domains in the free and ATP-bound structures of HtpG are drawn in forest green and dark blue, respectively. The middle domain in the free enzyme in Panel B is drawn in orange. (C) Superposition of catalytic domains in the structures of free and ATP-bound HtpG [30], showing the 119° rotation of the middle domain upon ATP binding. The middle domains in the free and ATP-bound HtpG models are drawn in orange and magenta, respectively.
The incompatibility of the ATP-bound conformation of the lid with the domain organisation of HtpG observed in the free state provided a structural rationale for the overall rearrangement of the enzyme in the presence of ATP (Figure 2C). The resulting clumping of HtpG due to ATP binding and the consequent lid switch is expected to allow HtpG to bind client proteins to facilitate their folding and then release them once folded through ATP binding [36,37,38,39].
3. HtpG Belongs to a Well-Orchestrated Chaperone Network
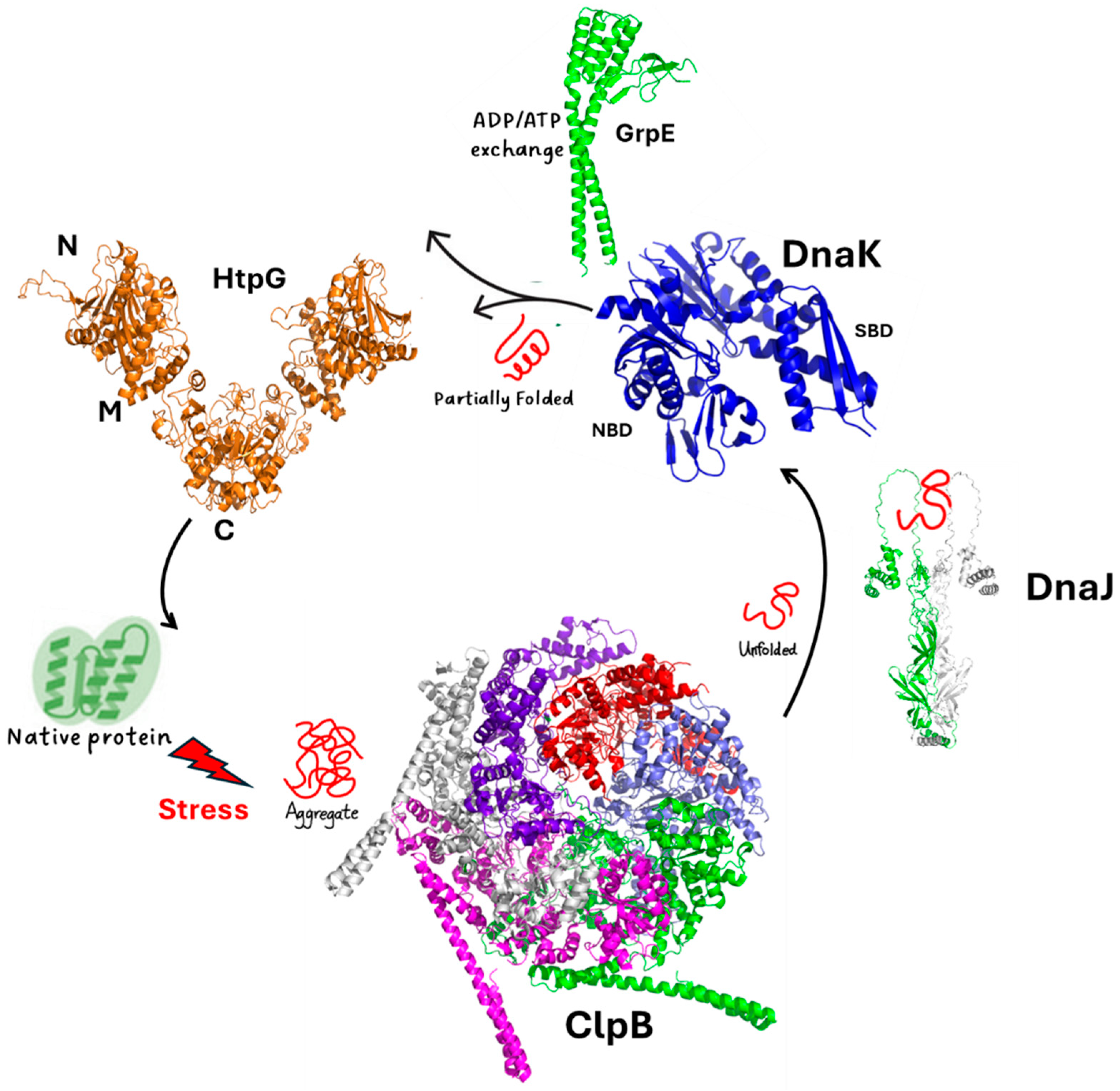
HtpG is part of a well-orchestrated and complex chaperone network, which includes the chaperone DnaK, its cofactors DnaJ1/J2, the release factor GrpE, and the protein disaggreases ClpB (Figure 3). In the complex scenario of protein rejuvenation after chemical stress, the disaggregase ClpB and the chaperone DnaK play an essential role in the early steps of protein recovery (Figure 3) [16,23,32,40].
