Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products
Abstract
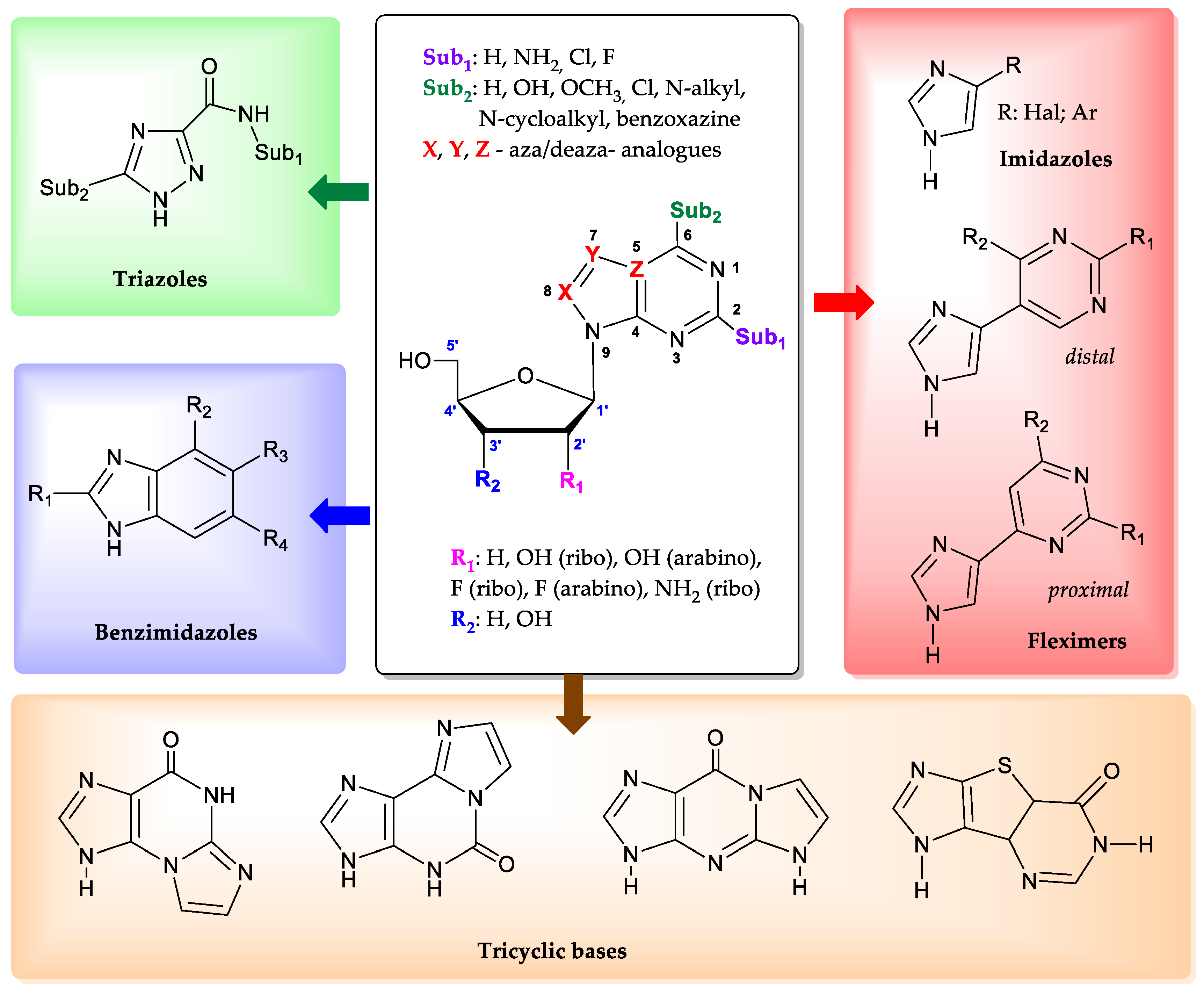
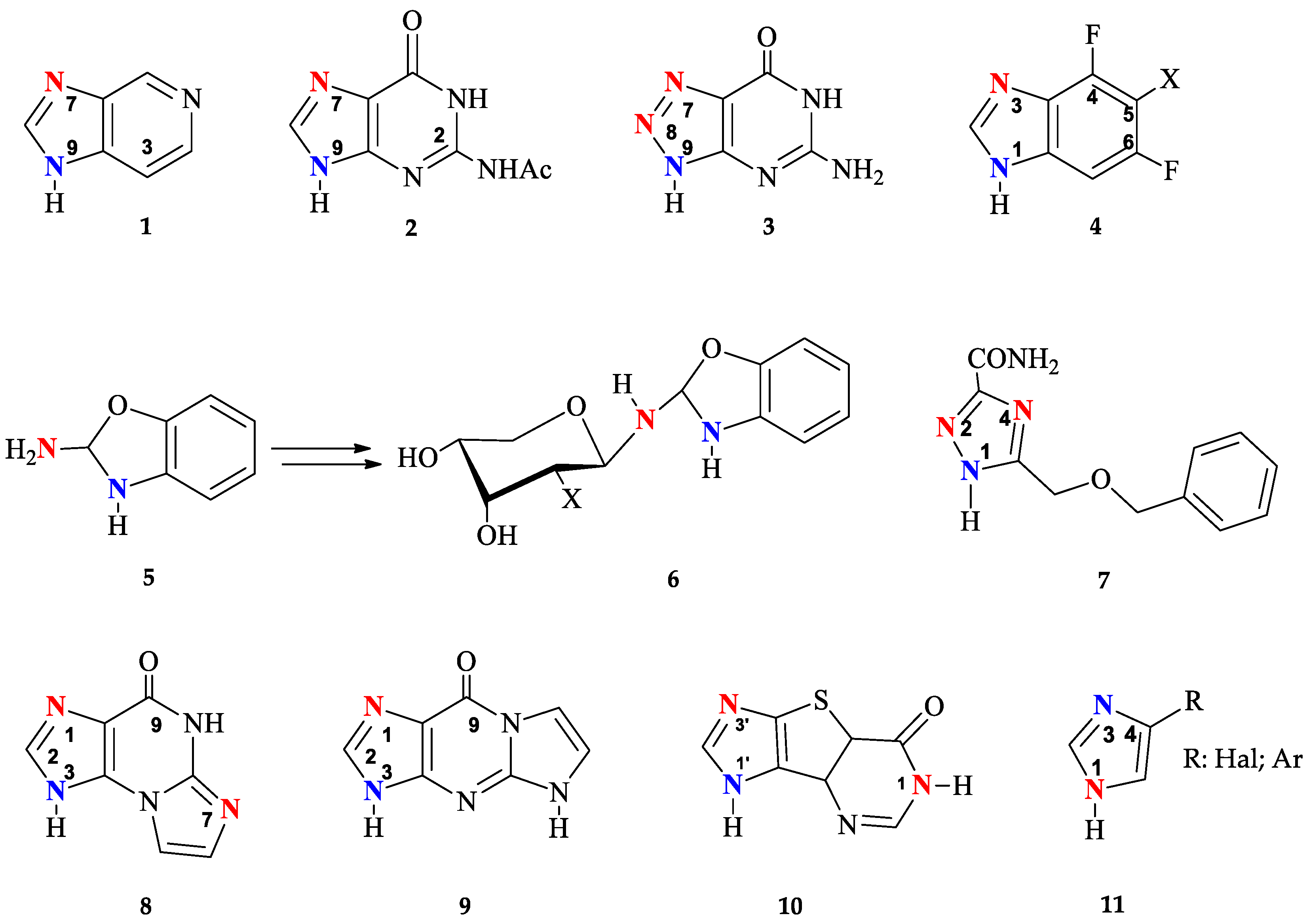
1. Introduction
2. Materials and Methods
2.1. General Information
2.2. Enzymatic Reactions
2.3. Isolation of Enzymatic Glycosylation Products
2.4. Modeling of the Active Site E. coli PNP
3. Results and Discussion
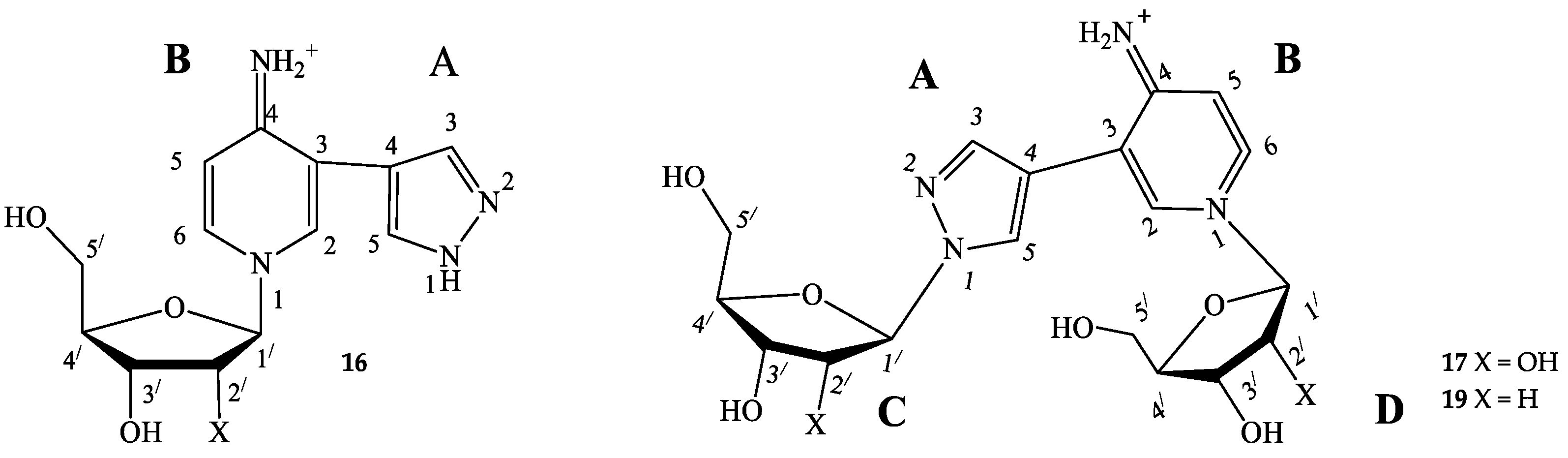
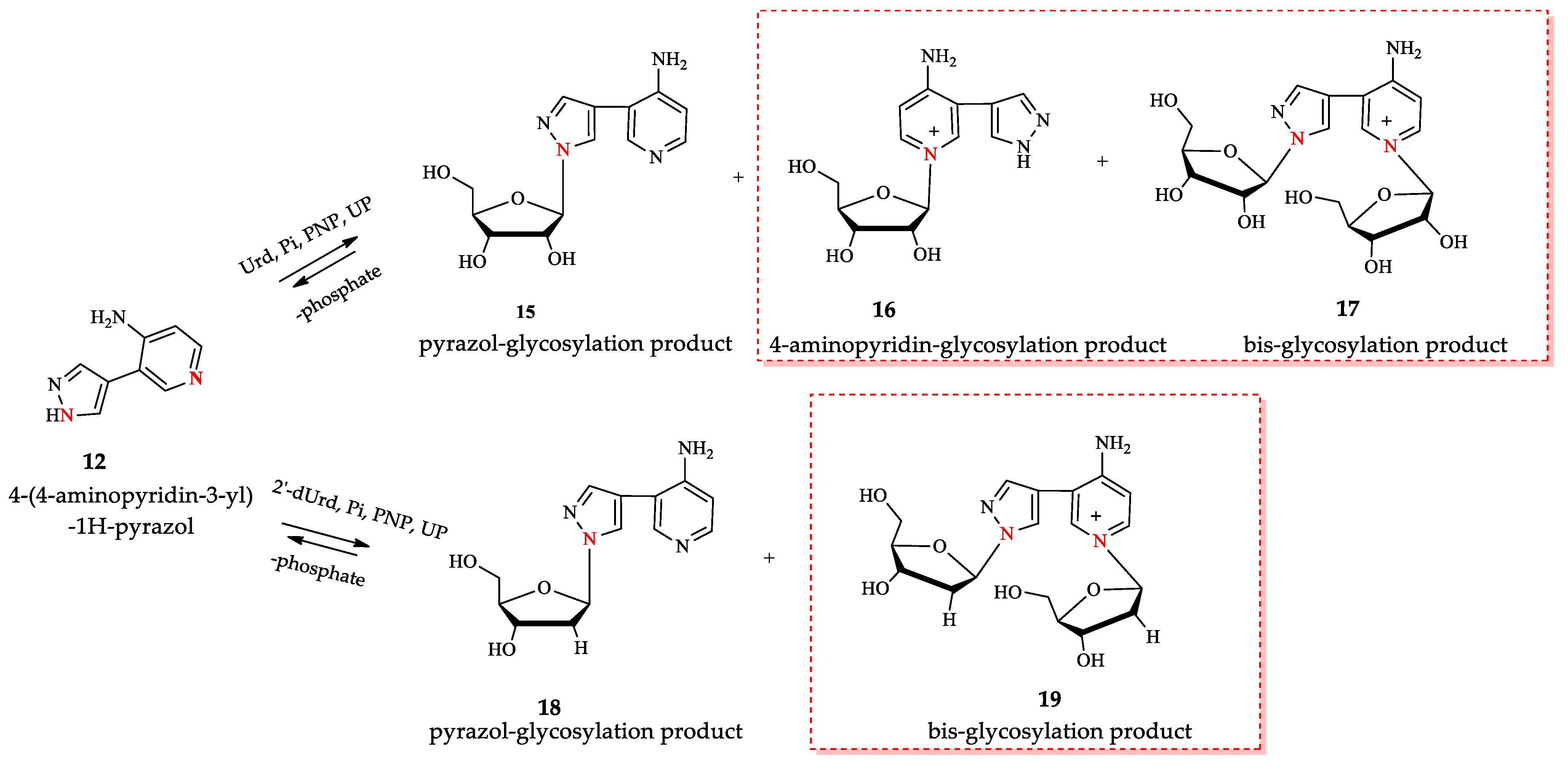
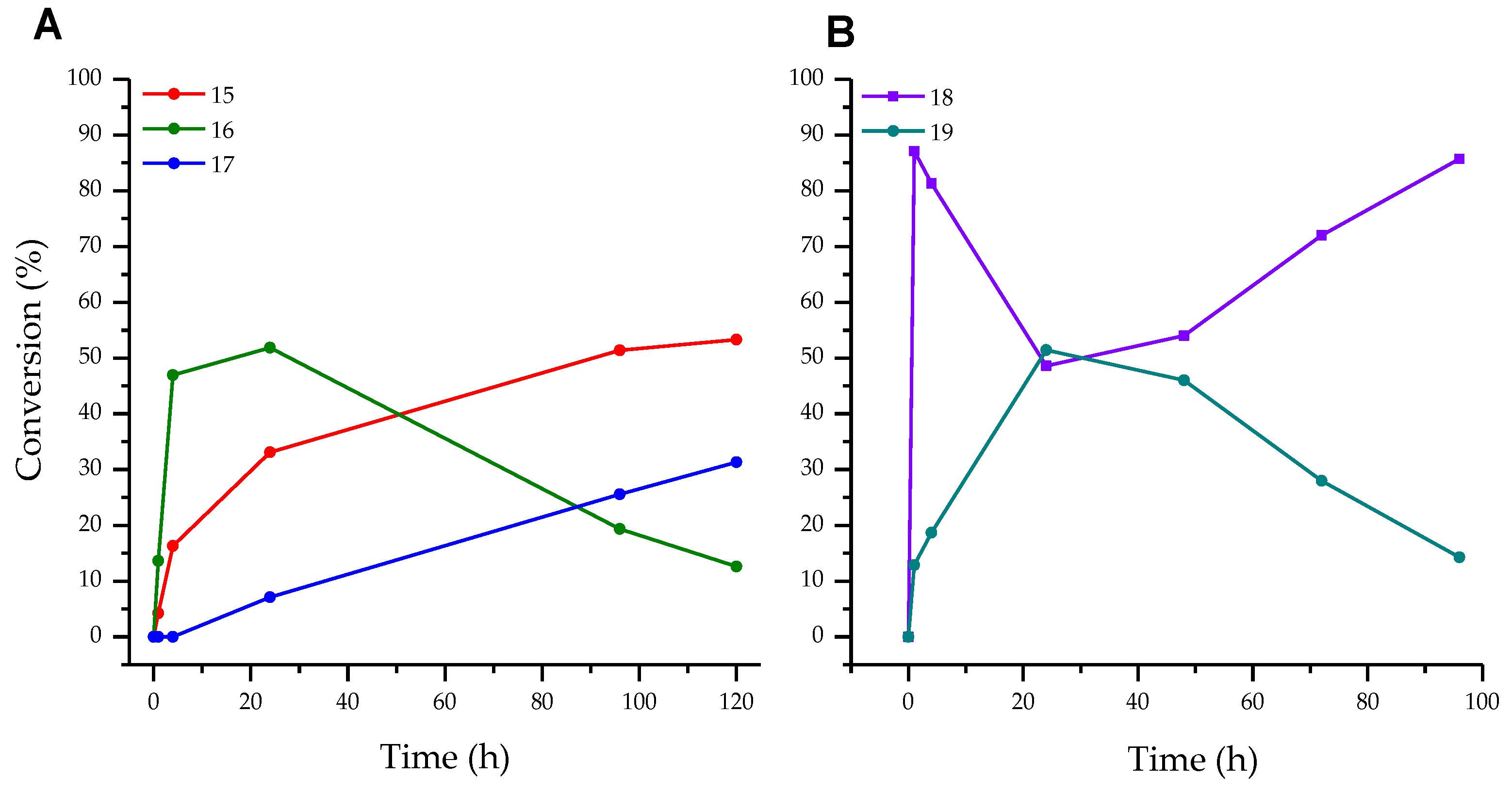
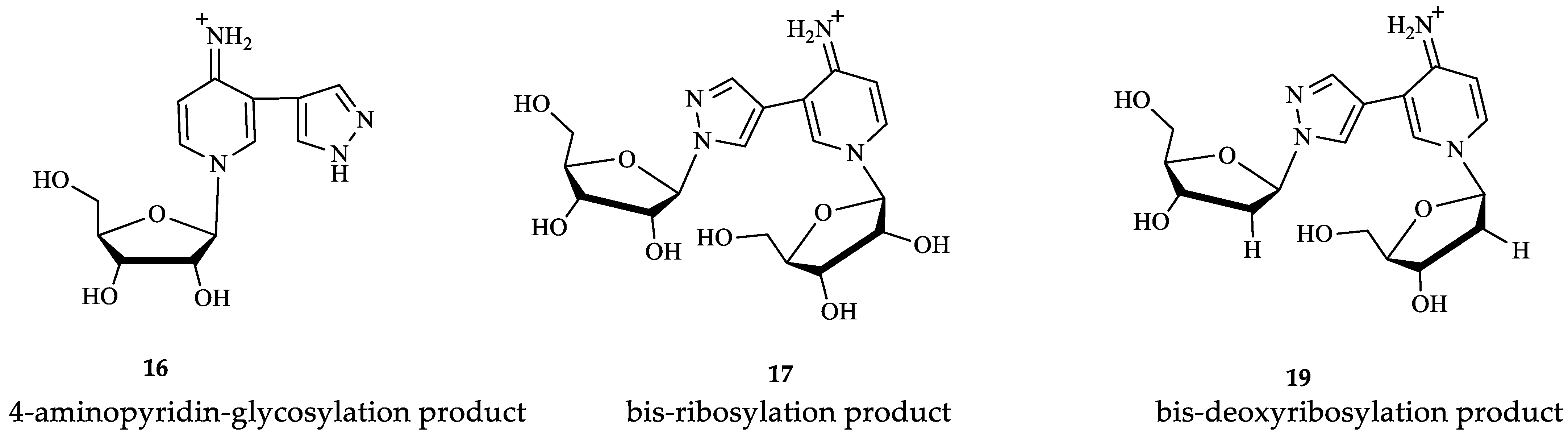
3.1. Enzymatic Transglycosylation of the Fleximer Heterocycle with the Formation of Isomer Products
3.2. Quantum Chemical Analysis
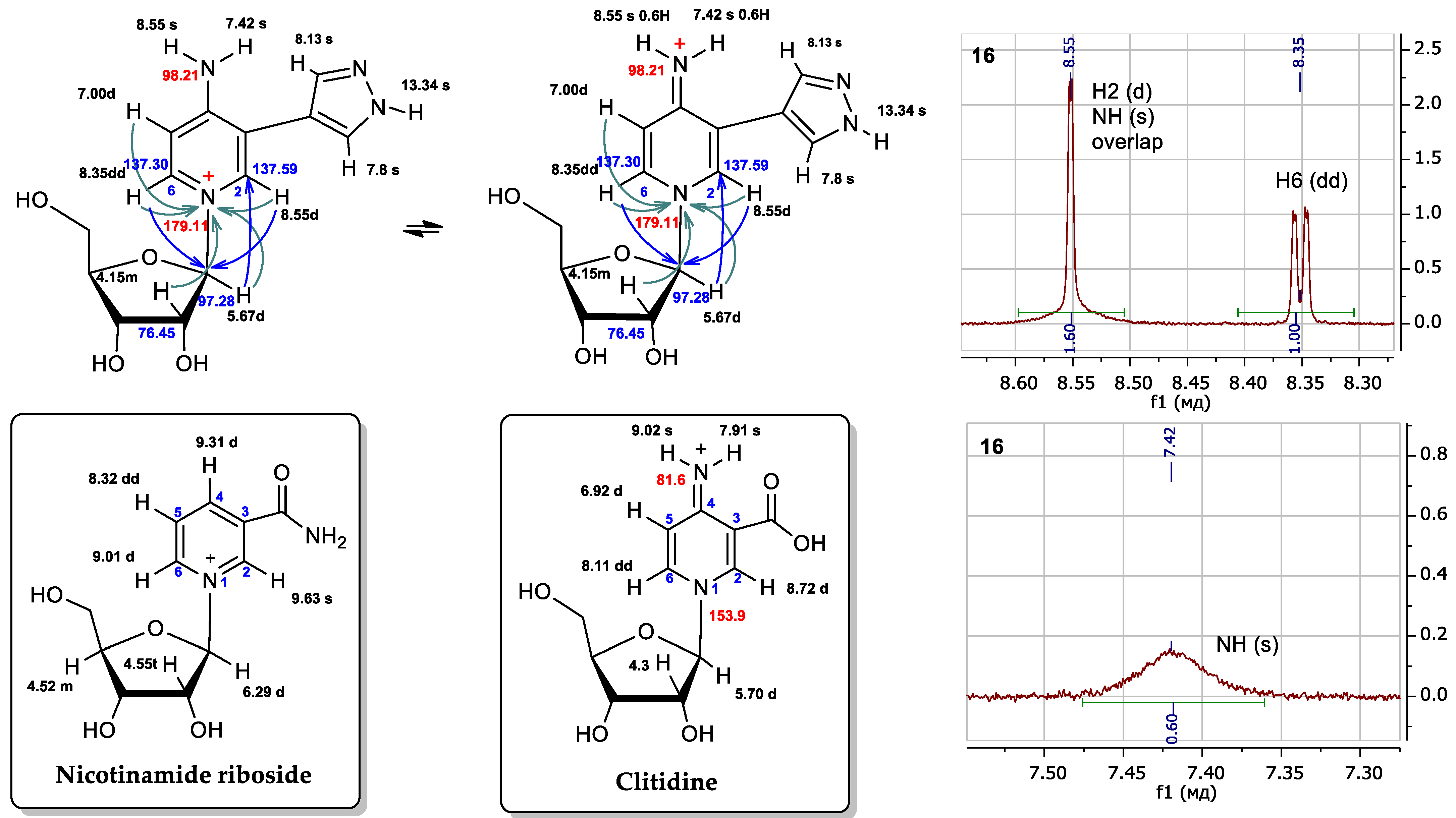
3.3. Structure Determination of Isomeric Nucleosides: Comparison of NMR Data of a Pyridinium Derivative with Known Compounds
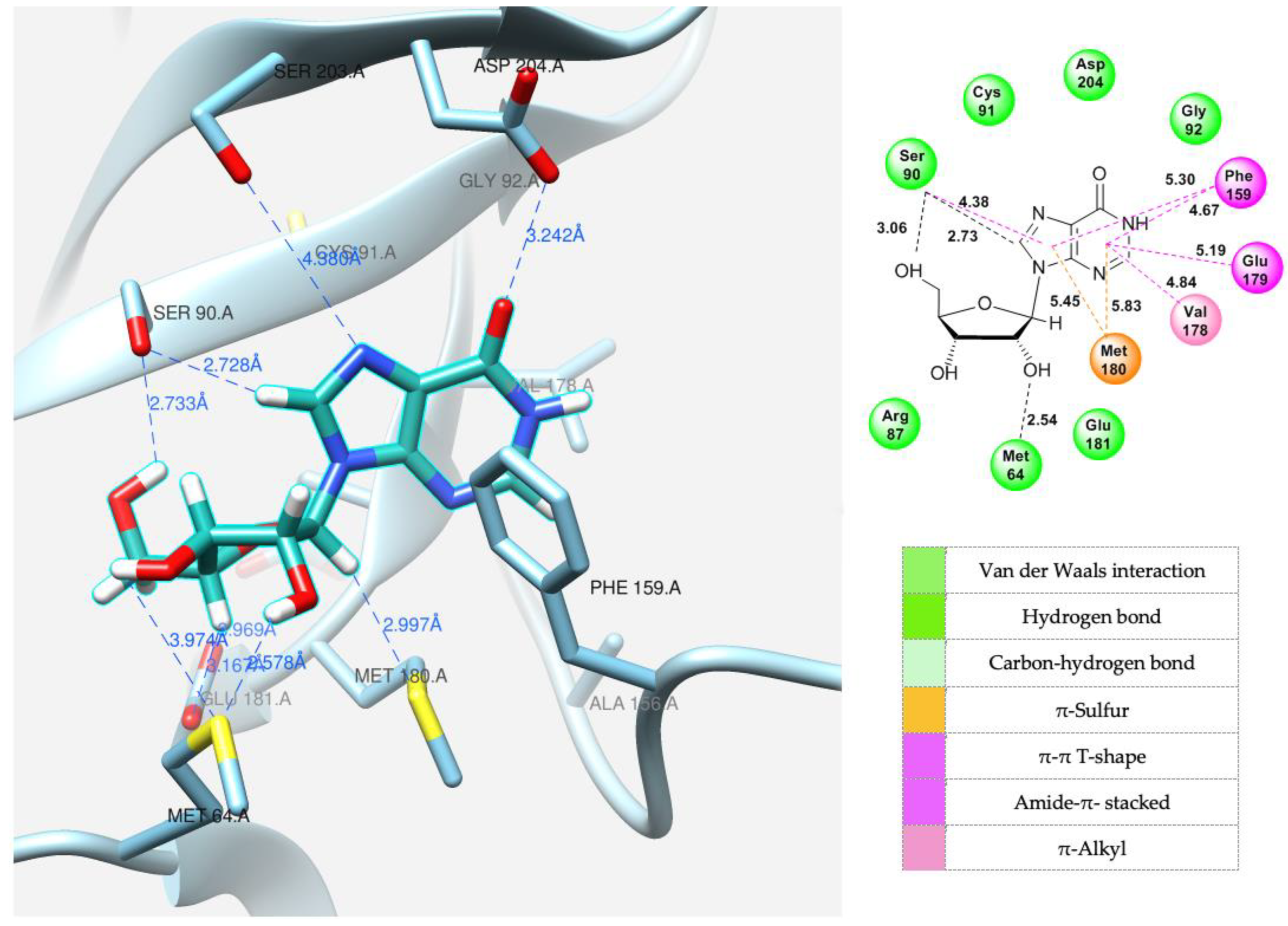
3.4. Modeling the Interaction of Fleximers in the Active Site of the E. coli PNP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinaldi, F.; Fernández-Lucas, J.; de La Fuente, D.; Zheng, C.; Bavaro, T.; Peters, B.; Massolini, G.; Annunziata, F.; Conti, P.; de la Mata, I.; et al. Immobilized enzyme reactors based on nucleoside phosphorylases and 2′-deoxyribosyltransferase for the in-flow synthesis of pharmaceutically relevant nucleoside analogues. Bioresour. Technol. 2020, 307, 123258. [Google Scholar] [CrossRef]
- Del Arco, J.; Alcántara, A.R.; Fernández-Lafuente, R.; Fernández-Lucas, J. Magnetic micro-macro biocatalysts applied to industrial bioprocesses. Bioresour. Technol. 2021, 322, 124547. [Google Scholar] [CrossRef]
- Calleri, E.; Cattaneo, G.; Rabuffetti, M.; Serra, I.; Bavaro, T.; Massolini, G.; Speranza, G.; Ubiali, D. Flow-synthesis of nucleosides catalyzed by an immobilized purine nucleoside phosphorylase from Aeromonas hydrophila: Integrated systems of reaction control and product purification. Adv. Synth. Catal. 2015, 357, 2520–2528. [Google Scholar] [CrossRef]
- Furihata, T.; Kishida, S.; Sugiura, H.; Kamiichi, A.; Iikura, M.; Chiba, K. Functional analysis of purine nucleoside phosphorylase as a key enzyme in ribavirin metabolism. Drug Metab. Pharmacokinet. 2014, 29, 211–214. [Google Scholar] [CrossRef]
