The Next Chapter in Cancer Diagnostics: Advances in HPV-Positive Head and Neck Cancer
Abstract
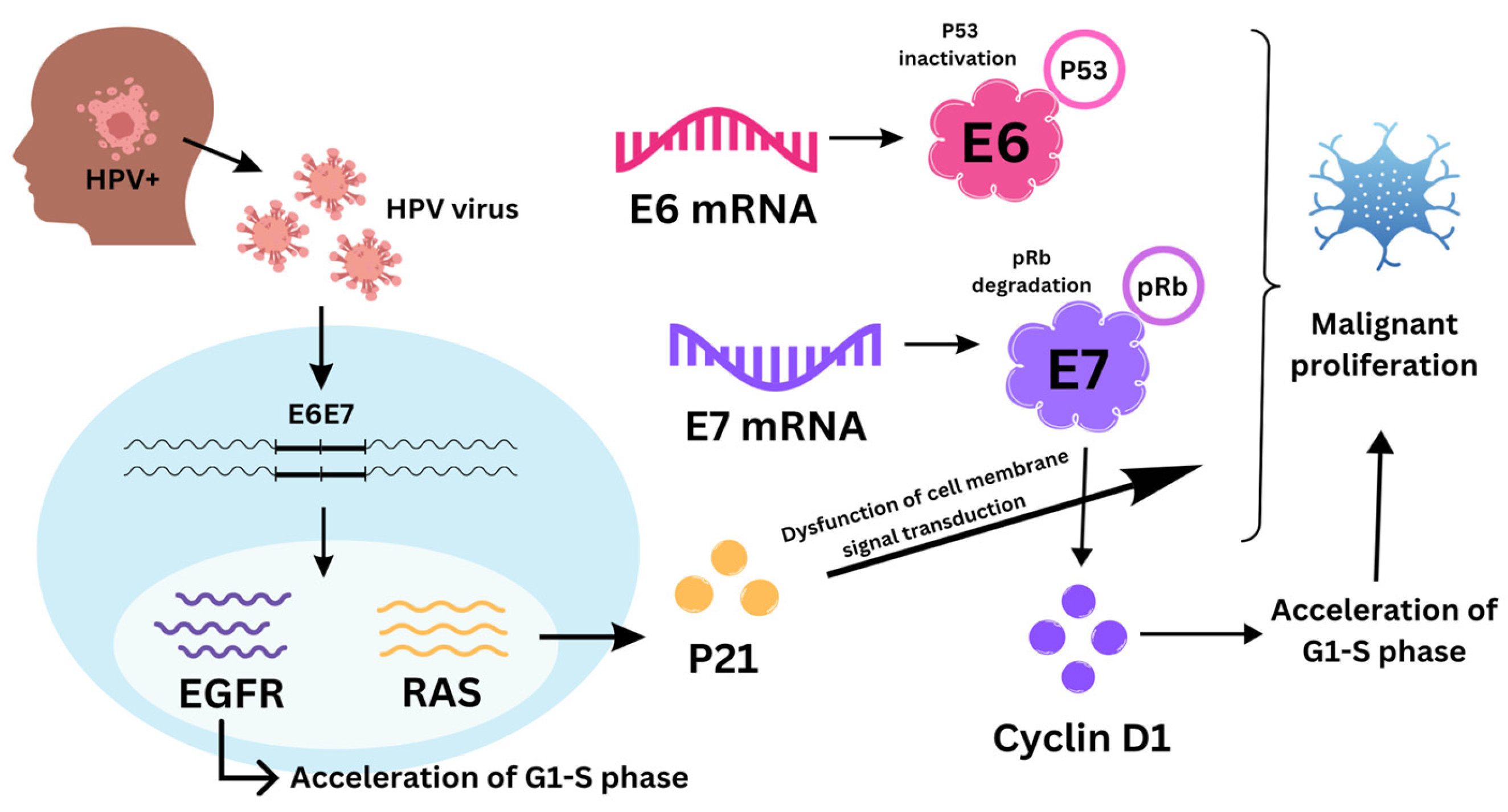
1. Introduction
2. Direct HPV Examination in Tumor Tissue
2.1. DNA-Based Detection
2.1.1. Polymerase Chain Reaction (PCR)
2.1.2. Next-Generation Sequencing (NGS)
2.2. RNA-Based Detection
2.2.1. MicroRNAs
2.2.2. E6/E7 Detection: Blood- and Saliva-Based HPV Biomarkers and Viral Integration
3. Indirect HPV Testing in Tumor Tissue
3.1. Protein-Based Markers
3.1.1. p16 Immunohistochemistry (IHC) and p16 Overexpression
3.1.2. Cyclin D1
3.1.3. Estrogen Receptor Positivity
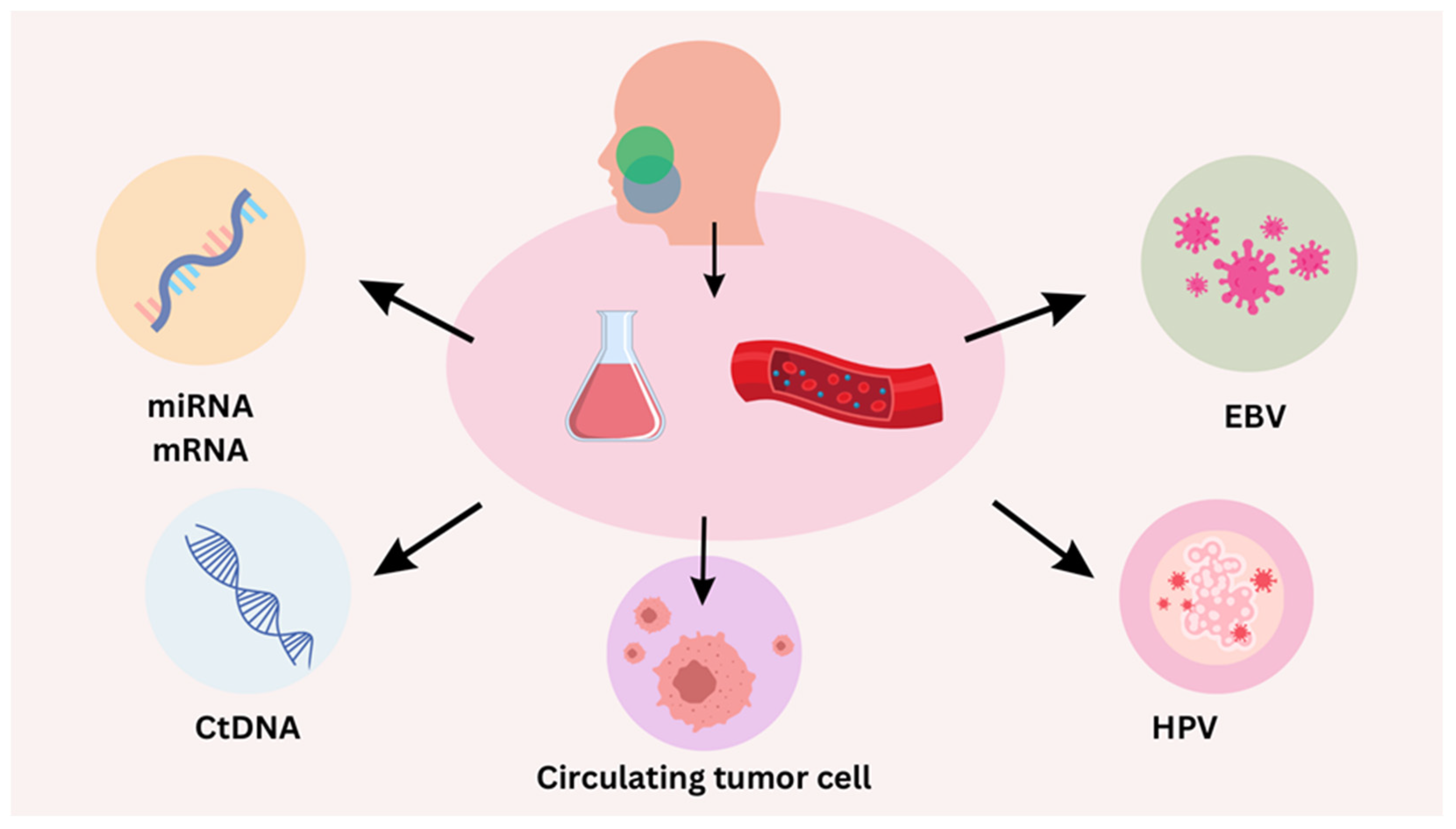
4. Liquid Biopsy for HPV-Positive HNSCC
4.1. Circulating Tumor DNA (ctDNA)
4.2. Droplet Digital PCR (ddPCR)
5. Role and Prognostic Value of PD-L1 and Emerging Immune Checkpoint Markers in HPV-Positive Head and Neck Cancers
6. Epigenetic Markers
7. Future Directions and Emerging Technologies
7.1. Development of Novel Biosensors and Biomarkers
7.2. Graphitic Nano-Onion/Molybdenum Disulfide Nanosheet
7.3. Potential of CRISPR-Based Diagnostics
7.4. Single Gene Markers and Emerging Biomarkers
8. Sensitivity and Specificity of HPV Detection Techniques
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Khoo, A.; Boyer, M.; Jafri, Z.; Makeham, T.; Pham, T.; Khachigian, L.M.; Floros, P.; Dowling, E.; Fedder, K.; Shonka, D., Jr.; et al. Human Papilloma Virus Positive Oropharyngeal Squamous Cell Carcinoma and the Immune System: Pathogenesis, Immunotherapy and Future Perspectives. Int. J. Mol. Sci. 2024, 25, 2798. [Google Scholar] [CrossRef]
- Shield, K.D.; Ferlay, J.; Jemal, A.; Sankaranarayanan, R.; Chaturvedi, A.K.; Bray, F.; Soerjomataram, I. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J. Clin. 2017, 67, 51–64. [Google Scholar] [CrossRef]
- Balk, M.; Rupp, R.; Sievert, M.; Mantsopoulos, K.; Allner, M.; Grundtner, P.; Mueller, S.K.; Eckstein, M.; Iro, H.; Hecht, M.; et al. A comparison between p16-positive head and neck cancer of unknown primary (HPV-HNCUP) and oropharyngeal squamous cell carcinoma (HPV-OPSCC): Are they the same disease? Eur. Arch. Otorhinolaryngol. 2023, 280, 5489–5497. [Google Scholar] [CrossRef]
- Wittekindt, C.; Wagner, S.; Bushnak, A.; Prigge, E.S.; von Knebel, D.M.; Würdemann, N.; Bernhardt, K.; Pons-Kühnemann, J.; Maulbecker-Armstrong, C.; Klussmann, J.P. Increasing incidence rates of oropharyngeal squamous cell carcinoma in Germany and significance of disease burden attributed to human papillomavirus. Cancer Prev. Res. 2019, 12, 375–382. [Google Scholar] [CrossRef]
- Zamani, M.; Grønhøj, C.; Jensen, D.H.; Carlander, A.F.; Agander, T.; Kiss, K.; Olsen, C.; Baandrup, L.; Nielsen, F.C.; Andersen, E.; et al. The current epidemic of HPV-associated oropharyngeal cancer: An 18-year Danish population-based study with 2169 patients. Eur. J. Cancer 2020, 134, 52–59. [Google Scholar] [CrossRef]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Syrjänen, K.J.; Pyrhönen, S.; Syrjänen, S.M.; Lamberg, M.A. Immunohistochemical demonstration of Human papilloma virus (HPV) antigens in oral squamous cell lesions. Br. J. Oral Surg. 1983, 21, 147–153. [Google Scholar] [CrossRef]
- Ferreira, C.C. The relation between human papillomavirus (HPV) and oropharyngeal cancer: A review. PeerJ 2023, 11, e15568. [Google Scholar] [CrossRef]
- Welters, M.J.P.; Santegoets, S.J.; van der Burg, S.H. The Tumor Microenvironment and Immunotherapy of Oropharyngeal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 545385. [Google Scholar] [CrossRef]
- Yarbrough, W.G.; Schrank, T.P.; Burtness, B.A.; Issaeva, N. De-Escalated Therapy and Early Treatment of Recurrences in HPV-Associated Head and Neck Cancer: The Potential for Biomarkers to Revolutionize Personalized Therapy. Viruses 2024, 16, 536. [Google Scholar] [CrossRef]
- Gillison, M.L.; Broutian, T.; Pickard, R.K.; Tong, Z.Y.; Xiao, W.; Kahle, L.; Graubard, B.I.; Chaturvedi, A.K. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA 2012, 307, 693–703. [Google Scholar] [CrossRef]
- Posner, M.R.; Lorch, J.H.; Goloubeva, O.; Tan, M.; Schumaker, L.M.; Sarlis, N.J.; Haddad, R.I.; Cullen, K.J. Survival and human papillomavirus in oropharynx cancer in TAX 324: A subset analysis from an international phase III trial. Ann. Oncol. 2011, 22, 1071–1077. [Google Scholar] [CrossRef]
