Diapause and Anoxia-Induced Quiescence Are Unique States in Embryos of the Annual Killifish, Austrofundulus limnaeus
Abstract
1. Introduction
2. Materials and Methods
2.1. Embryo Rearing and Staging
2.2. Anoxic Exposure and Aerobic Recovery
2.3. Sampling Embryos for RNA Sequencing and qPCR
2.4. RNA Extraction
2.5. Poly-A RNA Sequencing
2.6. Analysis of Sequence Data
2.7. Differential Gene Expression Analysis
2.8. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.9. Ortholog Mapping and Gene Ontology Analysis
2.10. Transcription Factor Analysis Using TFLink
2.11. Data Visualization
3. Results
3.1. Sequencing Library Quality and Coverage
3.2. Characterization of the Normoxic Diapause II Transcriptome Profile
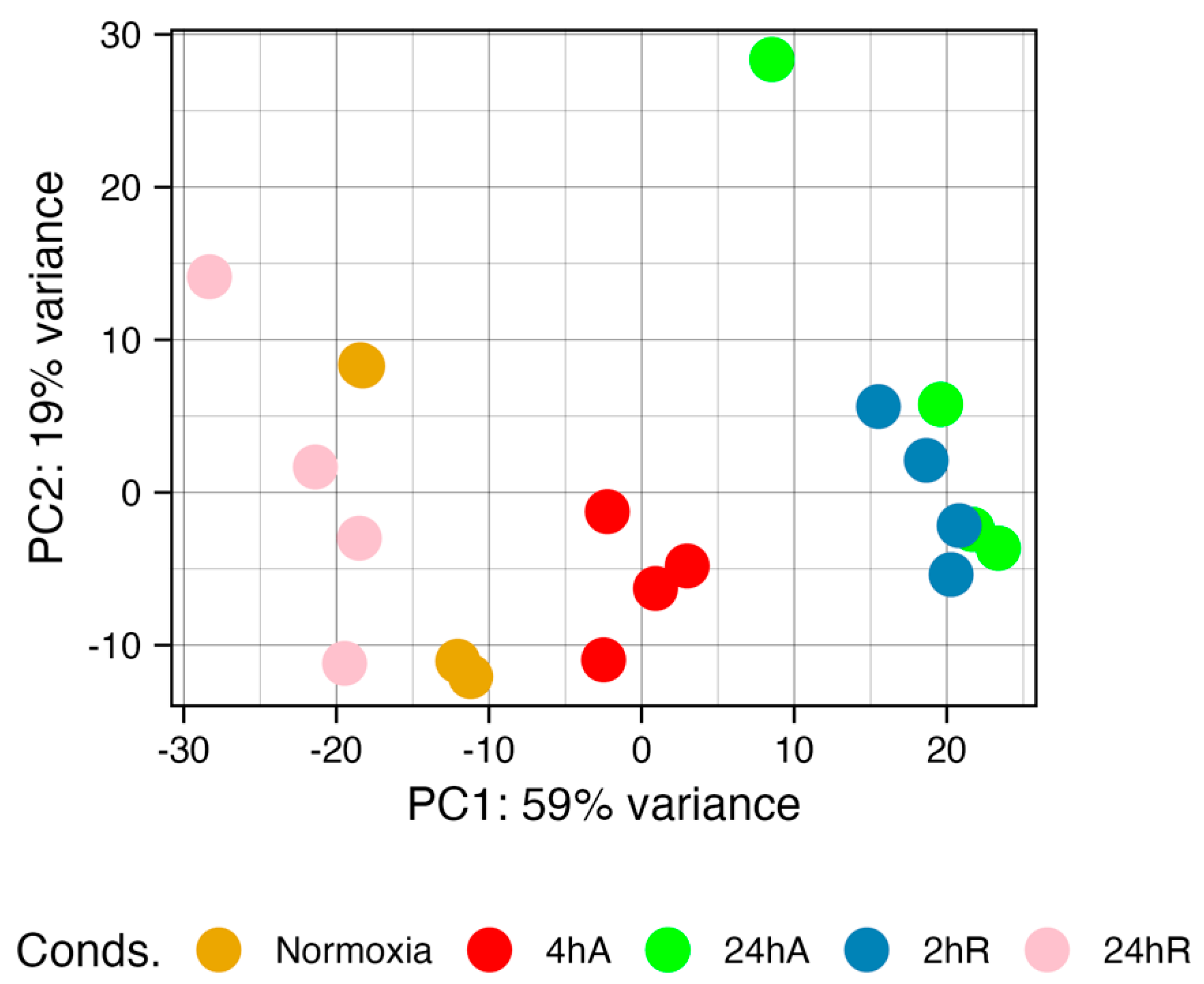
3.3. Overall Characterization of Experimental Treatments
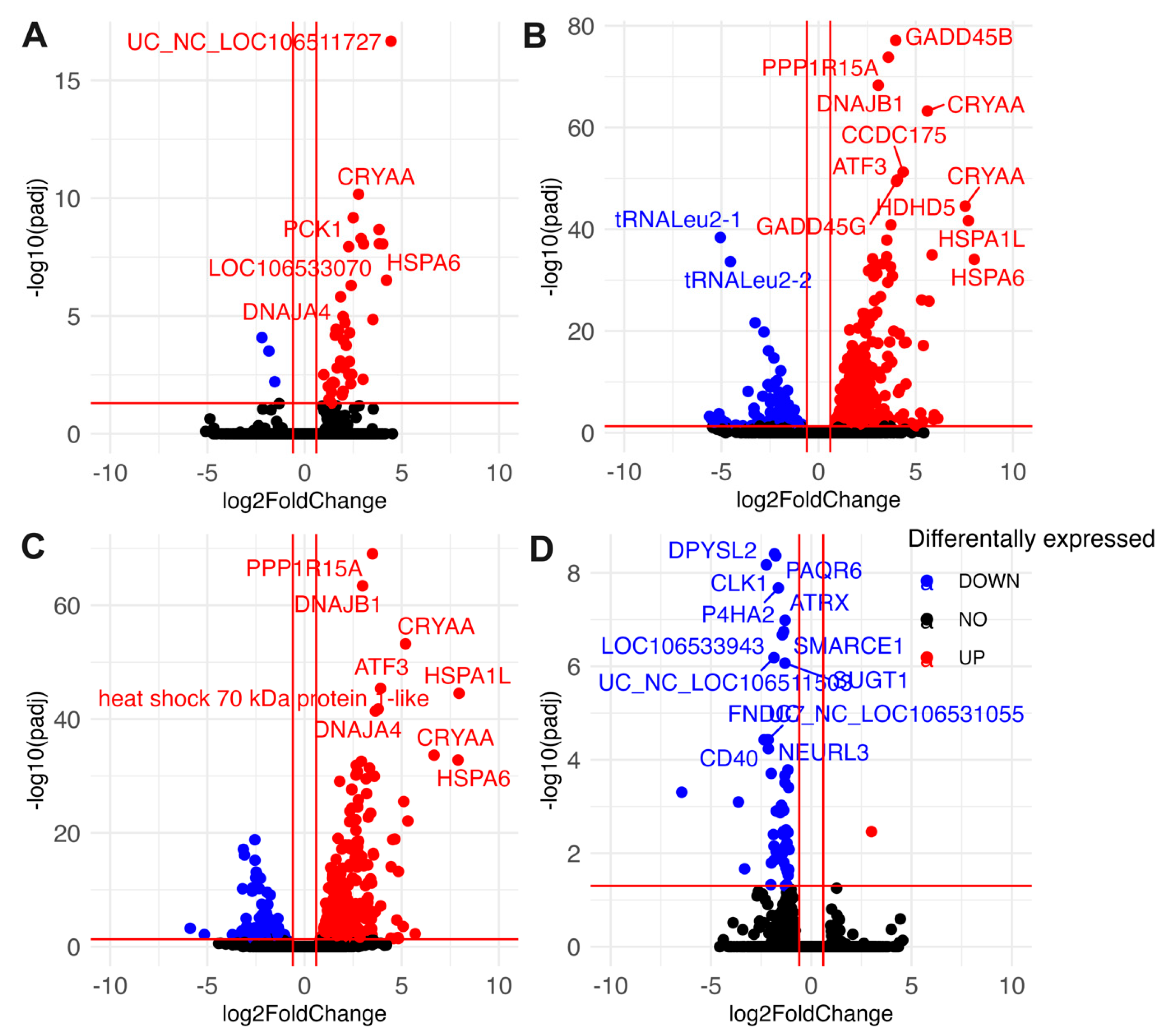
3.4. Differential Gene Expression
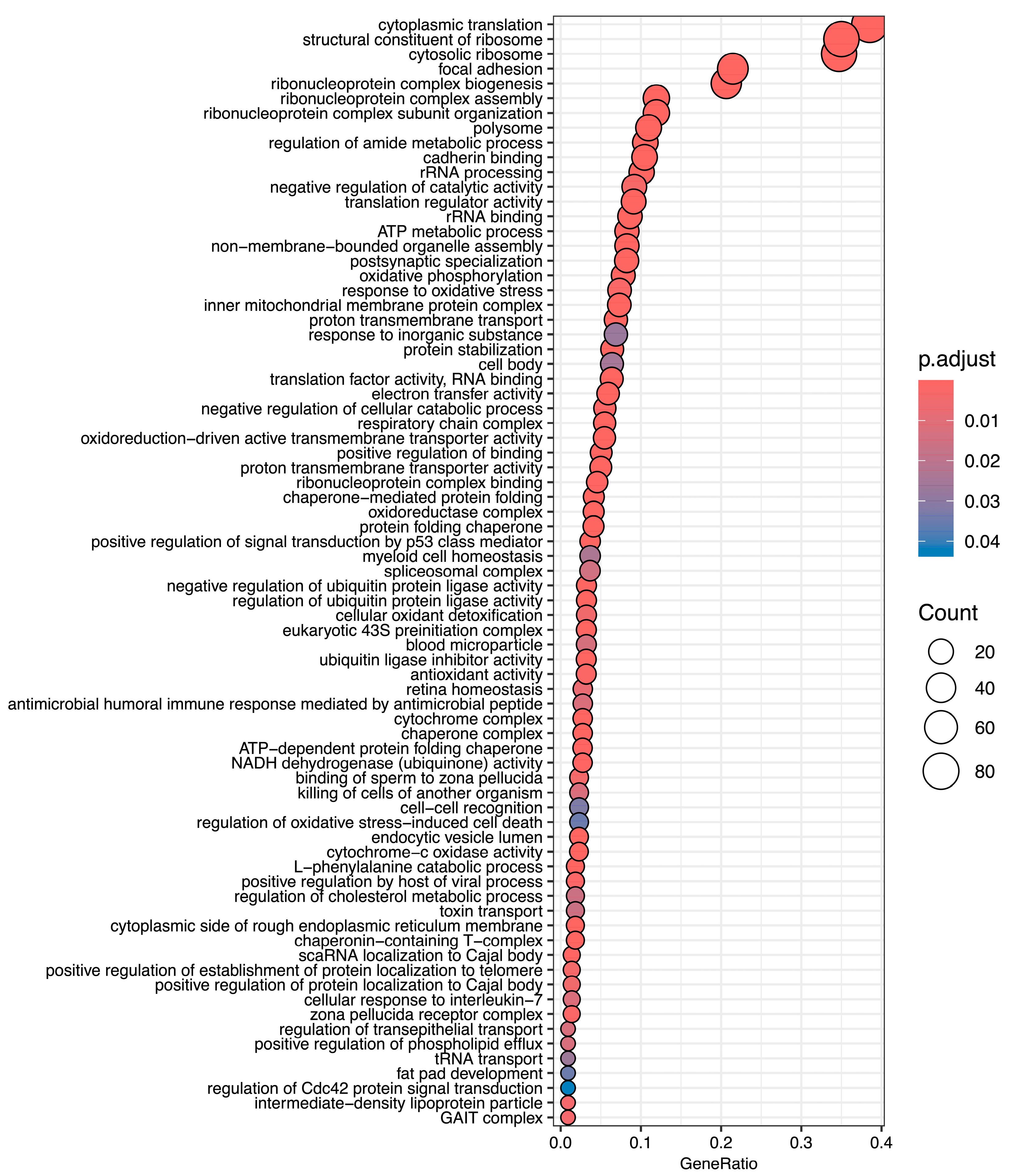
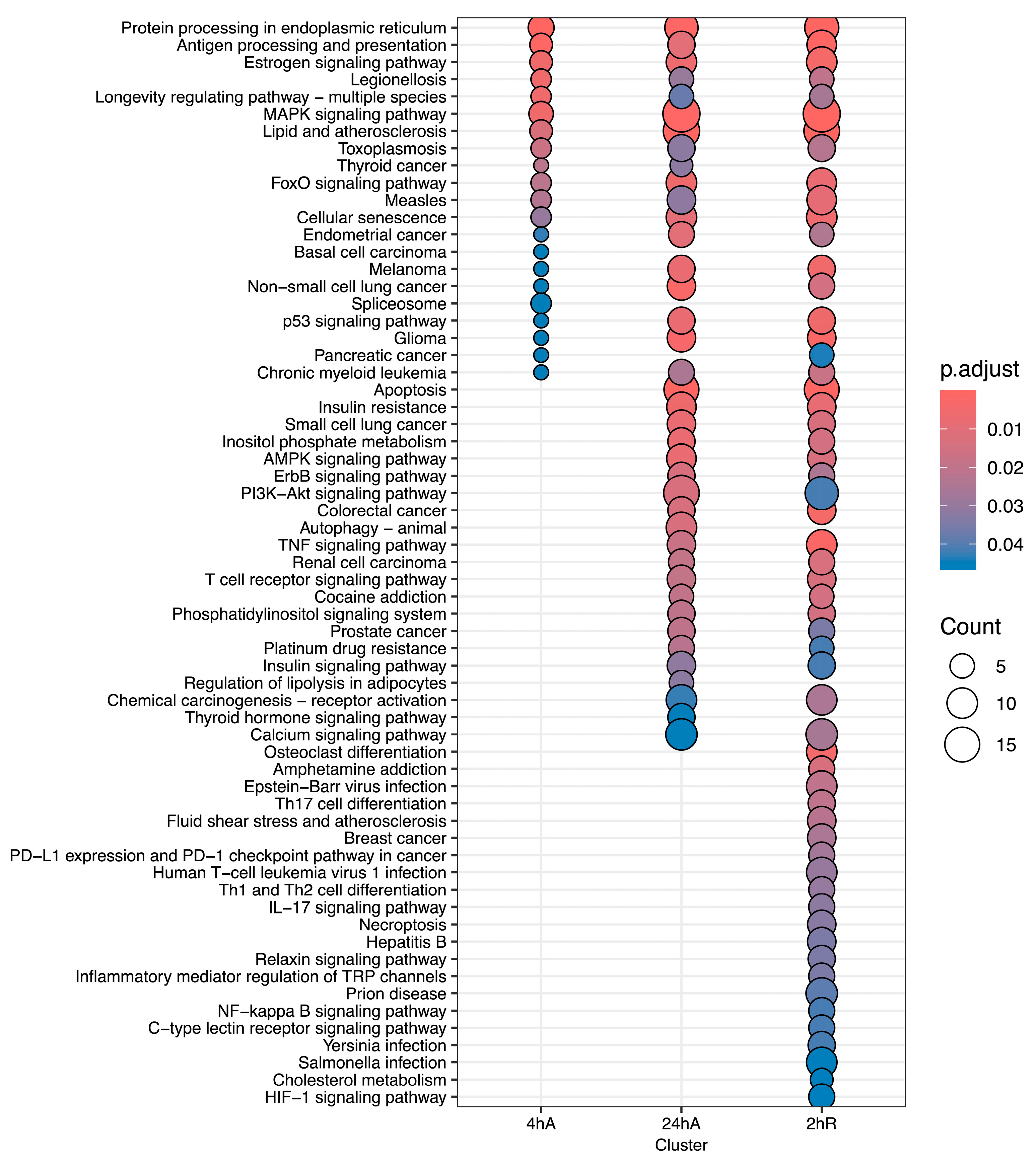
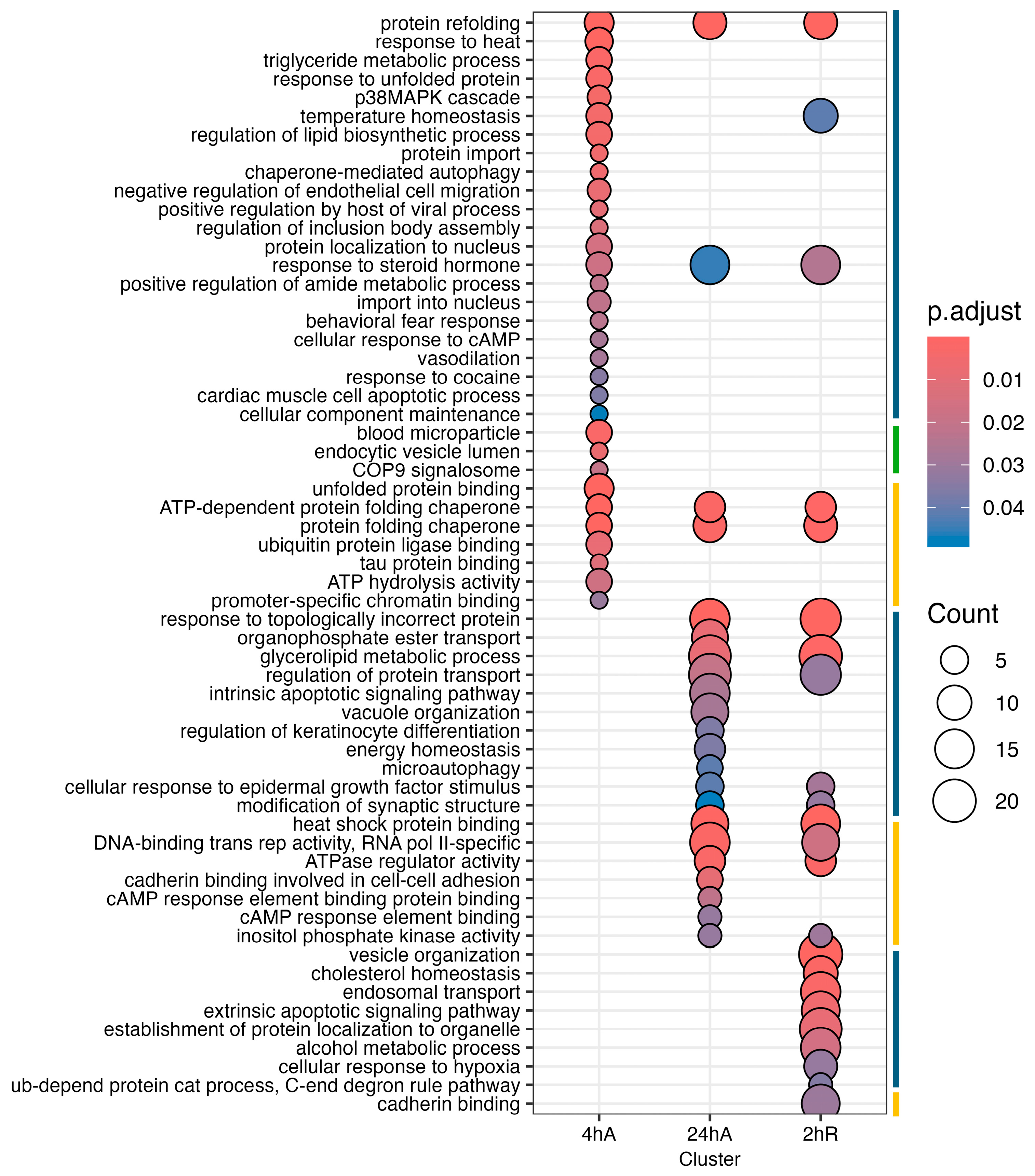
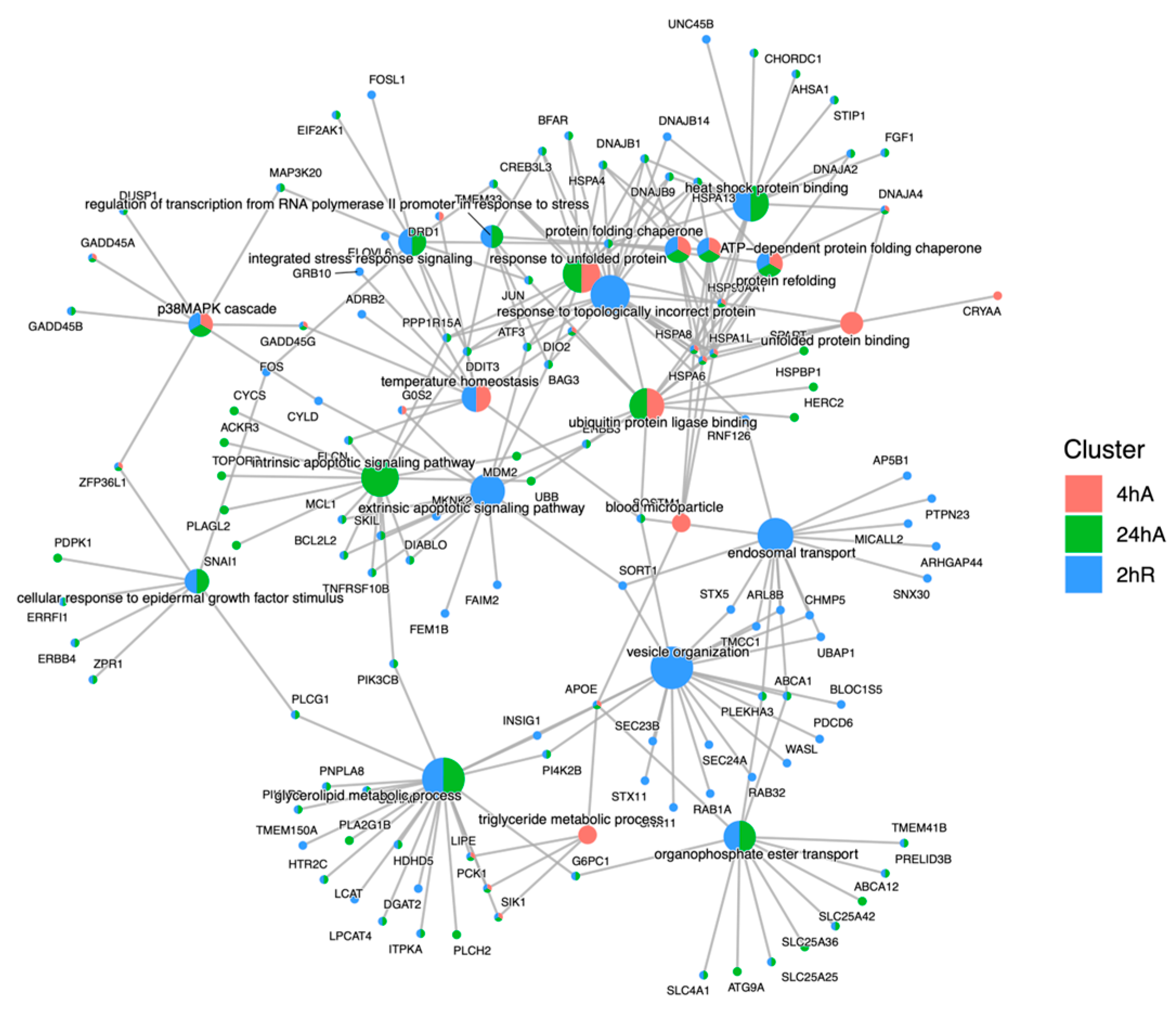
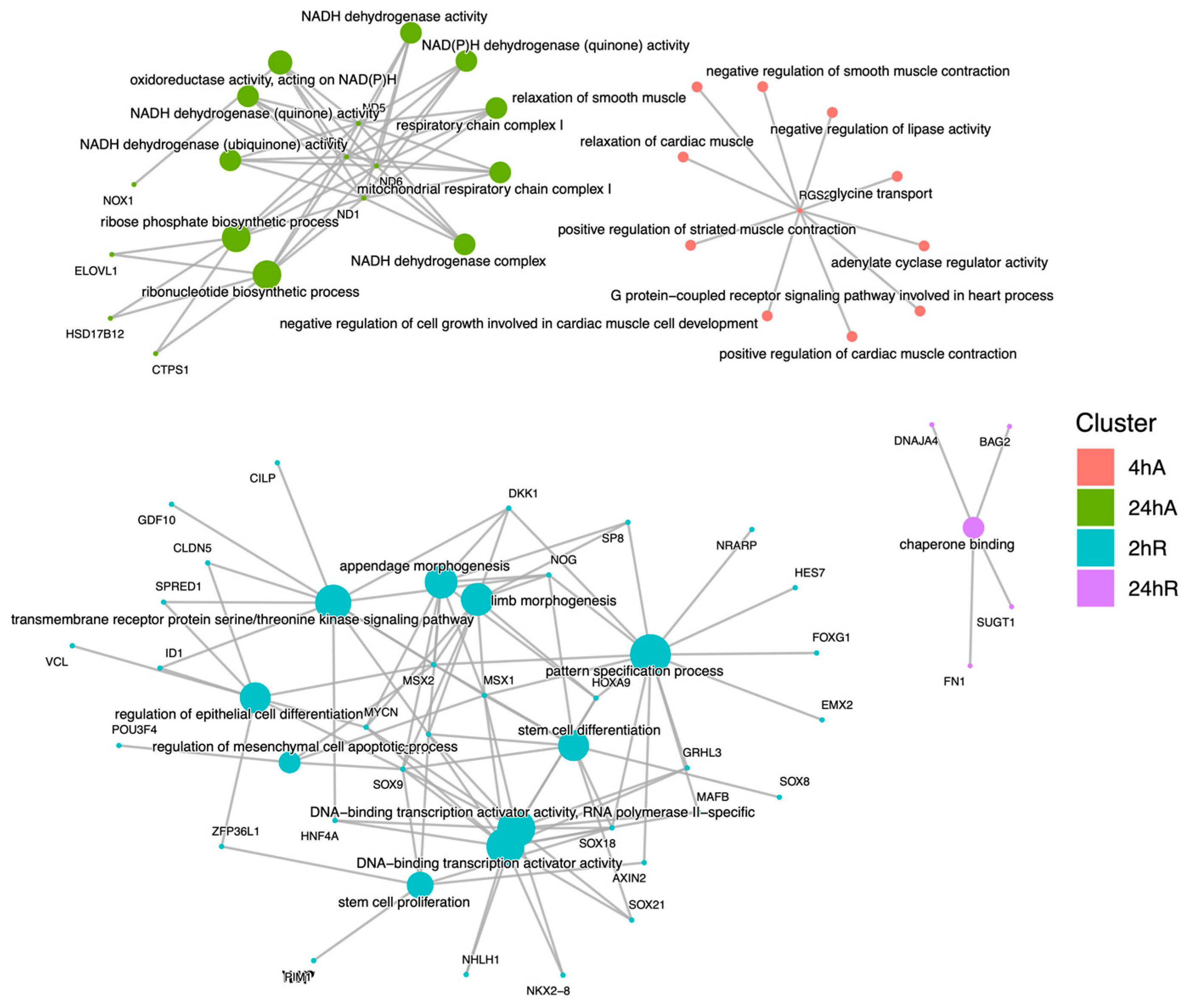
3.5. Gene Ontology Overrepresentation Analysis of Differentially Abundant Transcripts
3.6. Transcription Factor Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| dpf | days post fertilization |
| NCBI | The National Center for Biotechnology Information |
| ORA | overrepresentation analysis |
| TFs | transcription factors |
| ER | endoplasmic reticulum |
| UPR | unfolded protein response |
| ISR | integrated stress response |
References
- Storey, K.B.; Storey, J.M. Metabolic rate depression and biochemical adaptation in anaerobiosis, hibernation and estivation. Q. Rev. Biol. 1990, 65, 145–174. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B. Suspended animation: The molecular basis of metabolic depression. Can. J. Zool. 1988, 66, 124–132. [Google Scholar] [CrossRef]