- Kamel, S.; Thiele, I.; Neubauer, P.; Wagner, A. Thermophilic nucleoside phosphorylases: Their properties, characteristics and applications. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140304. [Google Scholar] [CrossRef]
- Liu, G.; Cheng, T.; Chu, J.; Li, S.; He, B. Efficient synthesis of purine nucleoside analogs by a new trimeric purine nucleoside phosphorylase from Aneurinibacillus migulanus AM007. Molecules 2020, 25, 100. [Google Scholar] [CrossRef]
- Mahor, D.; Priyanka, A.; Prasad, G.S.; Thakur, K.G. Functional and structural characterization of purine nucleoside phosphorylase from Kluyveromyces lactis and its potential applications in reducing purine content in food. PLoS ONE 2016, 11, e0164279. [Google Scholar] [CrossRef]
- Tomoike, F.; Kuramitsu, S.; Masui, R. Unique substrate specificity of purine nucleoside phosphorylases from Thermus thermophilus. Extremophiles 2013, 17, 505–514. [Google Scholar] [CrossRef]
- Xie, X.; Xia, J.; He, K.; Lu, L.; Xu, Q.; Chen, N. Low-molecular-mass purine nucleoside phosphorylase: Characterization and application in enzymatic synthesis of nucleoside antiviral drugs. Biotechnol. Lett. 2011, 33, 1107–1112. [Google Scholar] [CrossRef]
- Zhu, S.; Song, D.; Gong, C.; Tang, P.; Li, X.; Wang, J.; Zheng, G. Biosynthesis of nucleoside analogues via thermostable nucleoside phosphorylase. Appl. Microbiol. Biotechnol. 2013, 97, 6769–6778. [Google Scholar] [CrossRef]
- Doskočil, J.; Holý, A. Specificity of purine nucleoside phosphorylase from Escherichia coli. Collect. Czechoslov. Chem. Commun. 1977, 42, 370–383. [Google Scholar] [CrossRef]
- Koellner, G.; Bzowska, A.; Wielgus-Kutrowska, B.; Luić, M.; Steiner, T.; Saenger, W.; Stepiński, J. Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism. J. Mol. Biol. 2002, 315, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Kamel, S.; Yehia, H.; Neubauer, P.; Wagner, A. Enzymatic synthesis of nucleoside analogues by nucleoside phosphorylases. In Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives; Fernandez-Lucas, J., Camarasa-Rius, M.J., Eds.; Wiley-VCH: New York, NY, USA, 2019; Chapter 1. [Google Scholar]
- Roivainen, J.; Elizarova, T.; Lapinjoki, S.; Mikhailopulo, I.A.; Esipov, R.S.; Miroshnikov, A.I. An enzymatic transglycosylation of purine bases. Nucleosides Nucleotides Nucleic Acids 2007, 26, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Lapponi, M.J.; Rivero, C.W.; Zinni, M.A.; Britos, C.N.; Trelles, J.A. New developments in nucleoside analogues biosynthesis: A review. J. Mol. Catal. B Enzym. 2016, 133, 218–233. [Google Scholar] [CrossRef]