- O’Sullivan, B. Head and neck tumours. UICC TNM Classif. Malig. Tumours 2017, 8, 17–54. [Google Scholar]
- Morris, L.G.; Sikora, A.G.; Patel, S.G.; Hayes, R.B.; Ganly, I. Second primary cancers after an index head and neck cancer: Subsite-specific trends in the era of human papillomavirus-associated oropharyngeal cancer. J. Clin. Oncol. 2011, 29, 739–746. [Google Scholar] [CrossRef]
- Chen, A.M. De-Escalation Treatment for Human Papillomavirus–Related Oropharyngeal Cancer: Questions for Practical Consideration. Oncology 2023, 37, 281–287. [Google Scholar]
- Silver, J.A.; Turkdogan, S.; Roy, C.F.; Subramaniam, T.; Henry, M.; Sadeghi, N. De-Escalation Strategies for Human Papillomavirus-Associated Oropharyngeal Squamous Cell Carcinoma—Where Are We Now? Curr. Oncol. 2022, 29, 3668–3697. [Google Scholar] [CrossRef]
- Elhalawani, H.; Mohamed, A.S.R.; Elgohari, B.; Lin, T.A.; Sikora, A.G.; Lai, S.Y.; Abusaif, A.; Phan, J.; Morrison, W.H.; Gunn, G.B.; et al. Tobacco exposure as a major modifier of oncologic outcomes in human papillomavirus (HPV) associated oropharyngeal squamous cell carcinoma. BMC Cancer 2020, 20, 912. [Google Scholar] [CrossRef]
- Lai, Y.H.; Su, C.C.; Wu, S.Y.; Hsueh, W.T.; Wu, Y.H.; Chen, H.H.W.; Hsiao, J.R.; Liu, C.H.; Tsai, Y.S. Impact of Alcohol and Smoking on Outcomes of HPV-Related Oropharyngeal Cancer. J. Clin. Med. 2022, 11, 6510. [Google Scholar] [CrossRef]
- Venuti, A.; Paolini, F. HPV detection methods in head and neck cancer. Head Neck Pathol. 2012, 6, S63–S74. [Google Scholar] [CrossRef]
- van Doorn, L.J.; Molijn, A.; Kleter, B.; Quint, W.; Colau, B. Highly effective detection of human papillomavirus 16 and 18 DNA by a testing algorithm combining broad-spectrum and type-specific PCR. J. Clin. Microbiol. 2006, 44, 3292–3298. [Google Scholar] [CrossRef]
- Yeudall, W.A.; Campo, M.S. Human papillomavirus DNA in biopsies of oral tissues. J. Gen. Virol. 1991, 72, 173–176. [Google Scholar] [CrossRef]
- Nuovo, G.J. In situ detection of human papillomavirus DNA after PCR-amplification. Methods Mol. Biol. 2011, 688, 35–46. [Google Scholar] [CrossRef]
- Augustin, J.G.; Lepine, C.; Morini, A.; Brunet, A.; Veyer, D.; Brochard, C.; Mirghani, H.; Péré, H.; Badoual, C. HPV Detection in Head and Neck Squamous Cell Carcinomas: What Is the Issue? Front. Oncol. 2020, 10, 1751. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Maxam, A.M.; Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 1977, 74, 560–564. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.W.; Park, Y.-S. The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers. Life 2020, 10, 228. [Google Scholar] [CrossRef]
- Freedman, A.N.; Klabunde, C.N.; Wiant, K.; Enewold, L.; Gray, S.W.; Filipski, K.K.; Keating, N.L.; Leonard, D.G.B.; Lively, T.; McNeel, T.S.; et al. Use of next-generation sequencing tests to guide cancer treatment: Results from a nationally representative survey of oncologists in the United States. JCO Precis. Oncol. 2018, 2, 1–13. [Google Scholar] [CrossRef]
- Colomer, R.; Mondejar, R.; Romero-Laorden, N.; Alfranca, A.; Sanchez-Madrid, F.; Quintela-Fandino, M. When should we order a next generation sequencing test in a patient with cancer? eClinicalMedicine 2020, 25, 100487. [Google Scholar] [CrossRef]
- List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools); US Food and Drug Administration: Washington, DC, USA, 2024. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools (accessed on 8 June 2024).
- Bièche, I.; Kamal, M.; Tourneau, C.L. Multigene Sequencing for Treatment Selection: Esmo Biomarker Factsheet; Oncology Pro: Lugano, Switzerland, 2017; Available online: https://oncologypro.esmo.org/education-library/factsheets-on-biomarkers (accessed on 8 June 2024).
- Hodzic, J.; Gurbeta, L.; Omanovic-Miklicanin, E.; Badnjevic, A. Overview of Next-generation Sequencing Platforms Used in Published Draft Plant Genomes in Light of Genotypization of Immortelle Plant (Helichrysium arenarium). Med. Arch. 2017, 71, 288–292. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Oxford Nanopore bests PacBio. Nat. Biotechnol. 2019, 37, 336.
- Cui, J.; Shen, N.; Lu, Z.; Xu, G.; Wang, Y.; Jin, B. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 2020, 16, 85. [Google Scholar] [CrossRef] [PubMed]