- Hand, S.C. Metabolic dormancy in aquatic invertebrates. In Advances in Comparative and Environmental Physiology; Gilles, R., Ed.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 1–50. [Google Scholar] [CrossRef]
- MacRae, T.H. Gene expression, metabolic regulation and stress tolerance during diapause. Cell. Mol. Life Sci. 2010, 67, 2405–2424. [Google Scholar] [CrossRef]
- Denlinger, D.L. Insect diapause: From a rich history to an exciting future. J. Exp. Biol. 2023, 226, jeb245329. [Google Scholar] [CrossRef]
- Sim, C.; Kang, D.S.; Kim, S.; Bai, X.; Denlinger, D.L. Identification of FOXO targets that generate diverse features of the diapause phenotype in the mosquito Culex pipiens. Proc. Natl. Acad. Sci. USA 2015, 112, 3811–3816. [Google Scholar] [CrossRef]
- Rinehart, J.P.; Yocum, G.D.; Denlinger, D.L. Developmental upregulation of inducible hsp70 transcripts, but not the cognate form, during pupal diapause in the flesh fly, Sarcophaga crassipalpis. Insect Biochem. Mol. Biol. 2000, 30, 515–521. [Google Scholar] [CrossRef]
- Rinehart, J.P.; Denlinger, D.L. Heat-shock protein 90 is down-regulated during pupal diapause in the flesh fly, Sarcophaga crassipalpis, but remains responsive to thermal stress. Insect Mol. Biol. 2000, 9, 641–645. [Google Scholar] [CrossRef]
- Tammariello, S.P.; Rinehart, J.P.; Denlinger, D.L. Desiccation elicits heat shock protein transcription in the flesh fly, Sarcophaga crassipalpis, but does not enhance tolerance to high or low temperatures. J. Insect Physiol. 1999, 45, 933–938. [Google Scholar] [CrossRef]