ClpB is a large protein forming hexameric structures. ClpB proteins form a family of ATPases that disaggregate and solubilise aggregated proteins [41,42,43,44]. Similar to other prokaryotic ClpBs, ClpB of Mtb is composed of an N-terminal domain (NTD) and two nucleotide-binding domains (NBD1 and NBD2), which form a ring-shaped hexamer (Figure 3). DnaK is a highly conserved protein in prokaryotes. It is composed of an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD) that are connected by a flexible linker [32,33]. It collaborates with the two cochaperones, DnaJ1 and DnaJ2 (Figure 3), which deliver non-native client proteins to DnaK and accelerate ATP hydrolysis by DnaK [45]. The NEF GrpE helps the release of tightly bound ADP through binding in 1:2 stoichiometry and promoting substrate release [33] (Figure 3). Importantly, an association of DnaK and ClpB at the aggregate surface stimulates ATP hydrolysis, which mediates substrate remodelling [16,23,40,46,47,48,49]. Consistently, Mtb cells lacking ClpB are sensitive to oxidants and show fault in recovery after achieving a stationary growth phase [14].
In many organisms, DnaK interacts with HtpG and ClpB [50,51,52], whereas in Mtb, DnaK interactions with ClpB [32] and with HtpG [14], but not directly between ClpB and HptG, have been reported. This suggests that ClpB and HtpG are part of a broader network of ATPases, all connected by DnaK. Intriguingly, while both HtpG and ClpB are not essential in Mtb [16,53], cells lacking both nonessential chaperones are hypersensitive to host-like stresses that induce a nonreplicating state [14]. This further suggests that HtpG acts cooperatively with the other chaperones (Figure 3).

Figure 3.
A scheme of the chaperone proteins assisting protein folding in Mtb. Cartoon representations are drawn for the experimental structures of ClpB (pdb code 7l6n) and DnaK (pdb code 8gb3). For the other proteins, we determined their structures using AI and the program AlphaFold2 [54], which provided a reliable model with pLDDT > 90.
Figure 3.
A scheme of the chaperone proteins assisting protein folding in Mtb. Cartoon representations are drawn for the experimental structures of ClpB (pdb code 7l6n) and DnaK (pdb code 8gb3). For the other proteins, we determined their structures using AI and the program AlphaFold2 [54], which provided a reliable model with pLDDT > 90.
4. A Structural Model of HtpG Interactions in the Chaperone Machinery of Mtb
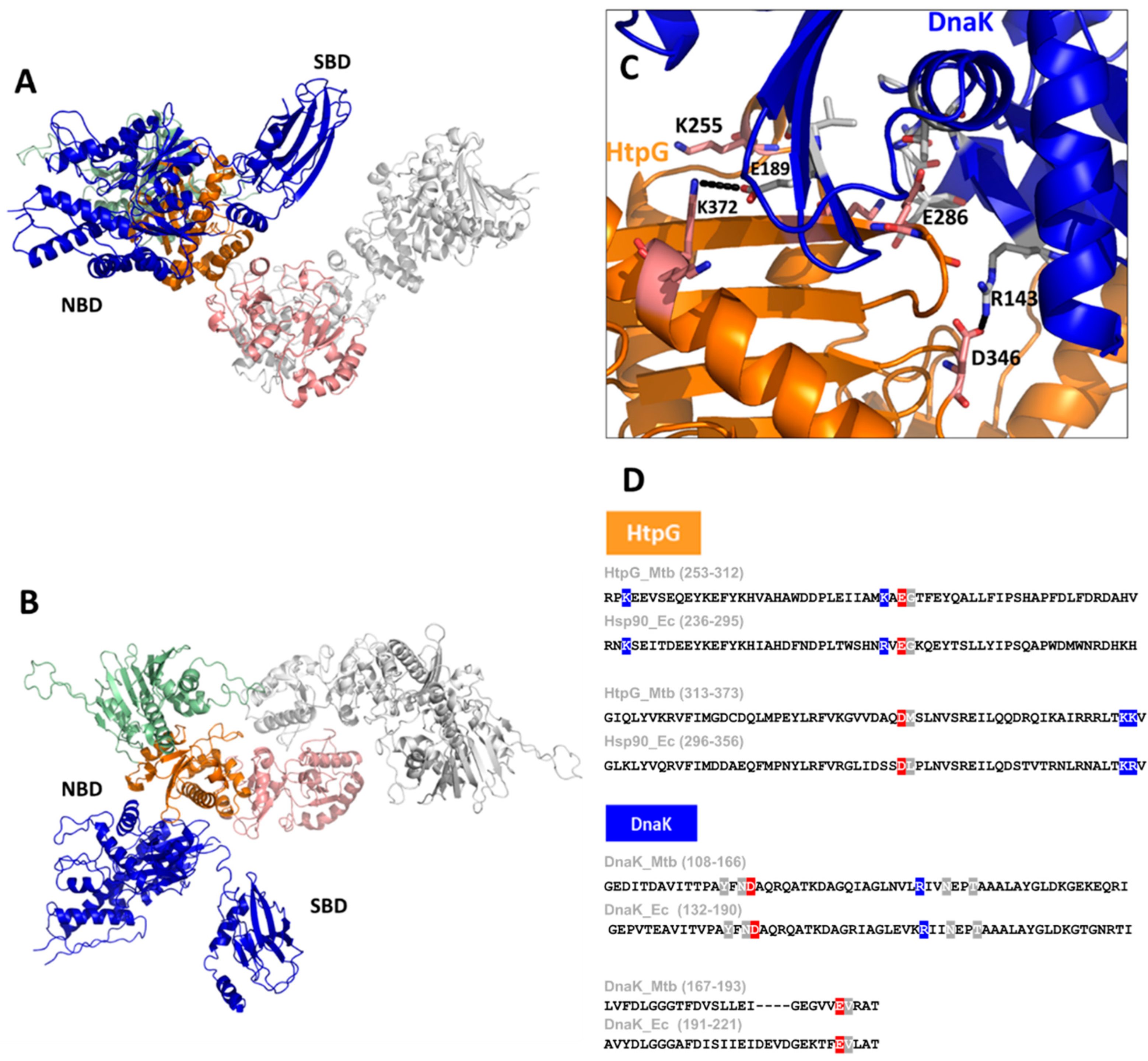