- Lechner, A.; Kumbrink, J.; Walz, C.; Jung, A.; Baumeister, P.; Flach, S. Molecular characterization of the evolution of premalignant lesions in the upper aerodigestive tract. Front. Oncol. 2024, 14, 1364958. [Google Scholar] [CrossRef]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. A comparative study of ddPCR and sanger sequencing for quantitative detection of low-frequency mutation rate. IOP Conf. Ser. Earth Environ. Sci. 2019, 332, 032023. [Google Scholar] [CrossRef]
- Lin, M.-T.; Mosier, S.L.; Thiess, M.; Beierl, K.F.; Debeljak, M.; Tseng, L.-H.; Chen, G.; Yegnasubramanian, S.; Ho, H.; Cope, L.; et al. Clinical Validation of KRAS, BRAF, and EGFR Mutation Detection Using Next-Generation Sequencing. Am. J. Clin. Pathol. 2014, 141, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Gabusi, A.; Gissi, D.B.; Tarsitano, A.; Asioli, S.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Intratumoral Heterogeneity in Recurrent Metastatic Squamous Cell Carcinoma of the Oral Cavity: New Perspectives Afforded by Multiregion DNA Sequencing and mtDNA Analysis. J. Oral Maxillofac. Surg. 2020, 77, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Gabusi, A.; Gissi, D.B.; Montebugnoli, L.; Asioli, S.; Tarsitano, A.; Marchetti, C.; Balbi, T.; Helliwell, T.R.; Foschini, M.P.; Morandi, L. Prognostic impact of intra-field heterogeneity in oral squamous cell carcinoma. Virchows Arch. 2019, 476, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Chau, N.G.; Li, Y.Y.; Jo, V.Y.; Rabinowits, G.; Lorch, J.H.; Tishler, R.B.; Margalit, D.N.; Schoenfeld, J.D.; Annino, D.J.; Goguen, L.A.; et al. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin. Cancer Res. 2016, 22, 2939–2949. (In English) [Google Scholar] [CrossRef]
- Hostetter, M.K. The Third Component of Complement: New Functions for an Old Friend. J. Lab. Clin. Med. 1993, 122, 491–496. [Google Scholar]
- Ludwig, S.; Sharma, P.; Wise, P.; Sposto, R.; Hollingshead, D.; Lamb, J.; Lang, S.; Fabbri, M.; Whiteside, T.L. mRNA and miRNA Profiles of Exosomes from Cultured Tumor Cells Reveal Biomarkers Specific for HPV16-Positive and HPV16-Negative Head and Neck Cancer. Int. J. Mol. Sci. 2020, 21, 8570. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.; Unger, K.; Maihoefer, C.; Schüttrumpf, L.; Wintergerst, L.; Heider, T.; Weber, P.; Marschner, S.; Braselmann, H.; Samaga, D.; et al. A Five-MicroRNA Signature Predicts Survival and Disease Control of Patients with Head and Neck Cancer Negative for HPV Infection. Clin. Cancer Res. 2019, 25, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.; Unger, K.; Maihoefer, C.; Schüttrumpf, L.; Weber, P.; Marschner, S.; Wintergerst, L.; Pflugradt, U.; Baumeister, P.; Walch, A.; et al. Integration of P16/HPV DNA Status with a 24-miRNA-Defined Molecular Phenotype Improves Clinically Relevant Stratification of Head and Neck Cancer Patients. Cancers 2022, 14, 3745. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.-K.; Müer, A.; Mairinger, F.D.; Weichert, W.; Stenzinger, A.; Hummel, M.; Budach, V.; Tinhofer, I. MiR-200b and miR-155 as Predictive Biomarkers for the Efficacy of Chemoradiation in Locally Advanced Head and Neck Squamous Cell Carcinoma. Eur. J. Cancer 2017, 77, 3–12. [Google Scholar] [CrossRef]
- Rettig, E.M.; Waterboer, T.; Sim, E.; Faden, D.L.; Butt, J.; Hanna, G.J.; Del Vecchio Fitz, C.; Kuperwasser, C.; Sroussi, H. Relationship of HPV16 E6 Seropositivity with Circulating Tumor Tissue Modified HPV16 DNA before Head and Neck Cancer Diagnosis. Oral Oncol. 2023, 141, 106417. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, A.R.; Ferreiro-Iglesias, A.; Nygard, M.; Bender, N.; Schroeder, L.; Hildesheim, A.; Robbins, H.A.; Pawlita, M.; Langseth, H.; Schlecht, N.F.; et al. Timing of HPV16-E6 Antibody Seroconversion before OPSCC: Findings from the HPVC3 Consortium. Ann. Oncol. 2019, 30, 1335–1343. [Google Scholar] [CrossRef]
- Holzinger, D.; Wichmann, G.; Baboci, L.; Michel, A.; Höfler, D.; Wiesenfarth, M.; Schroeder, L.; Boscolo-Rizzo, P.; Herold-Mende, C.; Dyckhoff, G.; et al. Sensitivity and Specificity of Antibodies against HPV16 E6 and Other Early Proteins for the Detection of HPV16-Driven Oropharyngeal Squamous Cell Carcinoma. Int. J. Cancer 2017, 140, 2748–2757. [Google Scholar] [CrossRef] [PubMed]