- Bennett, E.M.; Li, C.; Allan, P.W.; Parker, W.B.; Ealick, S.E. Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase. J. Biol. Chem. 2003, 278, 47110–47118. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ma, L. Enzymatic synthesis of fluorinated compounds. Appl. Microbiol. Biotechnol. 2021, 105, 8033–8058. [Google Scholar] [CrossRef]
- Hassan, A.E.; Abou-Elkhair, R.A.; Riordan, J.M.; Allan, P.W.; Parker, W.B.; Khare, R.; Waud, W.R.; Montgomery, J.A.; Secrist, J.A. Synthesis and evaluation of the substrate activity of C-6 substituted purine ribosides with E. coli purine nucleoside phosphorylase: Palladium mediated cross-coupling of organozinc halides with 6-chloropurine nucleosides. Eur. J. Med. Chem. 2012, 47, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.E.; Abou-Elkhair, R.A.; Parker, W.B.; Allan, P.W.; Secrist, J.A. 6-Methylpurine derived sugar modified nucleosides: Synthesis and evaluation of their substrate activity with purine nucleoside phosphorylases. Bioorg. Chem. 2016, 65, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Oslovsky, V.E.; Solyev, P.N.; Polyakov, K.M.; Alexeev, C.S.; Mikhailov, S.N. Chemoenzymatic synthesis of cytokinins from nucleosides: Ribose as a blocking group. Org. Biomol. Chem. 2018, 16, 2156–2163. [Google Scholar] [CrossRef]
- Eletskaya, B.Z.; Gruzdev, D.A.; Krasnov, V.P.; Levit, G.L.; Kostromina, M.A.; Paramonov, A.S.; Kayushin, A.L.; Muzyka, I.S.; Muravyova, T.I.; Esipov, R.S.; et al. Enzymatic synthesis of novel purine nucleosides bearing a chiral benzoxazine fragment. Chem. Biol. Dug Des. 2019, 93, 605–616. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J. Chemo-enzymatic generation of highly fluorescent nucleoside analogs using purine-nucleoside phosphorylase. Biomolecules 2024, 14, 701. [Google Scholar] [CrossRef] [PubMed]
- Matyugina, E.S.; Kochetkov, S.N.; Khandazhinskaya, A.L. Synthesis and biological activity of aza and deaza analogues of purine nucleosides. Russ. Chem. Rev. 2021, 90, 1454. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Bzowska, A.; Wielgus-Kutrowska, B. Site-selective ribosylation of fluorescent nucleobase analogs using purine-nucleoside phosphorylase as a catalyst effects of point mutations. Molecules 2016, 21, 44. [Google Scholar] [CrossRef]