Consistent with the observed interactions of HtpG with DnaK in Mtb [14], it was reported that an increased expression in the two cofactors of DnaK, DnaJ1, and DnaJ2, occurred in a ΔHtpG mutant strain [28]. Despite the importance of the HtpG–DnaK complex formation in Mtb, no structural models of this important interaction have been described so far. With the help of Artificial Intelligence (AI), we reliably model the HtpG–DnaK complex, as shown in Figure 4A,B. In addition to the typical parameters used to evaluate AI modelling reliability, we could validate this structure based on the available information from the homolog Hsp90 from E. coli [55]. Indeed, similar to Hsp90 of E. coli, HtpG interacts with DnaK exclusively through residues of its middle domain (Figure 4C), thus leaving the N-terminal domain free to exert its catalytic function. Importantly, we observe that the key residues involved in binding are strongly conserved between HtpG of Mtb and Hsp90 and between DnaK chaperones of the two bacteria, thus corroborating the similar organisation in Mtb (Figure 4D). As reported for Hsp90 [55], this structural organisation is mechanistically plausible, as it allows a client protein bound to the SBD of DnaK to readily interact with the client-binding site of HtpG (Figure 3). These studies provide insight into the interactions and collaboration between HtpG and DnaK.

Figure 4.
A cartoon representation of the AI-modelled structure of the HtpG–DnaK complex of Mtb (A,B). The structure was obtained using the software AlphaFold2.0 [54], which provided a reliable model with pLDDT > 90. A zoom of the interface between HtpG and DnaK is given in panel (C), with some important residues drawn in stick form. The sequence alignment of HtpG and DnaK of E. coli and Mtb are reported in panel (D), with the most important conserved residues highlighted (positives in blue, negative in red, uncharged in grey).
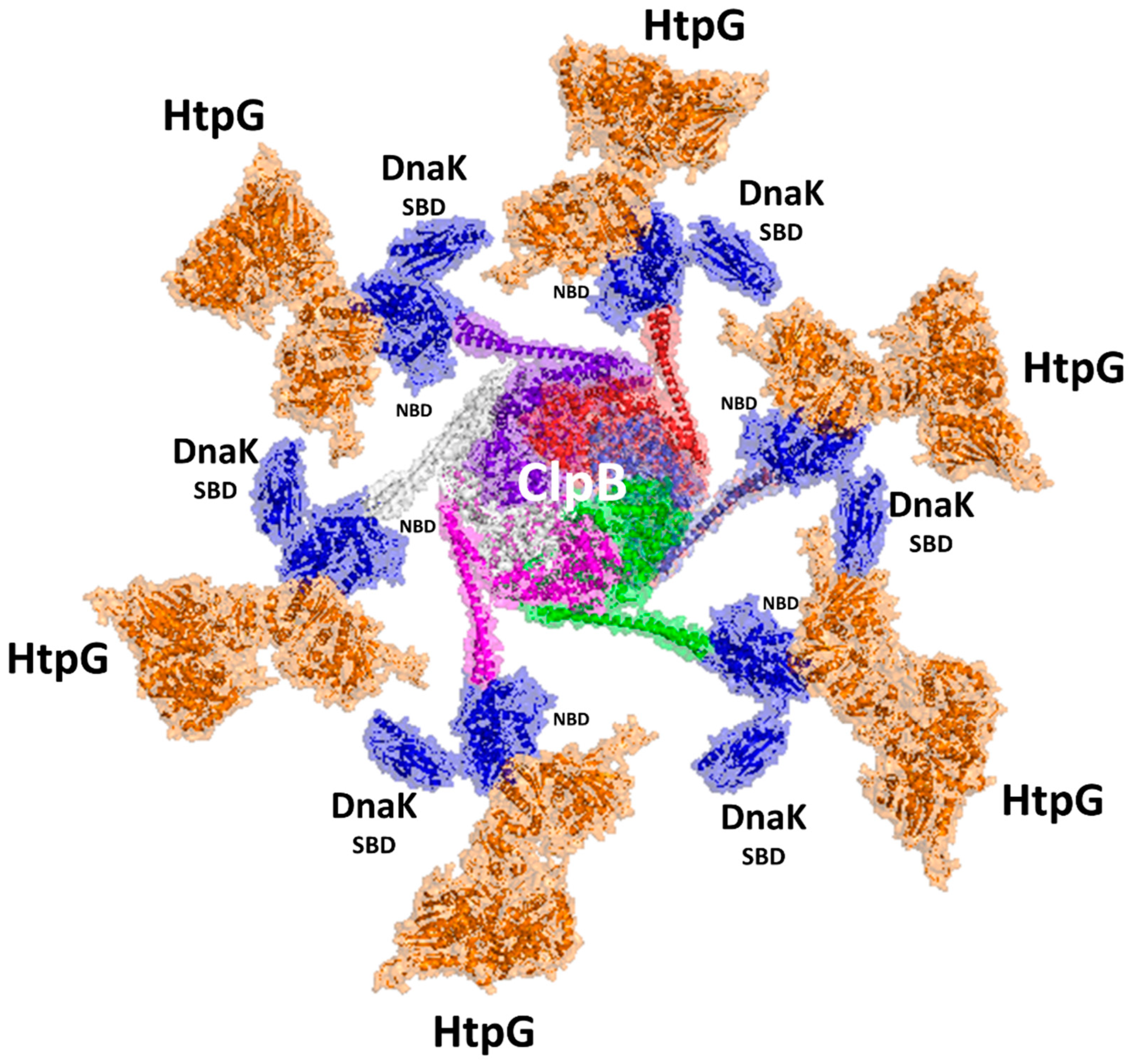
A large amount of structural information on DnaK complexes was released in the last three years [32], among which the structure of DnaK with ClpB is illuminating [32]. Up to four DnaK molecules were found to bind to a ClpB hexamer, although the exact stoichiometry is unknown, and it is clear whether DnaK functions processively on ClpB or hops on and off [32]. We combined the structural information of the HtpG–DnaK complex with that of the DnaK–ClpB complex to generate a hypothetical model of the entire machinery (Figure 5). Although there is no experimental proof, it is tempting to propose that ClpB may bind up to six DnaK–HtpG complexes in a carousel-like machinery (Figure 5). In this machinery, which is so far only a putative model, the refolding process may proceed radially from the disaggregation of proteins operated by ClpB to refolding by DnaK and HtpG. Possibly, proteins that are partially refolded by DnaK are shuttled from the SBD domain of DnaK to HtpG on an adjacent arm of the carousel, closest in space. A similar mechanism would allow a high efficacy of the entire refolding machine.