- Lang Kuhs, K.A.; Kreimer, A.R.; Trivedi, S.; Holzinger, D.; Pawlita, M.; Pfeiffer, R.M.; Gibson, S.P.; Schmitt, N.C.; Hildesheim, A.; Waterboer, T.; et al. Human Papillomavirus 16 E6 Antibodies Are Sensitive for Human Papillomavirus-Driven Oropharyngeal Cancer and Are Associated with Recurrence. Cancer 2017, 123, 4382–4390. [Google Scholar] [CrossRef] [PubMed]
- Eberly, H.W.; Sciscent, B.Y.; Lorenz, F.J.; Rettig, E.M.; Goyal, N. Current and Emerging Diagnostic, Prognostic, and Predictive Biomarkers in Head and Neck Cancer. Biomedicines 2024, 12, 415. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mima, M.; Okabe, A.; Hoshii, T.; Nakagawa, T.; Kurokawa, T.; Kondo, S.; Mizokami, H.; Fukuyo, M.; Fujiki, R.; Rahmutulla, B.; et al. Tumorigenic Activation around HPV Integrated Sites in Head and Neck Squamous Cell Carcinoma. Int. J. Cancer 2023, 152, 1847–1862. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Anderson, W.F.; Lortet-Tieulent, J.; Curado, M.P.; Ferlay, J.; Franceschi, S.; Rosenberg, P.S.; Bray, F.; Gillison, M.L. Worldwide Trends in Incidence Rates for Oral Cavity and Oropharyngeal Cancers. J. Clin. Oncol. 2013, 31, 4550–4559. [Google Scholar] [CrossRef] [PubMed]
- Mehanna, H.; Taberna, M.; von Buchwald, C.; Tous, S.; Brooks, J.; Mena, M.; Morey, F.; Grønhøj, C.; Rasmussen, J.H.; Garset-Zamani, M.; et al. Prognostic Implications of P16 and HPV Discordance in Oropharyngeal Cancer (HNCIG-EPIC-OPC): A Multicentre, Multinational, Individual Patient Data Analysis. Lancet Oncol. 2023, 24, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, S.F.; Salnikov, M.Y.; Zeng, P.Y.F.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. HPV16 Intratypic Variants in Head and Neck Cancers: A North American Perspective. Viruses 2023, 15, 2411. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Duijf, P.H.G.; Sriram, S.; Perera, G.; Vasani, S.; Kenny, L.; Leo, P.; Punyadeera, C. Circulating tumour DNA alterations: Emerging biomarker in head and neck squamous cell carcinoma. J. Biomed. Sci. 2023, 30, 65. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bartkova, J.; Lukas, J.; Strauss, M.; Bartek, J. Cyclin D1 Oncoprotein Aberrantly Accumulates in Malignancies of Diverse Histogenesis. Oncogene 1995, 10, 775–778. [Google Scholar]
- Patel, K.B.; Mroz, E.A.; Faquin, W.C.; Rocco, J.W. A Combination of Intra-Tumor Genetic Heterogeneity, Estrogen Receptor Alpha and Human Papillomavirus Status Predicts Outcomes in Head and Neck Squamous Cell Carcinoma Following Chemoradiotherapy. Oral Oncol. 2021, 120, 105421. [Google Scholar] [CrossRef]
- DE Oliveira Neto, C.P.; Brito, H.O.; DA Costa, R.M.G.; Brito, L.M.O. Is There a Role for Sex Hormone Receptors in Head-and-Neck Cancer? Links with HPV Infection and Prognosis. Anticancer Res. 2021, 41, 3707–3716. [Google Scholar] [CrossRef]
- Koenigs, M.B.; Lefranc-Torres, A.; Bonilla-Velez, J.; Patel, K.B.; Hayes, D.N.; Glomski, K.; Busse, P.M.; Chan, A.W.; Clark, J.R.; Deschler, D.G.; et al. Association of Estrogen Receptor Alpha Expression with Survival in Oropharyngeal Cancer Following Chemoradiation Therapy. J. Natl. Cancer Inst. 2019, 111, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; O’Boyle, C.J.; Varmeh, S.; Queenan, N.; Michel, A.; Stein, J.; Thierauf, J.; Sadow, P.M.; Faquin, W.C.; Perry, S.K.; et al. Cell-Free HPV DNA Provides an Accurate and Rapid Diagnosis of HPV-Associated Head and Neck Cancer. Clin. Cancer Res. 2022, 28, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, M.; Tanaka, H.; Takenaka, Y.; Suzuki, M.; Fukusumi, T.; Eguchi, H.; Watabe, T.; Kato, H.; Yachida, S.; Inohara, H.; et al. Association of circulating tumor HPV16DNA levels and quantitative PET parameters in patients with HPV-positive head and neck squamous cell carcinoma. Sci. Rep. 2024, 14, 3278. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferrandino, R.M.; Chen, S.; Kappauf, C.; Barlow, J.; Gold, B.S.; Berger, M.H.; Westra, W.H.; Teng, M.S.; Khan, M.N.; Posner, M.R.; et al. Performance of Liquid Biopsy for Diagnosis and Surveillance of Human Papillomavirus–Associated Oropharyngeal Cancer. JAMA Otolaryngol. Head Neck Surg. 2023, 149, 971. [Google Scholar] [CrossRef] [PubMed]