- Krenitsky, T.A.; Rideout, J.L.; Chao, E.Y.; Koszalka, G.W.; Gurney, F.; Crouch, R.C.; Cohn, T.N.; Wolberg, G.; Vinegar, R. Imidazo[4,5-c]pyridines (3-deazapurines) and their nucleosides as immunosuppressive and anti-inflammatory agents. J. Med. Chem. 1986, 29, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.S.; Berzina, M.Y.; Fateev, I.V.; Eletskaya, B.Z.; Kostromina, M.A.; Kaushin, A.L.; Paramonov, A.S.; Prutkov, A.N.; Matveev, A.V.; Grebenkina, L.E.; et al. Chemo-enzymatic synthesis of 5-substituted ribavirin analogs: Unexpected cooperative effect in the interaction of 5-alkyloxymethyl 1,2,4-triazol-3-carboxamides with E. coli purine nucleoside phosphorylase active site. Sustain. Chem. Pharm. 2022, 30, 100881. [Google Scholar] [CrossRef]
- El-Sebaey, S.A. Recent advances in 1,2,4-triazole scaffolds as antiviral agents. ChemistrySelect 2020, 5, 11654–11680. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A.; Kazimierczuk, Z.; Zinchenko, A.I.; Barai, V.N.; Romanova, V.V.; Eroshevskaya, L.A. Benzimidazoles in the reaction of enzymatic transglycosylation. Nucleosides Nucleotides 1995, 14, 3–5. [Google Scholar] [CrossRef]
- Kharitonova, M.I.; Fateev, I.V.; Kayushin, A.L.; Konstantinova, I.D.; Kotovskaya, S.K.; Andronova, V.L.; Galegov, G.A.; Charushin, V.N.; Miroshnikov, A.I. Chemoenzymatic synthesis and antiherpes activity of 5-substituted 4, 6-difluorobenzimidazoles ribo-and 2′-deoxyribonucleosides. Synthesis 2015, 48, 394–406. [Google Scholar] [CrossRef]
- Kharitonova, M.I.; Denisova, A.O.; Andronova, V.L.; Kayushin, A.L.; Konstantinova, I.D.; Kotovskaya, S.K.; Galegov, G.A.; Charushin, V.N.; Miroshnikov, A.I. New modified 2-aminobenzimidazole nucleosides: Synthesis and evaluation of their activity against herpes simplex virus type 1. Bioorg. Med. Chem. Lett. 2017, 27, 2484–2487. [Google Scholar] [CrossRef]
- Vichier-Guerre, S.; Dugué, L.; Bonhomme, F.; Pochet, S. An expedient synthesis of flexible nucleosides via a regio-controlled enzymatic glycosylation of functionalized imidazoles. Org. Biomol. Chem. 2017, 15, 8193–8203. [Google Scholar] [CrossRef]