Figure 5.
A model of the carousel-like chaperone machinery of Mtb. The model was generated upon superposition of the AI model of the HtpG–DnaK complex [54] to DnaK NBD in the cryoEM structure of the ClpB complex (pdb code 6w6e). Cartoon and surface representations are drawn for HtpG (orange), DnaK (blue), and the six chains of ClpB (white, prune, red, light blue, green, magenta).
5. HtpG as a Vaccine Antigen
As reported by the WHO, a new vaccine with better coverage than the current Mycobacterium bovis BCG vaccine is strongly needed [56]. There is extremely active research in the field, with a steep acceleration in the past decades due to the advance of technologies and more rational vaccine design strategies. Several vaccines against TB are in the pipeline, including live attenuated whole-cell vaccines, inactivated whole-cell vaccines, adjuvanted protein subunit vaccines, and viral-vectored vaccines [4]. Subunit vaccines contain purified parts of the pathogen that elicit immunogenic host response, and, as such, they overcome safety concerns typically associated with vaccines. Although they are specific and safe, they typically display a reduced capacity to stimulate a broad immune response compared to live attenuated or killed microorganisms. Therefore, these vaccines need to be engineered to enhance their immunogenicity. Structure-based antigen design is a rational approach that uses three-dimensional structural information to design novel and enhanced vaccine antigens. As previously mentioned, this strategy is crucial to the development of effective subunit antigens [57].
We have previously shown that HtpG is a promising vaccine antigen, as it activates dendritic cells (DCs), which can induce T-cell differentiation, resulting in inhibition of intracellular Mtb growth in macrophages [29,30,31]. The elucidation of the exact regions responsible for the immunoreactivity has helped the design of a more effective vaccine antigen [31]. Indeed, this step is an important tool for the design of improved antigens [57]. Crucial to the identification of epitope region is the prediction of the portions of the protein that are able to strongly bind the Major Histocompatibility Complex (MHC). The function of MHC molecules is indeed to bind peptide fragments derived from pathogens and display them on the cell surface for recognition by the appropriate T-cells. The human MHC class II is encoded by three different isotypes, HLA-DR, -DQ, and -DP (referred to as HLA in humans, which stands for human leukocyte antigens), all being highly polymorphic.