- Rettig, E.M.; Wang, A.A.; Tran, N.A.; Carey, E.; Dey, T.; Schoenfeld, J.D.; Sehgal, K.; Guenette, J.P.; Margalit, D.N.; Sethi, R.; et al. Association of Pretreatment Circulating Tumor Tissue-Modified Viral HPV DNA With Clinicopathologic Factors in HPV-Positive Oropharyngeal Cancer. JAMA Otolaryngol. Head Neck Surg. 2022, 148, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.; Sangal, N.R.; Aggarwal, A.; Rajasekaran, K.; Cannady, S.B.; Basu, D.; Chalian, A.; Weinstein, G.; Brody, R.M. Preoperative Circulating Tumor HPV DNA and Oropharyngeal Squamous Cell Disease. JAMA Otolaryngol. Head Neck Surg. 2024, 150, 444–450. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- O’Boyle, C.J.; Siravegna, G.; Varmeh, S.; Queenan, N.; Michel, A.; Pang, K.C.S.; Stein, J.; Thierauf, J.C.; Sadow, P.M.; Faquin, W.C.; et al. Cell-Free Human Papillomavirus DNA Kinetics after Surgery for Human Papillomavirus-Associated Oropharyngeal Cancer. Cancer 2022, 128, 2193–2204. [Google Scholar] [CrossRef] [PubMed]
- Routman, D.M.; Kumar, S.; Chera, B.S.; Jethwa, K.R.; Van Abel, K.M.; Frechette, K.; DeWees, T.; Golafshar, M.; Garcia, J.J.; Price, D.L.; et al. Detectable Postoperative Circulating Tumor Human Papillomavirus DNA and Association with Recurrence in Patients With HPV-Associated Oropharyngeal Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Rettig, E.M.; Faden, D.L.; Sandhu, S.; Wong, K.; Faquin, W.C.; Warinner, C.; Stephens, P.; Kumar, S.; Kuperwasser, C.; Richmon, J.D.; et al. Detection of Circulating Tumor Human Papillomavirus DNA before Diagnosis of HPV-Positive Head and Neck Cancer. Int. J. Cancer. 2022, 151, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Rettig, E.M.; Wentz, A.; Posner, M.R.; Gross, N.D.; Haddad, R.I.; Gillison, M.L.; Fakhry, C.; Quon, H.; Sikora, A.G.; Stott, W.J.; et al. Prognostic Implication of Persistent Human Papillomavirus Type 16 DNA Detection in Oral Rinses for Human Papillomavirus-Related Oropharyngeal Carcinoma. JAMA Oncol. 2015, 1, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.M.; Chan, J.Y.K.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and Plasma Quantitative Polymerase Chain Reaction-Based Detection and Surveillance of Human Papillomavirus-Related Head and Neck Cancer. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Chatfield-Reed, K.; Roche, V.P.; Pan, Q. cfDNA detection for HPV+ squamous cell carcinomas. Oral Oncol. 2021, 115, 104958. [Google Scholar] [CrossRef]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef]
- Woo, D.; Yu, M. Circulating tumor cells as “liquid biopsies” to understand cancer metastasis. Transl. Res. 2018, 201, 128–135. [Google Scholar] [CrossRef]
- Stremersch, S.; De Smedt, S.C.; Raemdonck, K. Therapeutic and diagnostic applications of extracellular vesicles. J. Control Release 2016, 244, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Métais, P. Les acides nucléiques du plasma sanguin chez l’Homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Nguyen, B.; Meehan, K.; Pereira, M.R.; Mirzai, B.; Lim, S.H.; Leslie, C.; Clark, M.; Sader, C.; Friedland, P.; Lindsay, A.; et al. A comparative study of extracellular vesicle-associated and cell-free DNA and RNA for HPV detection in oropharyngeal squamous cell carcinoma. Sci. Rep. 2020, 10, 6083. [Google Scholar] [CrossRef] [PubMed]
- Greytak, S.R.; Engel, K.B.; Parpart-Li, S.; Murtaza, M.; Bronkhorst, A.J.; Pertile, M.D.; Moore, H.M. Harmonizing Cell-Free DNA Collection and Processing Practices through Evidence-Based Guidance. Clin. Cancer Res. 2020, 26, 3104–3109. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, T.W.; Mazurek, A.M.; Śnietura, M.; Hejduk, B.; Jędrzejewska, M.; Bobek-Billewicz, B.; d’Amico, A.; Pigłowski, W.; Wygoda, A.; Składowski, K.; et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J. Transl. Med. 2020, 18, 167. [Google Scholar] [CrossRef]
- Corpman, D.W.; Masroor, F.; Carpenter, D.M.; Nayak, S.; Gurushanthaiah, D.; Wang, K.H. Posttreatment surveillance PET/CT for HPV-associated oropharyngeal cancer. Head Neck 2019, 41, 456–462. [Google Scholar] [CrossRef]
- Wang, K.; Wong, T.Z.; Amdur, R.J.; Mendenhall, W.M.; Sheets, N.C.; Green, R.; Thorp, B.D.; Patel, S.N.; Hackman, T.G.; Zanation, A.M.; et al. Pitfalls of post-treatment PET after de-intensified chemoradiotherapy for HPV-associated oropharynx cancer: Secondary analysis of a phase 2 trial. Oral Oncol. 2018, 78, 108–113. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J. Clin. Oncol. 2020, 38, 1050–1058. [Google Scholar] [CrossRef]