- Vichier-Guerre, S.; Ku, T.R.C.; Pochet, S.; Seley-Radtke, K.L. An expedient synthesis of flexible nucleosides through enzymatic glycosylation of proximal and distal fleximer bases. Chembiochem 2020, 21, 1412–1417. [Google Scholar] [CrossRef]
- Chudinov, M.V. Nucleoside analogs with fleximer nucleobase. Chem. Heterocycl. Compd. 2020, 56, 636–643. [Google Scholar] [CrossRef]
- Ye, W.; Paul, D.; Gao, L.; Seckute, J.; Sangaiah, R.; Jayaraj, K.; Zhang, Z.; Kaminski, P.A.; Ealick, S.E.; Gold, A.; et al. Ethenoguanines undergo glycosylation by nucleoside 2′-deoxyribosyltransferases at non-natural sites. PLoS ONE 2014, 9, e115082. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Górka, M.; Bzowska, A.; Wielgus-Kutrowska, B. Tri-cyclic nucleobase analogs and their ribosides as substrates of purine-nucleoside phosphorylases. II Guanine and isoguanine derivatives. Molecules 2019, 24, 1493. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Bzowska, A.; Wielgus-Kutrowska, B. Tricyclic nitrogen base 1, N6-ethenoadenine and its ribosides as substrates for purine-nucleoside phosphorylases: Spectroscopic and kinetic studies. Nucleosides Nucleotides Nucleic Acids 2018, 37, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Konstantinova, I.D.; Antonov, K.V.; Fateev, I.V.; Miroshnikov, A.I.; Stepchenko, V.A.; Baranovsky, A.V.; Mikhailopulo, I.A. A chemo-enzymatic synthesis of β-d-arabinofuranosyl purine nucleosides. Synthesis 2011, 2011, 1555–1560. [Google Scholar] [CrossRef][Green Version]
- Denisova, A.O.; Tokunova, Y.A.; Fateev, I.V.; Breslav, A.A.; Leonov, V.N.; Dorofeeva, E.V.; Lutonina, O.I.; Muzyka, I.S.; Esipov, R.S.; Kayushin, A.L.; et al. The chemoenzymatic synthesis of 2-chloro-and 2-fluorocordycepins. Synthesis 2017, 49, 4853–4860. [Google Scholar] [CrossRef]
- Fateev, I.V.; Antonov, K.V.; Konstantinova, I.D.; Muravyova, T.I.; Seela, F.; Esipov, R.S.; Miroshnikov, A.I.; Mikhailopulo, I.A. The chemoenzymatic synthesis of clofarabine and related 2′-deoxyfluoroarabinosyl nucleosides: The electronic and stereochemical factors determining substrate recognition by E. coli nucleoside phosphorylases. Beilstein J. Org. Chem. 2014, 10, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Cook, W.J.; Zhou, M.; Koszalka, G.W.; Krenitsky, T.A.; Ealick, S.E. The crystal structure of Escherichia coli purine nucleoside phosphorylase: A comparison with the human enzyme reveals a conserved topology. Structure 1997, 5, 1373–1383. [Google Scholar] [CrossRef]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Ther. 2000, 88, 349–425. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A. Biotechnology of nucleic acid constituents—State of the art and perspectives. Curr. Org. Chem. 2007, 11, 317–335. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Wielgus-Kutrowska, B.; Mikleušević, G. Enzymatic synthesis of highly fluorescent 8-azapurine ribosides using a purine nucleoside phosphorylase reverse reaction: Variable ribosylation sites. Molecules 2013, 18, 12587–12598. [Google Scholar] [CrossRef]
- Kharitonova, M.I.; Antonov, K.V.; Fateev, I.V.; Berzina, M.Y.; Kaushin, A.L.; Paramonov, A.S.; Kotovskaya, S.K.; Andronova, V.L.; Konstantinova, I.D.; Galegov, G.A.; et al. Chemoenzymatic synthesis of modified 2′-deoxy-2′-fluoro-β-d-arabinofuranosyl benzimidazoles and evaluation of their activity against herpes simplex virus type 1. Synthesis 2017, 49, 1043–1052. [Google Scholar] [CrossRef]
- Fateev, I.V.; Kharitonova, M.I.; Antonov, K.V.; Konstantinova, I.D.; Stepanenko, V.N.; Esipov, R.S.; Seela, F.; Temburnikar, K.W.; Seley-Radtke, K.L.; Stepchenko, V.A.; et al. Recognition of artificial nucleobases by E. coli purine nucleoside phosphorylase versus its Ser90Ala mutant in the synthesis of base-modified nucleosides. Chemistry 2015, 21, 13401–13419. [Google Scholar] [CrossRef] [PubMed]
- Khandazhinskaya, A.; Eletskaya, B.; Fateev, I.; Kharitonova, M.; Konstantinova, I.; Barai, V.; Azhayev, A.; Hyvonen, M.T.; Keinanen, T.A.; Kochetkov, S.; et al. Novel fleximer pyrazole-containing adenosine analogues: Chemical, enzymatic and highly efficient biotechnological synthesis. Org. Biomol. Chem. 2021, 19, 7379–7389. [Google Scholar] [CrossRef] [PubMed]
- Esipov, R.S.; Gurevich, A.I.; Chuvikovsky, D.V.; Chupova, L.A.; Muravyova, T.I.; Miroshnikov, A.I. Overexpression of Escherichia coli genes encoding nucleoside phosphorylases in the pET/Bl21 (DE3) system yields active recombinant enzymes. Protein Expr. Purif. 2002, 24, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Timofeev, V.I.; Zhukhlistova, N.E.; Abramchik, Y.A.; Fateev, I.I.; Kostromina, M.A.; Muravieva, T.I.; Esipov, R.S.; Kuranova, I.P. Crystal structure of Escherichia coli purine nucleoside phosphorylase in complex with 7-deazahypoxanthine. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Froimowitz, M. HyperChem: A software package for computational chemistry and molecular modeling. Biotechniques 1993, 14, 1010–1013. [Google Scholar] [PubMed]