- Yocum, G.; Joplin, K.; Denlinger, D. Upregulation of a 23 kDa small heat shock protein transcript during pupal diapause in the flesh fly, Sarcophaga crassipalpis. Insect Biochem. Mol. Biol. 1998, 28, 677–682. [Google Scholar] [CrossRef]
- Podrabsky, J.; Riggs, C.; Wagner, J. Tolerance of Environmental Stress. In Annual Fishes. Life History Strategy, Diversity, and Evolution; Berois, N., García, G., De Sá, R., Eds.; CRC Press, Taylor & Francis: Boca Raton, FL USA, 2016; pp. 159–184. [Google Scholar] [CrossRef]
- Podrabsky, J.; Romney, A.; Culpepper, K. Alternative Developmental Pathways. In Annual Fishes. Life History Strategy, Diversity, and Evolution; Berois, N., García, G., De Sá, R., Eds.; CRC Press, Taylor & Francis: Boca Raton, FL USA, 2016; pp. 63–73. [Google Scholar]
- Wourms, J.P. The developmental biology of annual fishes III. Pre-embryonic and embryonic diapause of variable duration in the eggs of annual fishes. J. Exp. Zool. 1972, 182, 389–414. [Google Scholar] [CrossRef]
- Podrabsky, J.; Riggs, C.; Romney, A.; Woll, S.; Wagner, J.; Culpepper, K.; Cleaver, T. Embryonic development of the annual killifish Austrofundulus limnaeus: An emerging model for ecological and evolutionary developmental biology research and instruction. Dev. Dyn. 2017, 246, 779–801. [Google Scholar] [CrossRef] [PubMed]
- Romney, A.; Davis, E.; Corona, M.; Wagner, J.; Podrabsky, J. Temperature dependent vitamin D signaling regulates developmental trajectory associated with diapause in an annual killifish. Proc. Natl. Acad. Sci. USA 2018, 115, 12763–12768. [Google Scholar] [CrossRef]
- Podrabsky, J.; Wilson, N. Hypoxia and anoxia tolerance in the annual killifish Austrofundulus limnaeus. Integr. Comp. Biol. 2016, 56, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Podrabsky, J.E.; Lopez, J.P.; Fan, T.W.M.; Higashi, R.; Somero, G.N. Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: Insights from a metabolomics analysis. J. Exp. Biol. 2007, 210, 2253–2266. [Google Scholar] [CrossRef] [PubMed]