- Lee, J.Y.; Garcia-Murillas, I.; Cutts, R.J.; De Castro, D.G.; Grove, L.; Hurley, T.; Wang, F.; Nutting, C.; Newbold, K.; Harrington, K.; et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br. J. Cancer 2017, 117, 876–883. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Jiang, J.; Pang, X.; Huang, M.; Tang, Y.; Liang, X.; Tang, Y. The Evolving Landscape of PD-1/PD-L1 Pathway in Head and Neck Cancer. Front. Immunol. 2020, 11, 1721. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.M.; Ferguson, P.; Dodds, T.; Jones, D.; Li, M.; Yang, J.; Scolyer, R.A. Significant association of PD-L1 expression with human papillomavirus positivity and its prognostic impact in oropharyngeal cancer. Oral Oncol. 2019, 92, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wylie, K.M.; Wyczalkowski, M.A.; Karpova, A.; Ley, J.; Sun, S.; Mashl, R.J.; Liang, W.W.; Wang, X.; Johnson, K.; et al. Dynamic host immune response in virus-associated cancers. Commun. Biol. 2019, 2, 109. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and Clinical Activity of Pembrolizumab for Treatment of Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-012): An Open-Label, Multicentre, Phase 1b Trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef]
- Miyamoto, R.; Uzawa, N.; Nagaoka, S.; Hirata, Y.; Amagasa, T. Prognostic Significance of Cyclin D1 Amplification and Overexpression in Oral Squamous Cell Carcinomas. Oral Oncol. 2003, 39, 610–618. [Google Scholar] [CrossRef]
- Ngamphaiboon, N.; Chureemas, T.; Siripoon, T.; Arsa, L.; Trachu, N.; Jiarpinitnun, C.; Pattaranutaporn, P.; Sirachainan, E.; Larbcharoensub, N. Characteristics and Impact of Programmed Death-Ligand 1 Expression, CD8+ Tumor-Infiltrating Lymphocytes, and P16 Status in Head and Neck Squamous Cell Carcinoma. Med. Oncol. 2019, 36, 21. [Google Scholar] [CrossRef] [PubMed]
- Roper, E.; Lum, T.; Palme, C.E.; Ashford, B.; Ch’ng, S.; Ranson, M.; Boyer, M.; Clark, J.; Gupta, R. PD-L1 Expression Predicts Longer Disease Free Survival in High Risk Head and Neck Cutaneous Squamous Cell Carcinoma. Pathology 2017, 49, 499–505. [Google Scholar] [CrossRef]
- Müller, T.; Braun, M.; Dietrich, D.; Aktekin, S.; Höft, S.; Kristiansen, G.; Göke, F.; Schröck, A.; Brägelmann, J.; Held, S.A.E.; et al. PD-L1: A Novel Prognostic Biomarker in Head and Neck Squamous Cell Carcinoma. Oncotarget 2017, 8, 52889–52900. [Google Scholar] [CrossRef]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef]
- Liouta, G.; Adamaki, M.; Tsintarakis, A.; Zoumpourlis, P.; Liouta, A.; Agelaki, S.; Zoumpourlis, V. DNA Methylation as a Diagnostic, Prognostic, and Predictive Biomarker in Head and Neck Cancer. Int. J. Mol. Sci. 2023, 24, 2996. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-H.; Hsia, S.-M.; Shih, Y.-H.; Shieh, T.-M. Association of Smoking, Alcohol Use, and Betel Quid Chewing with Epigenetic Aberrations in Cancers. Int. J. Mol. Sci. 2017, 18, 1210. [Google Scholar] [CrossRef] [PubMed]
- Camuzi, D.; de Almeida Simão, T.; Dias, F.; Ribeiro Pinto, L.F.; Soares-Lima, S.C. Head and Neck Cancers Are Not Alike When Tarred with the Same Brush: An Epigenetic Perspective from the Cancerization Field to Prognosis. Cancers 2021, 13, 5630. [Google Scholar] [CrossRef] [PubMed]
- Starzer, A.M.; Heller, G.; Tomasich, E.; Melchardt, T.; Feldmann, K.; Hatziioannou, T.; Traint, S.; Minichsdorfer, C.; Schwarz-Nemec, U.; Nackenhorst, M.; et al. DNA Methylation Profiles Differ in Responders versus Non-Responders to Anti-PD-1 Immune Checkpoint Inhibitors in Patients with Advanced and Metastatic Head and Neck Squamous Cell Carcinoma. J. Immunother. Cancer 2022, 10, e003420. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. (In English) [Google Scholar] [CrossRef] [PubMed]
- Available online: https://ecancer.org/en/news/23659-researchers-develop-novel-dna-biosensor-for-early-diagnosis-of-cervical-cancer (accessed on 8 June 2024).
- Lim, S.M.; Cho, S.H.; Hwang, I.G.; Choi, J.W.; Chang, H.; Ahn, M.J.; Park, K.U.; Kim, J.W.; Ko, Y.H.; Ahn, H.K.; et al. Investigating the feasibility of targeted next-generation sequencing to guide the treatment of head and neck squamous cell carcinoma. Cancer Res Treat. 2019, 51, 300–312. (In English) [Google Scholar] [CrossRef] [PubMed]
- Radiation Therapy with or without Cisplatin in Treating Patients with Stage III-IVA Squamous Cell Carcinoma of the Head and Neck Who Have Undergone Surgery. ClinicalTrials.gov. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02734537 (accessed on 8 June 2024).