- Wielgus-Kutrowska, B.; Kulikowska, E.; Wierzchowski, J.; Bzowska, A.; Shugar, D. Nicotinamide riboside, an unusual, non-typical, substrate of purified purine-nucleoside phosphorylases. Eur. J. Biochem. 1997, 243, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Shirahama, H.; Koshino, H.; Uzawa, J.; Yamano, K.; Konno, K.; Nakatsu, K. Tautomerism of clitidine, a pyridine nucleoside from the poisonous mushroom clitocybe acromelalga. Heterocycles 1998, 2, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Timofeev, V.I.; Zhukhlistova, N.E.; Abramchik, Y.A.; Muravieva, T.I.; Esipov, R.S.; Kuranova, I.P. Crystal structure of Escherichia coli purine nucleoside phosphorylase complexed with acyclovir. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 402–409. [Google Scholar] [CrossRef] [PubMed]


| pH 5–6 | pH 7 | pH 8–9 |
|---|---|---|
 |  |  |
 |  | 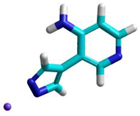 |
| ET = −329,907.8 kcal/mol | ET = −329,688.9 kcal/mol | ET = −430,902.4 kcal/mol; |
| N1 = −0.4000 e N2 = −0.2590 e | N1 = −0.4221 e N2 = −0.3061 e | N1 = −0.4150 e N2 = −0.4130 e |
| NH3 = −0.6600 e | NH2 = −0.7620 e | NH2 = −0.7610 e |
| Narom Py = −0.5010 e | Narom Py = −0.5759 e | Narom Py = −0.5803 e |
| - | - | - |
| C3-C4-C3Py-C4Py −61.7° | C3-C4-C3Py-C4Py −51.0° | C3-C4-C3Py-C4Py −46.2° |
| - | - | - |
| N1<—>NH2 4.302 Å | N1<—>NH2 4.830 Å | N1<—>NH2 4.828 Å |
| N2<—>Nar/para 5.714 Å | N2<—>Nar/para 5.690 Å | N2<—>NH2 4.316 Å |
| - | N1<—>Narom 5.676 Å | |
| - | N2<—>Narom 5.921 Å |
| Pyrazole Riboside 15 | Pyridine Riboside 16 |
|---|---|
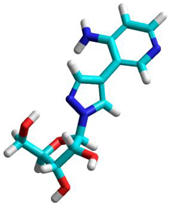 |  |
| ET = −639,329.0 kcal/mol | ET = −639,302.8 kcal/mol |
| ΔET = −26.2 kcal/mol | ΔET = +26.2 kcal/mol |
| C3′-exo; DM 4.316 Debye’s | C2′-endo/C1′-exo (twist); DM 8.833 Debye’s |
| Narom/para = −0.5757 e | N1 C1′Rib = −0.7586 e; NH = −0.6792 e; |
| N2<—>NH2 4.2685Å | NH = −0.4229 e; N = −0.3087 e |
| N2<—> NPy 5.9227 Å | O4′-C1′-N-C1″H +44° |
| O4′-C1′-N1-N2 +68.8° | O4′-C1′-N-C6″H −142° |
| C3-C4-C3Py-C4Py −48.6° | N3H-C4-C3″-C4″HNH −115° |
| - | N3H-C4-C3″-C2″HN +62° |