- Machado, B.E.; Podrabsky, J.E. Salinity tolerance in diapausing embryos of the annual killifish Austrofundulus limnaeus is supported by exceptionally low water and ion permeability. J. Comp. Physiol. B 2007, 177, 809–820. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Riggs, C.L.; Duerr, J.M. Anoxia Tolerance During Vertebrate Development—Insights from Studies on the Annual Killifish Austrofundulus limnaeus. In Anoxia; Padilla, P., Ed.; InTech: Rijeka, Croatia, 2012; pp. 3–24. [Google Scholar] [CrossRef]
- Hand, S.C.; Podrabsky, J.E. Bioenergetics of diapause and quiescence in aquatic animals. Thermochim. Acta 2000, 349, 31–42. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Hand, S.C. Depression of protein synthesis during diapause in embryos of the annual killifish Austrofundulus limnaeus. Physiol. Biochem. Zool. 2000, 73, 799–808. [Google Scholar] [CrossRef]
- Polačik, M.; García, D.; Arezo, M.J.; Papa, N.; Schlueb, H.; Blanco, D.; Podrabsky, J.E.; Vrtílek, M. Embryonic development of natural annual killifish populations of the genus Austrolebias: Evolutionary parallelism and the role of environment. Freshw. Biol. 2023, 68, 1726–1738. [Google Scholar] [CrossRef]
- Polačik, M.; Vrtílek, M.; Reichard, M.; Žák, J.; Blažek, R.; Podrabsky, J. Embryo ecology: Developmental synchrony and asynchrony in the embryonic development of wild annual fish populations. Ecol. Evol. 2021, 11, 4945–4956. [Google Scholar] [CrossRef]
- Zajic, D.; Nicholson, J.; Podrabsky, J. No water, no problem: Stage-specific metabolic responses to dehydration stress in annual killifish embryos. J. Exp. Biol. 2020, 223, jeb231985. [Google Scholar] [CrossRef]