HtpG is predicted to embed many T-cell epitopes with a strong affinity to the MHCII complex, including HLA-DR, HLA-DP, and HLA-DQ molecules [31]. These epitopes are mainly located in the C-terminal domain of HtpG and some in the middle domain. No epitopes were predicted in the catalytic N-terminal domain. Consistently, we found that the most immune-reactive region of the molecule is located on the C-terminal and middle domains, whereas the catalytic and nucleotide-binding N-terminal domain plays no role in the elicitation of the immune response [31]. This information was precious to designing vaccine antigens with enhanced biophysical properties, albeit conserved or enhanced antigenic properties [31]. We also adopted a combination of different immune stimulation mechanisms upon fusion of middle and C-terminal domains of HtpG, which stimulates DC cells, with ESAT6, known as a T cell activator. This fusion antigen possessed a higher anti-mycobacterial activity than the fusion protein embedding the entire HtpG [31]. These studies point to HtpG as an important target for vaccine development and are in line with the dual function of Hsps in humans, depending on their intracellular or extracellular location. Intracellular Hsps have a protective function, whereas extracellular located Hsps mediate immunological functions [58,59].
6. Conclusions
TB has undergone times when it was considered a thing of the past. The development of the first antibiotics in the 1940s and 1950s led to an important decrease in TB case fatality. However, WHO’s Global Tuberculosis Programme (GTB) declared TB a global emergency in 1993 due to the development of antibiotic-resistant mycobacterial strains. The prevention of the development of drug-resistant strains of TB was its main goal 30 years later. This goal has not yet been achieved, and the identification of important targets for drug and vaccine development is a precious tool for the advancement of therapeutic approaches against TB.
The chaperone HtpG has a central role in Mtb virulence, as demonstrated by its conservation in virulent strains. In this review, we have presented the complex and dual role of HtpG in both protecting Mtb from stress and activating an immune response. We highlight the synergic role of HtpG with a large number of chaperones, which all together are responsible for the recovery of proteins after host-induced stress. Indeed, HtpG interacts with DnaK, which in turn interacts with the disaggregase ClpB. It is tempting to propose that these proteins form a large macromolecular machinery where ClpB exerts its role in the dissolution of protein aggregates and then shuttles disaggregated molecules to DnaK and HtpG for proper refolding (Figure 5). These steps, which occur through the help of cofactors like DnaJ1 and DnaJ2 and the release factor GrpE, involve extensive ATP hydrolysis, a process that allows the large conformational changes needed to fold and then release client proteins. Although this is a speculative model, it is based on the most updated structural findings [32,33]. We believe that the understanding of further structural aspects associated with protein folding by this complex machinery will be precious to the understanding of this key biological process, which is the ace in the hole of Mtb and allows it to survive in extremely stressful conditions. Also, we are increasingly aware of the fact that the understanding of the structural basis of immunogenicity is key to the design of improved antigens with better biophysical properties and the ability to induce multiple immune-stimulating mechanisms.
Author Contributions
Conceptualisation, R.B. and A.R.; methodology and software, R.B., M.R., F.S., M.P., V.N. and A.R.; formal analysis, M.R., F.S., G.B., A.R. and R.B.; data curation, M.R., V.N., F.S., G.B., M.P., A.R. and R.B.; writing—original draft preparation, R.B. and A.R.; writing—review and editing, all authors; supervision, R.B. and A.R.; funding acquisition, R.B. and A.R. All authors have read and agreed to the published version of the manuscript.
Funding
A.R. acknowledges funding from PRIN 2022 PNRR “NextGen-erationEU”, project Prot. P2022JE8FN—FIGHT_TB “FIndinG High-grade anTigens towards an innovative TB vaccines” —D.D. MUR Prot. n. 0001363 of 1 September 2023. V.N. and R.B. were funded by the project INF-ACT “One Health Basic and Translational Research Actions addressing Unmet Needs on Emerging Infectious Diseases PE00000007”, PNRR Mission 4, EU “NextGen-erationEU”—D.D. MUR Prot. n. 0001554 of 11 October 2022.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Delogu, G.; Sali, M.; Fadda, G. The Biology of Mycobacterium Tuberculosis Infection. Mediterr. J. Hematol. Infect. Dis. 2013, 5, e2013070. [Google Scholar] [CrossRef] [PubMed]