| Model | Cluster | Energy (Kcal/mol) | ΔG (Kcal/mol) | Contacting Amino Acid Residues with Interaction Type | ||
|---|---|---|---|---|---|---|
| Hydrogen Bond | π–π Interaction | |||||
| Figure 10 | Inosine | 7 | 13.56 | −6.82 | Ser90, Met64 | Met180, Phe159, Val178, Glu179 |
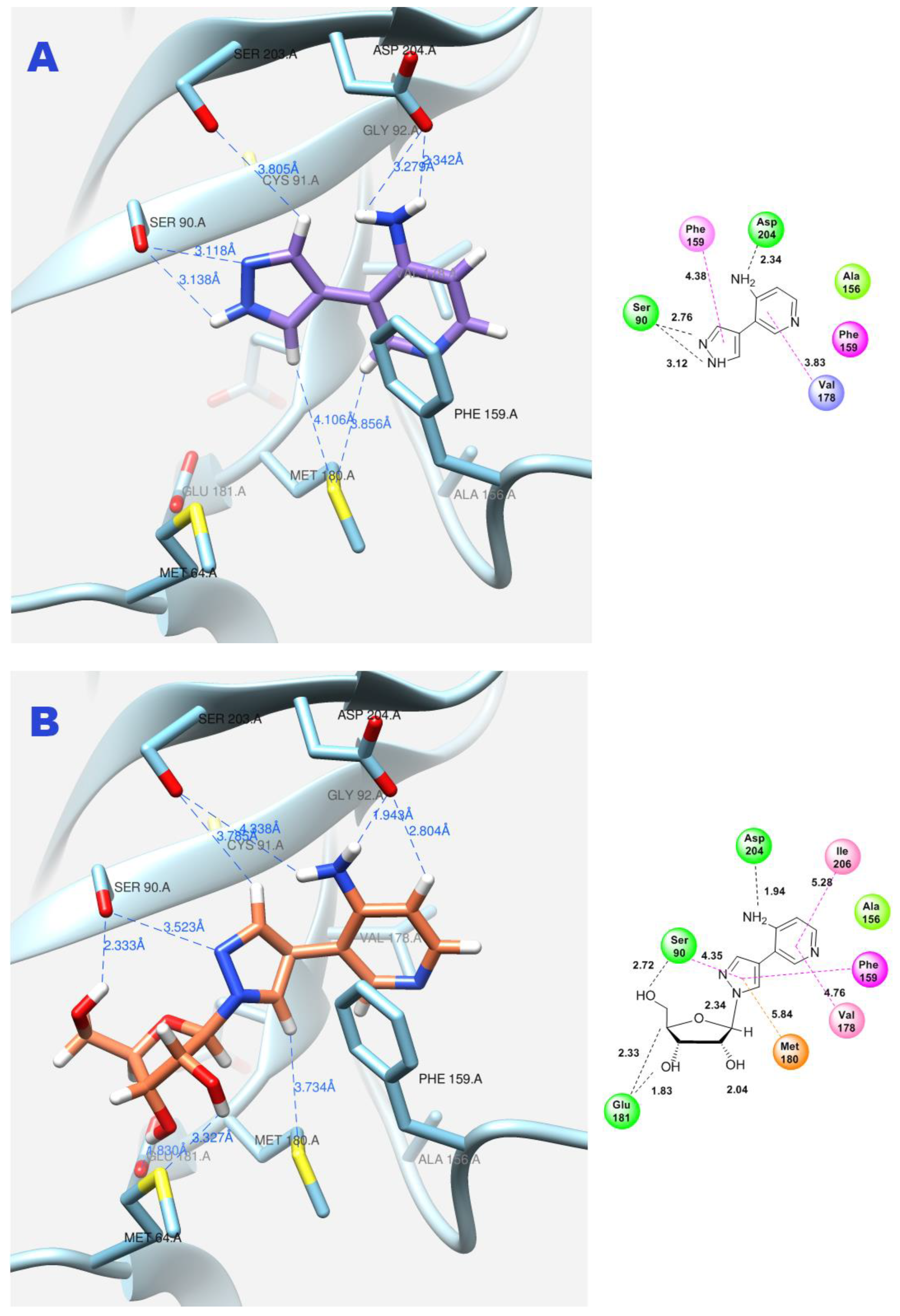
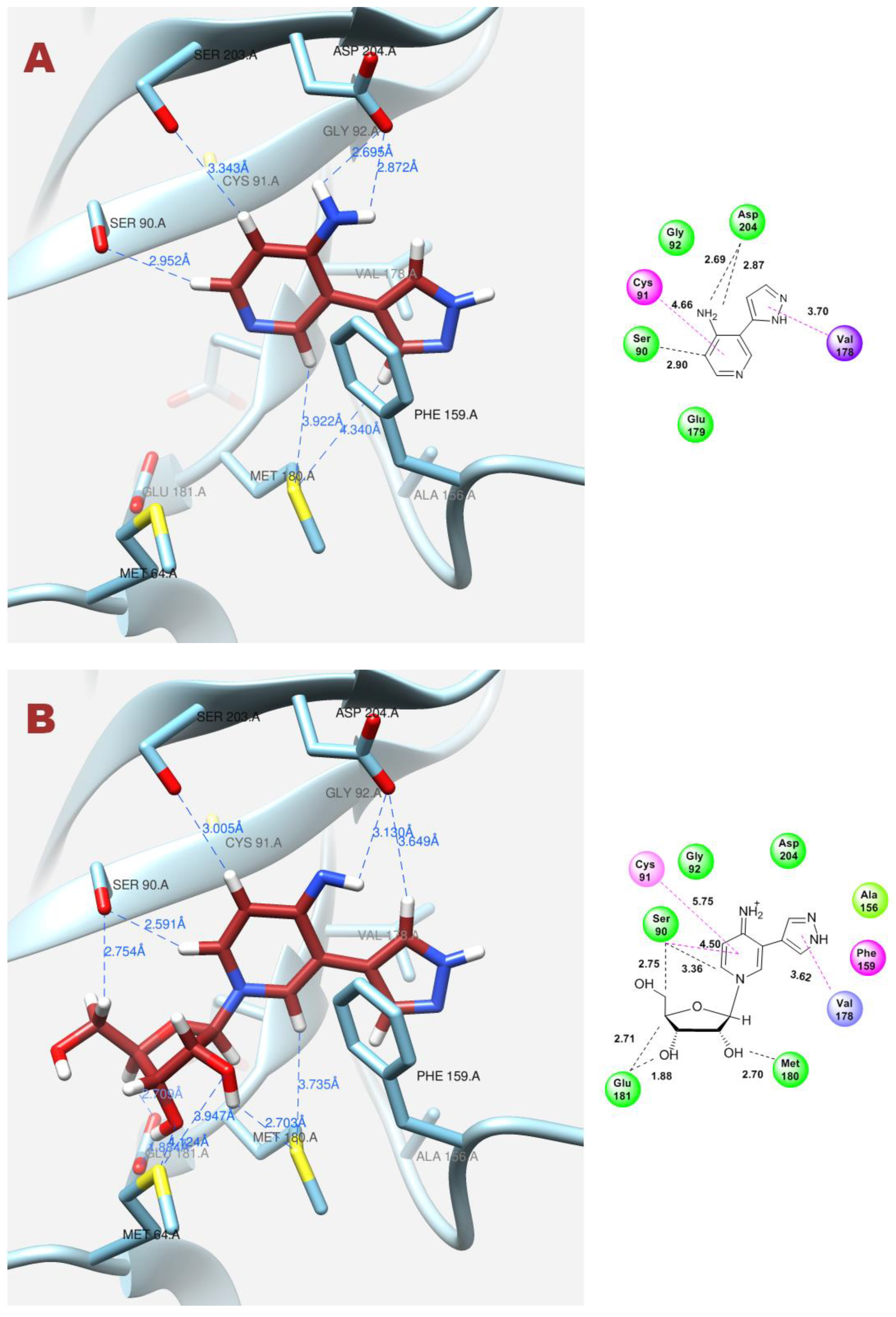
| Figure 11A | Fleximer base 12 | 3 | 3.76 | −6.55 | Ser90, Asp204 | Cys91, Val178 |
| Figure 11B | Pyrazole riboside 15 | 3 | 63.71 | −7.07 | Ser90, Cys91, Glu181, Asp204 | Phe159, Val179, Met 180, Ile206 |
| Figure 12A | Fleximer base 12 | 4 | 7.00 | −6.07 | Ser90, Asp204 | Cys91, Val178 |
| Figure 12B | Pyridinium riboside 16 | 6 | 12.99 | −7.80 | Ser90, Met180, Glu181 | Cys91, Val178 |
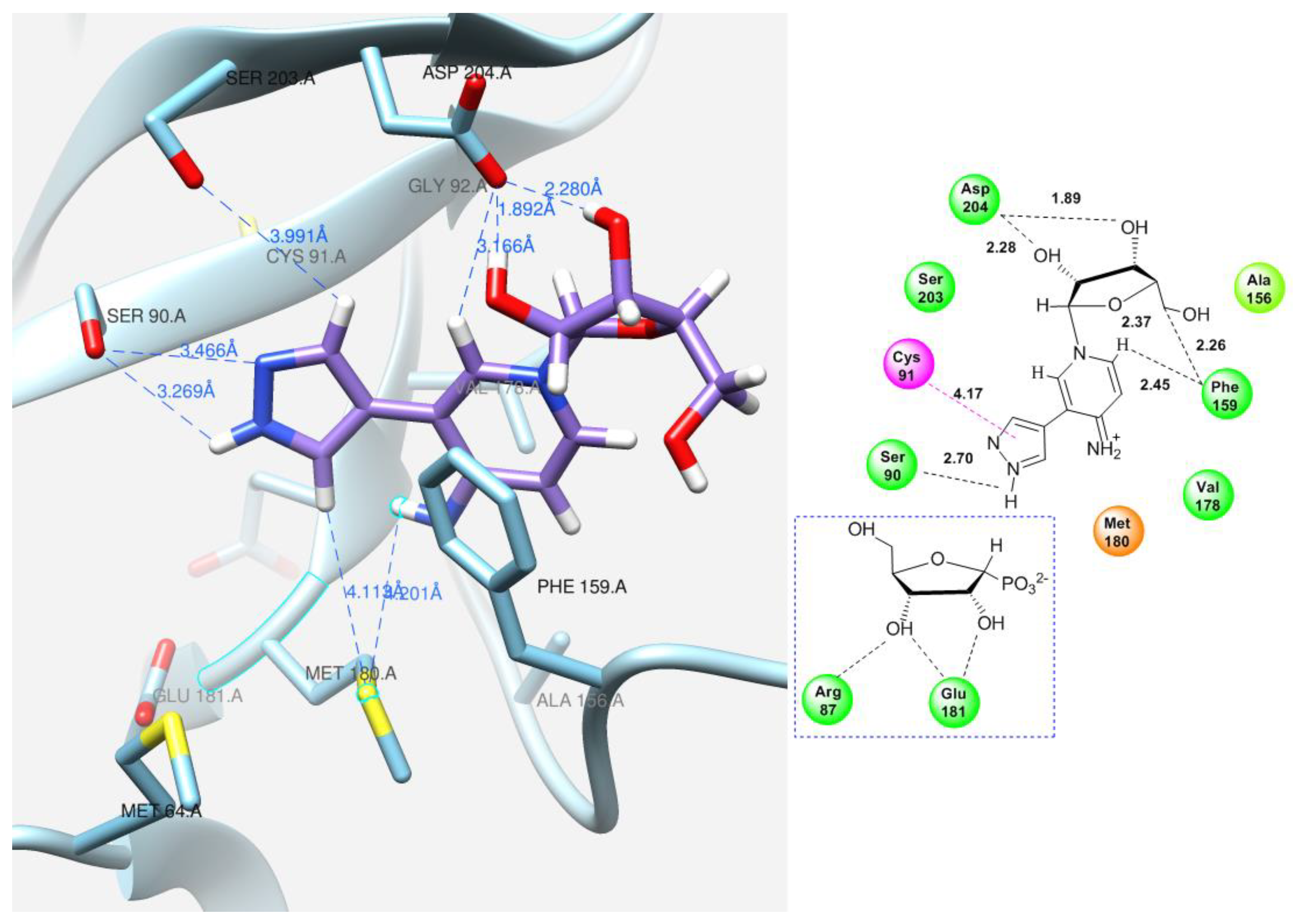
| Figure 13 | Pyridinium riboside 16 | 0 | 25.75 | −7.60 | Ser90, Phe159, Asp204 | Cys91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eletskaya, B.Z.; Mironov, A.F.; Fateev, I.V.; Berzina, M.Y.; Antonov, K.V.; Smirnova, O.S.; Zatsepina, A.B.; Arnautova, A.O.; Abramchik, Y.A.; Paramonov, A.S.; et al. Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products. Biomolecules 2024, 14, 798. https://doi.org/10.3390/biom14070798
Eletskaya BZ, Mironov AF, Fateev IV, Berzina MY, Antonov KV, Smirnova OS, Zatsepina AB, Arnautova AO, Abramchik YA, Paramonov AS, et al. Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products. Biomolecules. 2024; 14(7):798. https://doi.org/10.3390/biom14070798
Chicago/Turabian StyleEletskaya, Barbara Z., Anton F. Mironov, Ilya V. Fateev, Maria Ya. Berzina, Konstantin V. Antonov, Olga S. Smirnova, Alexandra B. Zatsepina, Alexandra O. Arnautova, Yulia A. Abramchik, Alexander S. Paramonov, and et al. 2024. "Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products" Biomolecules 14, no. 7: 798. https://doi.org/10.3390/biom14070798
APA StyleEletskaya, B. Z., Mironov, A. F., Fateev, I. V., Berzina, M. Y., Antonov, K. V., Smirnova, O. S., Zatsepina, A. B., Arnautova, A. O., Abramchik, Y. A., Paramonov, A. S., Kayushin, A. L., Khandazhinskaya, A. L., Matyugina, E. S., Kochetkov, S. N., Miroshnikov, A. I., Mikhailopulo, I. A., Esipov, R. S., & Konstantinova, I. D. (2024). Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products. Biomolecules, 14(7), 798. https://doi.org/10.3390/biom14070798