- Riggs, C.; Podrabsky, J. Small noncoding RNA expression during extreme anoxia tolerance of annual killifish (Austrofundulus limnaeus) embryos. Physiol. Genom. 2017, 49, 505–518. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Menze, M.A.; Hand, S.C. Rapid Communication: Long-term survival of anoxia despite rapid ATP decline in embryos of the annual killifish Austrofundulus limnaeus. J. Exp. Zool. A Ecol. Genet. Physiol. 2012, 317, 524–532. [Google Scholar] [CrossRef]
- Podrabsky, J.E. Husbandry of the annual killifish Austrofundulus limnaeus with special emphasis on the collection and rearing of embryos. Env. Biol. Fishes 1999, 54, 421–431. [Google Scholar] [CrossRef]
- Wourms, J.P. The developmental biology of annual fishes I. Stages in the normal development of Austrofundulus myersi Dahl. J. Exp. Zool. 1972, 182, 143–168. [Google Scholar] [CrossRef] [PubMed]
- Meller, C.L.; Meller, R.; Simons, R.P.; Podrabsky, J.E. Patterns of ubiquitylation and SUMOylation associated with exposure to anoxia in embryos of the annual killifish Austrofundulus limnaeus. J. Comp. Physiol. B 2014, 184, 235–247. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide-and proteoglycan-rich sources. Biotechniques 1995, 19, 942–945. [Google Scholar] [PubMed]
- Wagner, J.T.; Podrabsky, J.E. Gene expression patterns that support novel developmental stress buffering in embryos of the annual killifish Austrofundulus limnaeus. EvoDevo 2015, 6, 2. [Google Scholar] [CrossRef]
- Romney, A.; Podrabsky, J. Transcriptomic analysis of maternally provisioned cues for phenotypic plasticity in the annual killifish, Austrofundulus limnaeus. EvoDevo 2017, 8, 6. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 June 2023).