- Squeglia, F.; Ruggiero, A.; Berisio, R. Collagen Degradation in Tuberculosis Pathogenesis: The Biochemical Consequences of Hosting an Undesired Guest. Biochem. J. 2018, 475, 3123–3140. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; Karakousis, P.C. Latent Tuberculosis Infection: Myths, Models, and Molecular Mechanisms. Microbiol. Mol. Biol. Rev. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Romano, M.; Squeglia, F.; Kramarska, E.; Barra, G.; Choi, H.-G.; Kim, H.-J.; Ruggiero, A.; Berisio, R. A Structural View at Vaccine Development against M. Tuberculosis. Cells 2023, 12, 317. [Google Scholar] [CrossRef] [PubMed]
- Mukamolova, G.V.; Kaprelyants, A.S.; Young, M. Mycobacterial Dormancy and Its Relation to Persistence. In Parish; Parish, T., Ed.; Taylor & Francis: Abingdon, UK, 2005; pp. 265–320. ISBN 978-1-904933-14-4. [Google Scholar]
- Kana, B.D.; Mizrahi, V. Resuscitation-Promoting Factors as Lytic Enzymes for Bacterial Growth and Signaling. FEMS Immunol. Med. Microbiol. 2010, 58, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Tizzano, B.; Pedone, E.; Pedone, C.; Wilmanns, M.; Berisio, R. Crystal Structure of the Resuscitation-Promoting Factor (DeltaDUF)RpfB from M. Tuberculosis. J. Mol. Biol. 2009, 385, 153–162. [Google Scholar] [CrossRef]
- Nikitushkin, V.D.; Demina, G.R.; Shleeva, M.O.; Guryanova, S.V.; Ruggiero, A.; Berisio, R.; Kaprelyants, A.S. A Product of RpfB and RipA Joint Enzymatic Action Promotes the Resuscitation of Dormant Mycobacteria. FEBS J. 2015, 282, 2500–2511. [Google Scholar] [CrossRef]
- Rosser, A.; Stover, C.; Pareek, M.; Mukamolova, G.V. Resuscitation-Promoting Factors Are Important Determinants of the Pathophysiology in Mycobacterium Tuberculosis Infection. Crit. Rev. Microbiol. 2017, 43, 621–630. [Google Scholar] [CrossRef]
- Squeglia, F.; Ruggiero, A.; Berisio, R. Chemistry of Peptidoglycan in Mycobacterium Tuberculosis Life Cycle: An off-the-Wall Balance of Synthesis and Degradation. Chemistry 2018, 24, 2533–2546. [Google Scholar] [CrossRef]
- Ehrt, S.; Schnappinger, D. Mycobacterial Survival Strategies in the Phagosome: Defence against Host Stresses. Cell. Microbiol. 2009, 11, 1170–1178. [Google Scholar] [CrossRef]
- Baker, J.J.; Dechow, S.J.; Abramovitch, R.B. Acid Fasting: Modulation of Mycobacterium Tuberculosis Metabolism at Acidic pH. Trends Microbiol. 2019, 27, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Levitte, S.; Adams, K.N.; Berg, R.D.; Cosma, C.L.; Urdahl, K.B.; Ramakrishnan, L. Mycobacterial Acid Tolerance Enables Phagolysosomal Survival and Establishment of Tuberculous Infection In Vivo. Cell Host Microbe 2016, 20, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Harnagel, A.; Lopez Quezada, L.; Park, S.W.; Baranowski, C.; Kieser, K.; Jiang, X.; Roberts, J.; Vaubourgeix, J.; Yang, A.; Nelson, B.; et al. Nonredundant Functions of Mycobacterium Tuberculosis Chaperones Promote Survival under Stress. Mol. Microbiol. 2021, 115, 272–289. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.D.; Schroeder, K.; Jonas, K. Protein Aggregation in Bacteria. FEMS Microbiol. Rev. 2020, 44, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Vaubourgeix, J.; Lin, G.; Dhar, N.; Chenouard, N.; Jiang, X.; Botella, H.; Lupoli, T.; Mariani, O.; Yang, G.; Ouerfelli, O.; et al. Stressed Mycobacteria Use the Chaperone ClpB to Sequester Irreversibly Oxidized Proteins Asymmetrically within and between Cells. Cell Host Microbe 2015, 17, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Batyrshina, Y.; Schwartz, Y. New in Vitro Model of Mycobacterium Tuberculosis Dormancy. Eur. Respir. J. 2020, 56, 2810. [Google Scholar] [CrossRef]
- Starck, J.; Källenius, G.; Marklund, B.-I.; Andersson, D.I.; Åkerlund, T. Comparative Proteome Analysis of Mycobacterium Tuberculosis Grown under Aerobic and Anaerobic Conditions. Microbiology 2004, 150, 3821–3829. [Google Scholar] [CrossRef] [PubMed]
- Trutneva, K.A.; Shleeva, M.O.; Demina, G.R.; Vostroknutova, G.N.; Kaprelyans, A.S. One-Year Old Dormant, “Non-Culturable” Mycobacterium Tuberculosis Preserves Significantly Diverse Protein Profile. Front. Cell. Infect. Microbiol. 2020, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Albrethsen, J.; Agner, J.; Piersma, S.R.; Højrup, P.; Pham, T.V.; Weldingh, K.; Jimenez, C.R.; Andersen, P.; Rosenkrands, I. Proteomic Profiling of Mycobacterium Tuberculosis Identifies Nutrient-Starvation-Responsive Toxin-Antitoxin Systems. Mol. Cell. Proteom. 2013, 12, 1180–1191. [Google Scholar] [CrossRef]