- Spector, M.E.; Bellile, E.; Amlani, L.; Zarins, K.; Smith, J.; Brenner, J.C.; Rozek, L.; Nguyen, A.; Thomas, D.; McHugh, J.B.; et al. Prognostic value of tumor-infiltrating lymphocytes in head and neck squamous cell carcinoma. JAMA Otolaryngol. Head Neck Surg. 2019, 145, 1012–1019. (In English) [Google Scholar] [CrossRef] [PubMed]
- Heft Neal, M.E.; Smith, J.D.; Birkeland, A.C.; Haring, C.T.; Chinn, S.B.; Shuman, A.G.; Casper, K.A.; Malloy, K.M.; Stucken, C.L.; Mclean, S.A.; et al. Tumor-infiltrating lymphocytes in patients with advanced laryngeal cancer undergoing bioselection. Otolaryngol. Head Neck Surg. 2022, 166, 498–505. (In English) [Google Scholar] [CrossRef] [PubMed]
- Hadler-Olsen, E.; Wirsing, A.M. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2019, 120, 714–727. (In English) [Google Scholar] [CrossRef]
- Rao, C.N.R.; Gopalakrishnan, K.; Maitra, U. Comparative study of potential applications of graphene, MoS2, and other two-dimensional materials in energy devices, sensors, and related areas. ACS Appl. Mater. Inter. 2015, 7, 7809–7832. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, E. A graphitic nano-onion/molybdenum disulfide nanosheet composite as a platform for HPV-associated cancer-detecting DNA biosensors. J. Nanobiotechnol. 2023, 21, 187. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Huang, K.J.; Wu, X. Recent advances in transition-metal dichalcogenides based electrochemical biosensors: A review. Biosens. Bioelectron. 2017, 97, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Kukkar, M.; Mohanta, G.C.; Tuteja, S.K.; Kumar, P.; Bhadwal, A.S.; Samaddar, P.; Kim, K.H.; Deep, A. A comprehensive review on nano-molybdenum disulfide/DNA interfaces as emerging biosensing platforms. Biosens. Bioelectron. 2018, 107, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Wan, H.; Zhang, J.; Wang, Q.; Hu, X.; Xia, F. Electrochemical DNA sensors based on MoS2-AuNPs for polynucleotide kinase activity and inhibition assay. ACS Appl. Mater. Interfaces 2020, 12, 45814–45821. [Google Scholar] [CrossRef] [PubMed]
- Toto, E.; Botti, S.; Laurenzi, S.; Santonicola, M.G. UV-induced modification of PEDOT:PSS-based nanocomposite films investigated by Raman microscopy mapping. Appl. Surf. Sci. 2020, 513, 145839. [Google Scholar] [CrossRef]
- Kumar, M.; Wang, M.; Kumara Swamy, B.E.; Praveen, M.; Zhao, W. Poly (alanine)/NaOH/MoS2/MWCNTs modified carbon paste electrode for simultaneous detection of dopamine, ascorbic acid, serotonin and guanine. Colloids Surf. B Biointerfaces 2020, 196, 111299. [Google Scholar] [CrossRef] [PubMed]
- Majd, S.M.; Ghasemi, F.; Salimi, A.; Sham, T.K. Transport properties of a molybdenum disulfide and carbon dot nanohybrid transistor and its applications as a Hg2+ aptasensor. ACS Appl. Electron. Mater. 2020, 2, 635–645. [Google Scholar] [CrossRef]
- Eivazzadeh-Keihan, R.; Noruzi, E.B.; Chidar, E.; Jafari, M.; Davoodi, F.; Kashtiaray, A.; Gorab, M.G.; Hashemi, S.M.; Javanshir, S.; Cohan, R.A.; et al. Applications of carbon-based conductive nanomaterials in biosensors. Chem. Eng. J. 2022, 442, 136183. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Sandhyarani, N. Carbon nanostructures as immobilization platform for DNA: A review on current progress in electrochemical DNA sensors. Biosens. Bioelectron. 2017, 97, 226–237. [Google Scholar] [CrossRef]
- Kiransan, K.D.; Topcu, E. Conducting polymer-reduced graphene oxide sponge electrode for electrochemical detection based on dna hybridization. ACS Appl. Nano Mater. 2020, 3, 5449–5462. [Google Scholar] [CrossRef]
- Khodadadi, A.; Faghih-Mirzaei, E.; Karimi-Maleh, H.; Abbaspourrad, A.; Agarwal, S.; Gupta, V.K. A new epirubicin biosensor based on amplifying DNA interactions with polypyrrole and nitrogen-doped reduced graphene: Experimental and docking theoretical investigations. Sens. Actuators B-Chem. 2019, 284, 568–574. [Google Scholar] [CrossRef]
- Dong, Y.; Wan, L.; Lv, S.; Zhu, D.; Su, S.; Chao, J.; Wang, L. Construction of a molybdenum disulfide-based colorimetric sensor for label-free infectious disease analysis coupled with a catalyzed hairpin assembly reaction. Langmuir 2022, 38, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Niu, W.; Chen, S.; Xu, W.; Ji, X.; Yuan, L.; Zhao, H.; Geng, M.; Qiu, J.; Li, C. Target-inspired Pb(2+)-dependent DNAzyme for ultrasensitive electrochemical sensor based on MoS2-AuPt nanocomposites and hemin/G-quadruplex DNAzyme as signal amplifier. Biosens. Bioelectron. 2019, 144, 111560. [Google Scholar] [CrossRef] [PubMed]
- Chekin, F.; Bagga, K.; Subramanian, P.; Jijie, R.; Singh, S.K.; Kurungot, S.; Boukherroub, R.; Szunerits, S. Nucleic aptamer modified porous reduced graphene oxide/MoS2 based electrodes for viral detection: Application to human papillomavirus (HPV). Sens. Actuat B-Chem. 2018, 262, 991–1000. [Google Scholar] [CrossRef]
- Guerreiro, G.V.; Zaitouna, A.J.; Lai, R.Y. Characterization of an electrochemical mercury sensor using alternating current, cyclic, square wave and differential pulse voltammetry. Anal. Chim. Acta 2014, 810, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhao, Z.; Ma, X. Description of CRISPR-Cas9 development and its prospects in human papillomavirus-driven cancer treatment. Front. Immunol. 2022, 13, 1037124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, H.; Li, G.; Peng, X.; Deng, A.; Ye, L.; Shi, L.; Wang, T.; He, J. The Use of CRISPR/Cas9 as a Tool to Study Human Infectious Viruses. Front. Cell Infect. Microbiol. 2021, 11, 590989. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, C.; Wu, P.; Yu, L.; Liu, L.; Liu, H.; Tan, X.; Wang, L.; Huang, X.; Wang, H. The application of CRISPR/Cas9 system in cervical carcinogenesis. Cancer Gene Ther. 2022, 29, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Ghouneimy, A.; Mahas, A.; Marsic, T.; Aman, R.; Mahfouz, M. CRISPR-Based Diagnostics: Challenges and Potential Solutions toward Point-of-Care Applications. ACS Synth. Biol. 2023, 12, 1–16. [Google Scholar] [CrossRef]
- Lou, J.; Wang, B.; Li, J.; Ni, P.; Jin, Y.; Chen, S.; Xi, Y.; Zhang, R.; Duan, G. The CRISPR-Cas system as a tool for diagnosing and treating infectious diseases. Mol. Biol. Rep. 2022, 49, 11301–11311. [Google Scholar] [CrossRef]