- Bolger, A.A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Wagner, J.; Warren, W.; Minx, P.; Podrabsky, J. Austrofundulus limnaeus 1.0 Draft Genome Assembly with Annotation. Available online: http://www.ncbi.nlm.nih.gov/genome/?term=txid52670[orgn] (accessed on 23 June 2023).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Liska, O.; Bohár, B.; Hidas, A.; Korcsmáros, T.; Papp, B.; Fazekas, D.; Ari, E. TFLink: An integrated gateway to access transcription factor–target gene interactions for multiple species. Database 2022, 2022, baac083. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Somero, G.N. An inducible 70 kDa-class heat shock protein is constitutively expressed during early development and diapause in the annual killifish Austrofundulus limnaeus. Cell Stress Chaperones 2007, 12, 199–204. [Google Scholar] [CrossRef]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, X.; Wang, X.; Zhai, Y.; Li, M.; Pan, J.; Bai, Y.; Rong, X.; Zhou, J. Genetic deletion of hspa8 leads to selective tissue malformations in zebrafish embryonic development. J. Cell Sci. 2022, 135, jcs259734. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Hand, S.C. Physiological strategies during animal diapause: Lessons from brine shrimp and annual killifish. J. Exp. Biol. 2015, 218, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Podrabsky, J.E.; Hand, S.C. The bioenergetics of embryonic diapause in an annual killifish, Austrofundulus limnaeus. J. Exp. Biol. 1999, 202, 2567–2580. [Google Scholar] [CrossRef]
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Buttgereit, F.; Brand, M.D.; Muller, M. ConA induced changes in energy metabolism of rat thymocytes. Biosci. Rep. 1992, 12, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, D.F.S.; Brown, G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997, 77, 731–758. [Google Scholar] [CrossRef]
- Hardewig, I.; Anchordoguy, T.J.; Crawford, D.L.; Hand, S.C. Profiles of nuclear and mitochondrial encoded mRNAs in developing and quiescent embryos of Artemia franciscana. Mol. Cell Biochem. 1996, 158, 139–147. [Google Scholar] [CrossRef]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef]
- Milton, S.L.; Nayak, G.; Lutz, P.L.; Prentice, H.M. Gene transcription of neuroglobin is upregulated by hypoxia and anoxia in the brain of the anoxia-tolerant turtle Trachemys scripta. J. Biomed. Sci. 2006, 13, 509–514. [Google Scholar] [CrossRef]
- Bundgaard, A.; Ruhr, I.M.; Fago, A.; Galli, G.L. Metabolic adaptations to anoxia and reoxygenation: New lessons from freshwater turtles and Crucian carp. Curr. Opin. Endocr. Metab. Res. 2020, 11, 55–64. [Google Scholar] [CrossRef]
- Neill, G.; Masson, G.R. A stay of execution: ATF4 regulation and potential outcomes for the integrated stress response. Front. Mol. Neurosci. 2023, 16, 1112253. [Google Scholar] [CrossRef]
- Suragani, R.N.V.S.; Zachariah, R.S.; Velazquez, J.G.; Liu, S.; Sun, C.-W.; Townes, T.M.; Chen, J.-J. Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood 2012, 119, 5276–5284. [Google Scholar] [CrossRef] [PubMed]
- Crespillo-Casado, A.; Chambers, J.E.; Fischer, P.M.; Marciniak, S.J.; Ron, D. PPP1R15A-mediated dephosphorylation of eIF2α is unaffected by Sephin1 or Guanabenz. eLife 2017, 6, e26109. [Google Scholar] [CrossRef] [PubMed]
- Dharshini, P.L.C.; Vishnupriya, S.; Sakthivel, K.M.; Rasmi, R.R. Oxidative stress responsive transcription factors in cellular signalling transduction mechanisms. Cell Signal 2020, 72, 109670. [Google Scholar] [CrossRef]
- Ubeda, M.; Vallejo, M.; Habener, J.F. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell Biol. 1999, 19, 7589–7599. [Google Scholar] [CrossRef][Green Version]
- Yadav, S.; Kalra, N.; Ganju, L.; Singh, M. Activator protein-1 (AP-1): A bridge between life and death in lung epithelial (A549) cells under hypoxia. Mol. Cell Biochem. 2017, 436, 99–110. [Google Scholar] [CrossRef]
- Lin, S.-J.; Huang, Y.-C.; Chen, H.-Y.; Fang, J.-Y.; Hsu, S.-Y.; Shih, H.-Y.; Liu, Y.-C.; Cheng, Y.-C. RGS2 suppresses melanoma growth via inhibiting MAPK and AKT signaling pathways. Anticancer. Res. 2021, 41, 6135–6145. [Google Scholar] [CrossRef]
- Cho, J.; Min, H.-Y.; Lee, H.J.; Hyun, S.Y.; Sim, J.Y.; Noh, M.; Hwang, S.J.; Park, S.-H.; Boo, H.-J.; Lee, H.-J.; et al. RGS2-mediated translational control mediates cancer cell dormancy and tumor relapse. J. Clin. Investig. 2021, 131, e136779. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.H.; Ming, H.; Zhao, P.; Hugendubler, L.; Gros, R.; Kimball, S.R.; Chidiac, P. Translational control by RGS2. J. Cell Biol. 2009, 186, 755–765. [Google Scholar] [CrossRef]
- Tao, J.; Gu, P.; Lai, H.; Peng, H.; Guo, Z.; Yuan, Y.; Yu, X.; Shen, X.; Liu, J.; Xier, Z.; et al. TXLNG improves insulin resistance in obese subjects in vitro and in vivo by inhibiting ATF4 transcriptional activity. Mol. Cell Endocrinol. 2023, 568–569, 111928. [Google Scholar] [CrossRef]