- Shleeva, M.O.; Kudykina, Y.K.; Vostroknutova, G.N.; Suzina, N.E.; Mulyukin, A.L.; Kaprelyants, A.S. Dormant Ovoid Cells of Mycobacterium Tuberculosis Are Formed in Response to Gradual External Acidification. Tuberculosis 2011, 91, 146–154. [Google Scholar] [CrossRef]
- Schubert, O.T.; Ludwig, C.; Kogadeeva, M.; Zimmermann, M.; Rosenberger, G.; Gengenbacher, M.; Gillet, L.C.; Collins, B.C.; Röst, H.L.; Kaufmann, S.H.E.; et al. Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium Tuberculosis. Cell Host Microbe 2015, 18, 96–108. [Google Scholar] [CrossRef]
- Fay, A.; Glickman, M.S. An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding. PLoS Genet. 2014, 10, e1004516. [Google Scholar] [CrossRef]
- Kwiatkowska, J.; Matuszewska, E.; Kuczyńska-Wiśnik, D.; Laskowska, E. Aggregation of Escherichia coli Proteins during Stationary Phase Depends on Glucose and Oxygen Availability. Res. Microbiol. 2008, 159, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Ezraty, B.; Dukan, S. Protein Aggregates: An Aging Factor Involved in Cell Death. J. Bacteriol. 2008, 190, 6070. [Google Scholar] [CrossRef]
- Navarro Llorens, J.M.; Tormo, A.; Martínez-García, E. Stationary Phase in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef]
- Josefson, R.; Andersson, R.; Nyström, T. How and Why Do Toxic Conformers of Aberrant Proteins Accumulate during Ageing? Essays Biochem. 2017, 61, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Mangla, N.; Singh, R.; Agarwal, N. HtpG Is a Metal-Dependent Chaperone Which Assists the DnaK/DnaJ/GrpE Chaperone System of Mycobacterium Tuberculosis via Direct Association with DnaJ2. Microbiol. Spectr. 2023, 11, e0031223. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-G.; Choi, S.; Back, Y.W.; Paik, S.; Park, H.-S.; Kim, W.S.; Kim, H.; Cha, S.B.; Choi, C.H.; Shin, S.J.; et al. Rv2299c, a Novel Dendritic Cell-Activating Antigen of Mycobacterium Tuberculosis, Fused-ESAT-6 Subunit Vaccine Confers Improved and Durable Protection against the Hypervirulent Strain HN878 in Mice. Oncotarget 2017, 8, 19947–19967. [Google Scholar] [CrossRef]
- Moreira, M.; Ruggiero, A.; Esposito, L.; Choi, H.-G.; Kim, H.-J.; Berisio, R. Structural Features of HtpGMtb and HtpG-ESAT6Mtb Vaccine Antigens against Tuberculosis: Molecular Determinants of Antigenic Synergy and Cytotoxicity Modulation. Int. J. Biol. Macromol. 2020, 158, 305–317. [Google Scholar] [CrossRef]
- Ruggiero, A.; Choi, H.-G.; Barra, G.; Squeglia, F.; Back, Y.W.; Kim, H.-J.; Berisio, R. Structure Based Design of Effective HtpG-Derived Vaccine Antigens against M. Tuberculosis. Front. Mol. Biosci. 2022, 9, 964645. [Google Scholar] [CrossRef]
- Yin, Y.; Feng, X.; Yu, H.; Fay, A.; Kovach, A.; Glickman, M.S.; Li, H. Structural Basis for Aggregate Dissolution and Refolding by the Mycobacterium Tuberculosis ClpB-DnaK Bi-Chaperone System. Cell Rep. 2021, 35, 109166. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Fay, A.; Molina, P.S.; Kovach, A.; Glickman, M.S.; Li, H. Structure of the M. Tuberculosis DnaK−GrpE Complex Reveals How Key DnaK Roles Are Controlled. Nat. Commun. 2024, 15, 660. [Google Scholar] [CrossRef] [PubMed]
- Guillet, V.; Bordes, P.; Bon, C.; Marcoux, J.; Gervais, V.; Sala, A.J.; Dos Reis, S.; Slama, N.; Mares-Mejía, I.; Cirinesi, A.-M.; et al. Structural Insights into Chaperone Addiction of Toxin-Antitoxin Systems. Nat. Commun. 2019, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, H.; Fleming, J.; Wei, W.; Chen, H.; Dai, X.; Liu, Y.; Li, C.; Ran, F.; Wu, Z.; et al. EspB and HtpG Interact with the Type III-A CRISPR/Cas System of Mycobacterium Tuberculosis. Front. Mol. Biosci. 2023, 10, 1261613. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Prodromou, C.; Hu, B.; Vaughan, C.; Roe, S.M.; Panaretou, B.; Piper, P.W.; Pearl, L.H. Structural and Functional Analysis of the Middle Segment of Hsp90: Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions. Mol. Cell 2003, 11, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.U.; Roe, S.M.; Vaughan, C.K.; Meyer, P.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex. Nature 2006, 440, 1013–1017. [Google Scholar] [CrossRef]
- Shiau, A.K.; Harris, S.F.; Southworth, D.R.; Agard, D.A. Structural Analysis of E. coli Hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell 2006, 127, 329–340. [Google Scholar] [CrossRef]