- Puig-Serra, P.; Casado-Rosas, M.C.; Martinez-Lage, M.; Olalla-Sastre, B.; Alonso-Yanez, A.; Torres-Ruiz, R.; Rodriguez-Perales, S. CRISPR Approaches for the Diagnosis of Human Diseases. Int. J. Mol. Sci. 2022, 23, 1757. [Google Scholar] [CrossRef] [PubMed]
- Chehelgerdi, M.; Chehelgerdi, M.; Khorramian-Ghahfarokhi, M.; Shafieizadeh, M.; Mahmoudi, E.; Eskandari, F.; Rashidi, M.; Arshi, A.; Mokhtari-Farsani, A. Comprehensive review of CRISPR-based gene editing: Mechanisms, challenges, and applications in cancer therapy. Mol. Cancer 2024, 23, 9, Erratum in Mol. Cancer 2024, 23, 43. [Google Scholar] [CrossRef] [PubMed]
- Stefanoudakis, D.; Kathuria-Prakash, N.; Sun, A.W.; Abel, M.; Drolen, C.E.; Ashbaugh, C.; Zhang, S.; Hui, G.; Tabatabaei, Y.A.; Zektser, Y.; et al. The Potential Revolution of Cancer Treatment with CRISPR Technology. Cancers 2023, 15, 1813. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al Saihati, H.; Bukhamsin, R.; Bakhrebah, M.A.; Nassar, M.S.; Alsaleh, A.A.; Alhashem, Y.N.; Bukhamseen, A.Y.; Al-Ruhimy, K.; Alotaibi, M.; et al. Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction. Curr. Oncol. 2023, 30, 1954–1976. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schnoell, J.; Scheiflinger, A.; Al-Gboore, S.; Kadletz-Wanke, L.; Kenner, L.; Heiduschka, G.; Jank, B.J. The Prognostic Role of PSMD14 in Head and Neck Squamous Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 2483–2490. [Google Scholar] [CrossRef] [PubMed]
- Langer, C.J. Exploring Biomarkers in Head and Neck Cancer. Cancer 2012, 118, 3882–3892. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.; Kim, T.M.; Jeon, Y.K.; Kwon, T.-K.; Hah, J.H.; Lee, S.-H.; Kim, D.-W.; Wu, H.-G.; Rhee, C.-S.; Sung, M.-W.; et al. Class III Beta-Tubulin, but Not ERCC1, Is a Strong Predictive and Prognostic Marker in Locally Advanced Head and Neck Squamous Cell Carcinoma. Ann. Oncol. 2009, 20, 1414–1419. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Duan, Y.; Zhou, M.; Yue, K.; Zhuo, S.; Li, X.; Liu, D.; Ye, B.; Lai, Q.; Li, L.; et al. Blockade of Deubiquitinating Enzyme PSMD14 Overcomes Chemoresistance in Head and Neck Squamous Cell Carcinoma by Antagonizing E2F1/Akt/SOX2-Mediated Stemness. Theranostics 2021, 11, 2655–2669. [Google Scholar] [CrossRef]
- Liu, M.; Huang, L.; Liu, Y.; Yang, S.; Rao, Y.; Chen, X.; Nie, M.; Liu, X. Identification of the MMP Family as Therapeutic Targets and Prognostic Biomarkers in the Microenvironment of Head and Neck Squamous Cell Carcinoma. J. Transl. Med. 2023, 21, 208. [Google Scholar] [CrossRef]
- Campo, F.; Paolini, F.; Manciocco, V.; Moretto, S.; Pichi, B.; Moretti, C.; Blandino, G.; De Pascale, V.; Benevolo, M.; Pimpinelli, F.; et al. Circulating tumor HPV DNA in the management of HPV+ oropharyngeal cancer and its correlation with MRI. Head Neck 2024. [CrossRef] [PubMed]
- Kais, A.; Santiago, S.P.; Han, P.C.; Clump, D.A.; Stokes, W.A.; Fancy, T.; Cui, R.; Martin, E.; Turner, M.T. Human papillomavirus circulating tumor DNA: A diagnostic tool in squamous cell carcinoma of unknown primary-a pilot study. Front. Oncol. 2024, 14, 1376595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, F.; Snow, J.T.; Simsir, A.; Hernandez, O.; Levine, P.; Szeto, O.; Sun, W.; Givi, B.; Brandler, T.C. p16 immunostaining in fine-needle aspirations of the head and neck: Determining the optimal positivity threshold in HPV-related squamous cell cancer. J. Am. Soc. Cytopathol. 2021, 10, 592–600. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, G.; Clemens, G.; Troy, T.; Castillo, R.G.; Struijk, L.; Waterboer, T.; Bender, N.; Pierorazio, P.M.; Best, S.R.; Strickler, H.; et al. Evaluating the Utility and Prevalence of HPV Biomarkers in Oral Rinses and Serology for HPV-related Oropharyngeal Cancer. Cancer Prev. Res. 2019, 12, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Shinn, J.R.; Davis, S.J.; Lang-Kuhs, K.A.; Rohde, S.; Wang, X.; Liu, P.; Dupont, W.D.; Plummer, D., Jr.; Thorstad, W.L.; Chernock, R.D.; et al. Oropharyngeal Squamous Cell Carcinoma with Discordant p16 and HPV mRNA Results: Incidence and Characterization in a Large, Contemporary United States Cohort. Am. J. Surg. Pathol. 2021, 45, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; D’Souza, G.; Khan, Z.; Allen, H.; Henson, S.; Seiwert, T.Y.; Koch, W.; Pardoll, D.M.; Fakhry, C. Comparison of next generation sequencing, droplet digital PCR, and quantitative real-time PCR for the earlier detection and quantification of HPV in HPV-positive oropharyngeal cancer. Oral Oncol. 2022, 128, 105805. [Google Scholar] [CrossRef]
- Naegele, S.; Ruiz-Torres, D.A.; Zhao, Y.; Goss, D.; Faden, D.L. Comparing the Diagnostic Performance of Quantitative PCR, Digital Droplet PCR, and Next-Generation Sequencing Liquid Biopsies for Human Papillomavirus-Associated Cancers. J. Mol. Diagn. JMD 2024, 26, 179–190. [Google Scholar] [CrossRef]
| HPV Detection Technique | Sensitivity (%) | Specificity (%) |
|---|---|---|
| Serology (antibody detection) | 70–80 | 85–95 |
| p16 immunohistochemistry (IHC) | 85–90 | 85–90 |
| In situ hybridization (ISH) | 85–90 | 90–95 |
| Polymerase chain reaction (PCR) | 90–95 | 80–90 |
| RNA-based methods | 95–100 | 90–95 |
| Next-generation sequencing (NGS) | 95–100 | 95–100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krsek, A.; Baticic, L.; Braut, T.; Sotosek, V. The Next Chapter in Cancer Diagnostics: Advances in HPV-Positive Head and Neck Cancer. Biomolecules 2024, 14, 925. https://doi.org/10.3390/biom14080925
Krsek A, Baticic L, Braut T, Sotosek V. The Next Chapter in Cancer Diagnostics: Advances in HPV-Positive Head and Neck Cancer. Biomolecules. 2024; 14(8):925. https://doi.org/10.3390/biom14080925
Chicago/Turabian StyleKrsek, Antea, Lara Baticic, Tamara Braut, and Vlatka Sotosek. 2024. "The Next Chapter in Cancer Diagnostics: Advances in HPV-Positive Head and Neck Cancer" Biomolecules 14, no. 8: 925. https://doi.org/10.3390/biom14080925
APA StyleKrsek, A., Baticic, L., Braut, T., & Sotosek, V. (2024). The Next Chapter in Cancer Diagnostics: Advances in HPV-Positive Head and Neck Cancer. Biomolecules, 14(8), 925. https://doi.org/10.3390/biom14080925