- Hotokezaka, Y.; Katayama, I.; Nakamura, T. ATM-associated signalling triggers the unfolded protein response and cell death in response to stress. Commun. Biol. 2020, 3, 378. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Wang, J.; Guo, J.; Wu, J.; Lieberman, H.B.; Yin, Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol. Cancer Res. 2008, 6, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mader, M.M.; Toth, J.E.; Yu, X.; Jin, N.; Campbell, R.M.; Smallwood, J.K.; Christe, M.E.; Chatterjee, A.; Goodson, T.; et al. Complete inhibition of anisomycin and UV radiation but not cytokine induced JNK and p38 activation by an aryl-substituted dihydropyrrolopyrazole quinoline and mixed lineage kinase 7 small interfering RNA. J. Biol. Chem. 2005, 280, 19298–19305. [Google Scholar] [CrossRef] [PubMed]
- Milton, S.L.; J Dirk, L.; F Kara, L.; Prentice, H.M. Adenosine modulates ERK1/2, PI3K/Akt, and p38MAPK activation in the brain of the anoxia-tolerant turtle Trachemys scripta. J. Cereb. Blood Flow. Metab. 2008, 28, 1469–1477. [Google Scholar] [CrossRef]
- Whitaker, R.H.; Cook, J.G. Stress relief techniques: p38 MAPK determines the balance of cell cycle and apoptosis pathways. Biomolecules 2021, 11, 1444. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Gupta, M.; Gupta, S.K.; Hoffman, B.; Liebermann, D.A. Gadd45a and Gadd45b protect hematopoietic cells from UV-induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. J. Biol. Chem. 2006, 281, 17552–17558. [Google Scholar] [CrossRef]
- Joshi, S. Mnk kinase pathway: Cellular functions and biological outcomes. World J. Biol. Chem. 2014, 5, 321. [Google Scholar] [CrossRef]
- Sheikh, S.; Saxena, D.; Tian, X.; Amirshaghaghi, A.; Tsourkas, A.; Brem, S.; Dorsey, J.F. An integrated stress response agent that modulates DR5-dependent TRAIL synergy reduces patient-derived glioma stem cell viability. Mol. Cancer Res. 2019, 17, 1102–1114. [Google Scholar] [CrossRef]
- Culpepper, K.M.; Podrabsky, J.E. Cell cycle regulation during development and dormancy in embryos of the annual killifish Austrofundulus limnaeus. Cell Cycle 2012, 11, 1697–1704. [Google Scholar] [CrossRef]
- Meller, C.L.; Podrabsky, J.E. Avoidance of apoptosis in embryonic cells of the annual killifish Austrofundulus limnaeus exposed to anoxia. PLoS ONE 2013, 8, e75837. [Google Scholar] [CrossRef]
- Lesnefsky, E.J.; Chen, Q.; Tandler, B.; Hoppel, C.L. Mitochondrial dysfunction and myocardial ischemia-reperfusion: Implications for novel therapies. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 535–565. [Google Scholar] [CrossRef] [PubMed]
- Hochrainer, K.; Jackman, K.; Anrather, J.; Iadecola, C. Reperfusion rather than ischemia drives the formation of ubiquitin aggregates after middle cerebral artery occlusion. Stroke 2012, 43, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Braunersreuther, V.; Jaquet, V. Reactive oxygen species in myocardial reperfusion injury: From physiopathology to therapeutic approaches. Curr. Pharm. Biotechnol. 2012, 13, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell Mol. Biol. 2012, 298, 229–317. [Google Scholar] [CrossRef]
- Zhang, Y.; Takemori, H.; Wang, C.; Fu, J.; Xu, M.; Xiong, L.; Li, N.; Wen, X. Role of salt inducible kinase 1 in high glucose-induced lipid accumulation in HepG2 cells and metformin intervention. Life Sci. 2017, 173, 107–115. [Google Scholar] [CrossRef]
- Keenan, S.W.; Hill, C.A.; Kandoth, C.; Buck, L.T.; Warren, D.E. Transcriptomic responses of the heart and brain to anoxia in the Western Painted Turtle. PLoS ONE 2015, 10, e0131669. [Google Scholar] [CrossRef]
- Grober, J.; Lucas, S.; Sörhede-Winzell, M.; Zaghini, I.; Mairal, A.; Contreras, J.-A.; Besnard, P.; Holm, C.; Langin, D. Hormone-sensitive lipase is a cholesterol esterase of the intestinal mucosa. J. Biol. Chem. 2003, 278, 6510–6515. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72, 3–12. [Google Scholar] [CrossRef]
- Khalil, Y.A.; Rabès, J.-P.; Boileau, C.; Varret, M. APOE gene variants in primary dyslipidemia. Atherosclerosis 2021, 328, 11–22. [Google Scholar] [CrossRef]
- Nye, C.K.; Hanson, R.W.; Kalhan, S.C. Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J. Biol. Chem. 2008, 283, 27565–27574. [Google Scholar] [CrossRef]
- Chatterjee, S.; Choi, A.J.; Frankel, G. A systematic review of Sec24 cargo interactome. Traffic 2021, 22, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Linders, P.T.; Van Der Horst, C.; Ter Beest, M.; Van Den Bogaart, G. Stx5-mediated ER-Golgi transport in mammals and yeast. Cells 2019, 8, 780. [Google Scholar] [CrossRef]
- López-Hernández, T.; Haucke, V.; Maritzen, T. Endocytosis in the adaptation to cellular stress. Cell Stress 2020, 4, 230–247. [Google Scholar] [CrossRef]
- Park, J.; Kim, J.; Park, H.; Kim, T.; Lee, S. ESCRT-III: A versatile membrane remodeling machinery and its implications in cellular processes and diseases. Anim. Cells Syst. 2024, 28, 367–380. [Google Scholar] [CrossRef]
- Kumar, A.V.; Mills, J.; Lapierre, L.R. Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front. Cell Dev. Biol. 2022, 10, 793328. [Google Scholar] [CrossRef]