- Mader, S.L.; Lopez, A.; Lawatscheck, J.; Luo, Q.; Rutz, D.A.; Gamiz-Hernandez, A.P.; Sattler, M.; Buchner, J.; Kaila, V.R.I. Conformational Dynamics Modulate the Catalytic Activity of the Molecular Chaperone Hsp90. Nat. Commun. 2020, 11, 1410. [Google Scholar] [CrossRef]
- Lupoli, T.J.; Fay, A.; Adura, C.; Glickman, M.S.; Nathan, C.F. Reconstitution of a Mycobacterium Tuberculosis Proteostasis Network Highlights Essential Cofactor Interactions with Chaperone DnaK. Proc. Natl. Acad. Sci. USA 2016, 113, E7947–E7956. [Google Scholar] [CrossRef]
- Doyle, S.M.; Wickner, S. Hsp104 and ClpB: Protein Disaggregating Machines. Trends Biochem. Sci. 2009, 34, 40–48. [Google Scholar] [CrossRef]
- Maurizi, M.R.; Xia, D. Protein Binding and Disruption by Clp/Hsp100 Chaperones. Structure 2004, 12, 175–183. [Google Scholar] [CrossRef]
- Shorter, J.; Southworth, D.R. Spiraling in Control: Structures and Mechanisms of the Hsp104 Disaggregase. Cold Spring Harb. Perspect. Biol. 2019, 11, a034033. [Google Scholar] [CrossRef]
- Lee, S.; Sowa, M.E.; Choi, J.-M.; Tsai, F.T.F. The ClpB/Hsp104 Molecular Chaperone—A Protein Disaggregating Machine. J. Struct. Biol. 2004, 146, 99–105. [Google Scholar] [CrossRef]
- Nelson, B.; Hong, S.H.; Lupoli, T.J. Protein Cofactor Mimics Disrupt Essential Chaperone Function in Stressed Mycobacteria. ACS Infect. Dis. 2022, 8, 901–910. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, M.A.; Gerrick, E.R.; Xu, W.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive Essentiality Analysis of the Mycobacterium Tuberculosis Genome via Saturating Transposon Mutagenesis. mBio 2017, 8, e02133-16. [Google Scholar] [CrossRef]
- Lupoli, T.J.; Vaubourgeix, J.; Burns-Huang, K.; Gold, B. Targeting the Proteostasis Network for Mycobacterial Drug Discovery. ACS Infect. Dis. 2018, 4, 478–498. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes Required for Mycobacterial Growth Defined by High Density Mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Yu, H.; Lupoli, T.J.; Kovach, A.; Meng, X.; Zhao, G.; Nathan, C.F.; Li, H. ATP Hydrolysis-Coupled Peptide Translocation Mechanism of Mycobacterium Tuberculosis ClpB. Proc. Natl. Acad. Sci. USA 2018, 115, E9560–E9569. [Google Scholar] [CrossRef] [PubMed]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 Chaperones: Collaborators in Protein Remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, R.J.; Hansen, W.J.; Freeman, B.C.; Alnemri, E.; Litwack, G.; Toft, D.O. Cooperative Action of Hsp70, Hsp90, and DnaJ Proteins in Protein Renaturation. Biochemistry 1996, 35, 14889–14898. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the Hub of Protein Homeostasis: Emerging Mechanistic Insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Lopez Quezada, L.; Smith, R.; Lupoli, T.J.; Edoo, Z.; Li, X.; Gold, B.; Roberts, J.; Ling, Y.; Park, S.W.; Nguyen, Q.; et al. Activity-Based Protein Profiling Reveals That Cephalosporins Selectively Active on Non-Replicating Mycobacterium Tuberculosis Bind Multiple Protein Families and Spare Peptidoglycan Transpeptidases. Front. Microbiol. 2020, 11, 1248. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and Its Applications in the Fields of Biology and Medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Kravats, A.N.; Doyle, S.M.; Hoskins, J.R.; Genest, O.; Doody, E.; Wickner, S. Interaction of E. coli Hsp90 with DnaK Involves the DnaJ Binding Region of DnaK. J. Mol. Biol. 2017, 429, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Flores-Valdez, M.A. After 100 Years of BCG Immunization against Tuberculosis, What Is New and Still Outstanding for This Vaccine? Vaccines 2021, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Schijns, V.; Majhen, D.; van der Ley, P.; Thakur, A.; Summerfield, A.; Berisio, R.; Nativi, C.; Fernández-Tejada, A.; Alvarez-Dominguez, C.; Gizurarson, S.; et al. Rational Vaccine Design in Times of Emerging Diseases: The Critical Choices of Immunological Correlates of Protection, Vaccine Antigen and Immunomodulation. Pharmaceutics 2021, 13, 501. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.; Gehrmann, M.; Brunet, M.; Multhoff, G.; Garrido, C. Intracellular and Extracellular Functions of Heat Shock Proteins: Repercussions in Cancer Therapy. J. Leukoc. Biol. 2007, 81, 15–27. [Google Scholar] [CrossRef]
- Zhang, Z.; Jing, J.; Ye, Y.; Chen, Z.; Jing, Y.; Li, S.; Hong, W.; Ruan, H.; Liu, Y.; Hu, Q.; et al. Characterization of the Dual Functional Effects of Heat Shock Proteins (HSPs) in Cancer Hallmarks to Aid Development of HSP Inhibitors. Genome Med. 2020, 12, 101. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).