- Hu, X.; Wang, L.; Wang, Y.; Ji, J.; Li, J.; Wang, Z.; Li, C.; Zhang, Y.; Zhang, Z.-R. RNF126-mediated reubiquitination is required for proteasomal degradation of p97-extracted membrane proteins. Mol. Cell 2020, 79, 320–331.e329. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.-Y.; Sun, Q.-Y.; Ding, L.-W.; Mayakonda, A.; Venkatachalam, N.; Yeo, M.-S.; Silva, T.C.; Xiao, J.-F.; Doan, N.B.; Said, J.W.; et al. RNA-binding protein ZFP36L1 suppresses hypoxia and cell-cycle signaling. Cancer Res. 2020, 80, 219–233. [Google Scholar] [CrossRef] [PubMed]
- She, Z.-Y.; Yang, W.-X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015, 94, 547–563. [Google Scholar] [CrossRef]
- Svingen, T.; Tonissen, K.F. Hox transcription factors and their elusive mammalian gene targets. Heredity 2006, 97, 88–96. [Google Scholar] [CrossRef]
- Robert-Moreno, À.; Naranjo, S.; De La Calle-Mustienes, E.; Gómez-Skarmeta, J.L.; Alsina, B. Characterization of new otic enhancers of the Pou3f4 gene reveal distinct signaling pathway regulation and spatio-temporal patterns. PLoS ONE 2010, 5, e15907. [Google Scholar] [CrossRef]
- Bell, S.M.; Schreiner, C.M.; Waclaw, R.R.; Campbell, K.; Potter, S.S.; Scott, W.J. Sp8 is crucial for limb outgrowth and neuropore closure. Proc. Natl. Acad. Sci. USA 2003, 100, 12195–12200. [Google Scholar] [CrossRef] [PubMed]
- Maier, C.J.; Maier, R.H.; Rid, R.; Trost, A.; Hundsberger, H.; Eger, A.; Hintner, H.; Bauer, J.W.; Onder, K. PIM-1 kinase interacts with the DNA binding domain of the vitamin D receptor: A further kinase implicated in 1,25-(OH)2D3 signaling. BMC Mol. Biol. 2012, 13, 18. [Google Scholar] [CrossRef]
- Gu, J.J.; Wang, Z.; Reeves, R.; Magnuson, N.S. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene 2009, 28, 4261–4271. [Google Scholar] [CrossRef] [PubMed]
- König, B.; Rauer, C.; Rosenbaum, S.; Brandsch, C.; Eder, K.; Stangl, G.I. Fasting upregulates PPARα target genes in brain and influences pituitary hormone expression in a PPARα dependent manner. PPAR Res. 2009, 2009, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mandard, S.; Müller, M.; Kersten, S. Peroxisome proliferator-activated receptor a target genes. Cell. Mol. Life Sci. 2004, 61, 393–416. [Google Scholar] [CrossRef]
- Kim, D.; Ha, S.K.; Gonzalez, F.J. CBFA2T3 is PPARA sensitive and attenuates fasting-induced lipid accumulation in mouse liver. Cells 2024, 13, 831. [Google Scholar] [CrossRef]



| Transcription Factor | Number of Associated Genes | Gene List | p Value | p Adjust |
|---|---|---|---|---|
| 4 h anoxia | ||||
| PPARA | 5 | 25 | 2.13 × 10−8 | 2.00 × 10-5 |
| POU4F1 | 1 | 25 | 1.52 × 10−6 | 4.74 × 10-4 |
| ZNF746 | 1 | 25 | 1.52 × 10−6 | 4.74 × 10-4 |
| HSF2 | 4 | 25 | 7.71 × 10−6 | 1.83 × 10-3 |
| POU3F1 | 1 | 25 | 1.51 × 10−5 | 2.84 × 10-3 |
| TFAP2B | 1 | 25 | 2.67 × 10−5 | 3.54 × 10-3 |
| DICER1 | 2 | 25 | 4.43 × 10−5 | 7.28 × 10-3 |
| 24 h anoxia | ||||
| TFAP4 | 366 | 409 | 4.80 × 10−14 | 1.17 × 10−11 |
| TBP | 376 | 409 | 1.23x 10−13 | 2.50 × 10−11 |
| KLF5 | 347 | 409 | 2.18 × 10−12 | 3.82 × 10−10 |
| SMAD1 | 323 | 409 | 5.72 × 10−12 | 8.74 × 10−10 |
| E2F1 | 383 | 409 | 8.09 × 10−12 | 1.11 × 10−9 |
| SNAI2 | 302 | 409 | 1.07 × 10−11 | 1.30 × 10−9 |
| SP5 | 351 | 409 | 1.75 × 10−11 | 1.94 × 10−9 |
| SMARCA4 | 401 | 409 | 1.98 × 10−11 | 2.01 × 10−9 |
| EED | 348 | 409 | 2.30 × 10−11 | 2.16 × 10−9 |
| CEBPA | 381 | 409 | 3.19 × 10−11 | 2.79 × 10−9 |
| 2 h recovery | ||||
| SIN3A | 375 | 405 | 1.06 × 10−14 | 2.58 × 10−12 |
| TAF1 | 369 | 405 | 1.92 × 10−14 | 3.72 × 10−12 |
| TBP | 374 | 405 | 2.15 × 10−14 | 3.72 × 10−12 |
| CHD2 | 339 | 405 | 3.54 × 10−14 | 5.35 × 10−12 |
| SP4 | 322 | 405 | 3.45 × 10−13 | 4.63 × 10−11 |
| KLF9 | 339 | 405 | 4.06 × 10−13 | 4.91 × 10−11 |
| SP1 | 390 | 405 | 4.87 × 10−13 | 5.35 × 10−11 |
| TFAP4 | 360 | 405 | 7.99 × 10−13 | 8.05 × 10−11 |
| MYCN | 381 | 405 | 9.70 × 10−13 | 9.02 × 10−11 |
| E2F1 | 381 | 405 | 1.19 × 10−12 | 1.03 × 10−10 |
| 24 h recovery | ||||
| XRCC6 | 1 | 42 | 4.35 × 10−6 | 4.24 × 10−3 |
| TRAF6 | 1 | 42 | 1.30 × 10−5 | 6.36 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clouser, P.R.; Riggs, C.L.; Romney, A.L.T.; Podrabsky, J.E. Diapause and Anoxia-Induced Quiescence Are Unique States in Embryos of the Annual Killifish, Austrofundulus limnaeus. Biomolecules 2025, 15, 515. https://doi.org/10.3390/biom15040515
Clouser PR, Riggs CL, Romney ALT, Podrabsky JE. Diapause and Anoxia-Induced Quiescence Are Unique States in Embryos of the Annual Killifish, Austrofundulus limnaeus. Biomolecules. 2025; 15(4):515. https://doi.org/10.3390/biom15040515
Chicago/Turabian StyleClouser, Patrick R., Claire L. Riggs, Amie L. T. Romney, and Jason E. Podrabsky. 2025. "Diapause and Anoxia-Induced Quiescence Are Unique States in Embryos of the Annual Killifish, Austrofundulus limnaeus" Biomolecules 15, no. 4: 515. https://doi.org/10.3390/biom15040515
APA StyleClouser, P. R., Riggs, C. L., Romney, A. L. T., & Podrabsky, J. E. (2025). Diapause and Anoxia-Induced Quiescence Are Unique States in Embryos of the Annual Killifish, Austrofundulus limnaeus. Biomolecules, 15(4), 515. https://doi.org/10.3390/biom15040515






